10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(9):997-1006. doi:10.7150/ijbs.9058 This issue Cite
Review
microRNAs in Spinal Cord Injury: Potential Roles and Therapeutic Implications
1. Department Spinal Surgery, Jinan Central Hospital affiliated to Shandong University, Jinan, Shandong, China;
2. Department of Neurobiology, Shandong Provincial Key Laboratory of Mental Disorders, School of Medicine, Shandong University, Jinan, Shandong, China;
3. School of Medicine, Shandong University, Jinan, Shandong, China.
Received 2014-3-9; Accepted 2014-8-1; Published 2014-9-6
Abstract
microRNAs (miRNAs) are a novel class of small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. miRNAs can modulate gene expression and thus play important roles in diverse neurobiological processes, such as cell differentiation, growth, proliferation and neural activity, as well as the pathogenic processes of spinal cord injury (SCI) like inflammation, oxidation, demyelination and apoptosis. Results from animal studies have revealed the temporal alterations in the expression of a large set of miRNAs following SCI in adult rats, and the expressional changes in miRNAs following SCI is bidirectional (increase or decrease). In addition, several miRNAs have distinct roles in prognosis of SCI (protective, detrimental and varied). Taken together, the existing evidence suggests that abnormal miRNA expression following SCI contributes to the pathogenesis of SCI, and miRNAs may become potential targets for the therapy of SCI.
Keywords: microRNA, spinal cord injury, therapy, diagnosis.
Introduction
microRNAs (miRNAs) are endogenous, small (18-25 nucleotides), non-coding RNAs that are negative regulators of gene expression at the post-transcriptional level [1-3]. With sequences partially complementary to their target mRNAs, miRNAs play vital roles in regulating (mostly inhibiting) gene expression in various organisms [1, 4]. miRNAs are transcribed in the nucleus by RNA polymerase II and processed by the endonuclease, DROSHA, in the nucleus. The pre-miRNA is then exported to the cytoplasm and processed by DICER. The resulting mature miRNA sequence is incorporated into the RNA silencing complex (RISC). Within the RISC, the miRNA guides the complex to its RNA target, thereby mediating its repression. The target selectivity is dictated by the miRNA “seed” sequence located within nts 2-8 at the 5'-end of the mature miRNA sequence. The seed sequence binds through Watson-crick base pairing within the 3' untranslated region (3' UTR) of the target genes, leading to translational repression of the target mRNA by either transcript destabilization, translational inhibition or both. Since the first miRNA, lin4, was discovered 10 years ago [5], thousands of miRNAs have been identified in organisms, and studies have confirmed that they participate in a variety of biological processes (such as development, proliferation, cell growth, and metabolism) largely through inhibiting mRNA translation [4, 6]. Recent studies suggest that the expression of approximately 20-30% of human genes is regulated by miRNAs [1].
Two different mechanisms have been proposed for the pathogenesis of spinal cord injury (SCI): a primary mechanical injury and a secondary injury induced by multiple biological processes, including inflammation, excitotoxicity, apoptosis, and demyelination [7-10]. Alterations in the expression of many genes have been shown to play vital roles in the pathogenesis of secondary SCI [7, 8]. In addition, demyelination, a process inducing neuronal death, gradually increases after SCI and during the secondary injury, and may perpetuate the nerve conduction deficits [11]. These pathophysiological changes may continue for several years after injury and cause strong suppression of spontaneous neuronal precursor neurogenesis, finally leading to the paralysis. To develop effective treatments is a major focus in studies on SCI, and great efforts have been made to improve the functional deficits of SCI patients, including paralysis [12]. Several therapeutic approaches have been used to improve the microenvironment of the injured spinal cord and/or to stimulate the endogenous repair in an attempt to ameliorate secondary injury [9, 11, 13, 14]. Although a majority of studies has focused on compounds with neuroprotection against secondary injury in SCI, their therapeutic efficacy remains controversial [15].
A more recent review summarizes the miRNA dysregulation in the SCI [10]. Bioinformatics analysis suggests that the altered expression of these miRNAs may contribute to the pathogenesis of secondary SCI [16]. These findings are exciting in that they might reveal potential targets for the therapy of SCI [16-18].
Central nervous system (CNS) distributions and functions of miRNAs
To determine the expression patterns of miRNAs in adult mice, 13 different areas were dissected from the central nervous system (CNS) of adult mice (spinal cord, cerebellum, medulla oblongata, pons, mesencephalon, thalamus, hypothalamus, hippocampus, amygdala, neocortex, olfactory bulb, eye and pituitary gland) and the miRNA profiling was performed [19]. Of interest, many miRNAs show extremely high enrichment in specific regions, suggesting that they might be associated with certain functions. In agreement with some previous reports, mouse brain was rich in miR-9, miR-124a, miR-125b, miR-127, miR-128, and members of the let-7 family. The medulla oblongata exhibited the enrichment of miR-34a, miR-451, miR-219, miR-338, miR-10a, and miR-10b, and miR-195, miR-497 and miR-30b were highly expressed in the cerebellum [20]. The hypothalamus showed accumulation of miR-7 and miR-7b [21]. miR-218, miR-221, miR-222, miR-26a, miR-128a/b, miR-138, and let-7c were enriched in the hippocampus. miR-375, miR-141, miR-200a, miR-375, miR-7, and miR-7b were highly expressed in the pituitary [22]. miR-10a and miR-10b were enriched in the spinal cord [23].
To investigate the specific roles of miRNAs in the CNS, many studies have ablated Dicer, which is critical for the miRNA biosynthesis because its loss can result in embryonic lethality [24, 25]. Studies have shown that, during the early CNS development, deletion of Dicer in the neural crest (NC) may lead to the cell loss in the sensory, enteric, and sympathetic nervous systems [26]. During the late embryonic stage, Dicer affects the survival and differentiation of cortical neural progenitor cells, leading to the deviant migration of cortical neurons [27]. Ago, a part of the RNA-induced silencing complex [27, 28], also plays important roles in the CNS development. For example, neural tube closure is prevented in mice with Ago mutation [29]. Existing evidence indicates that miRNAs are important for the CNS development and function.
miRNAs in the spinal cord
To understand changes in the expression levels and the corresponding regulatory functions of miRNAs following SCI, it is necessary to elucidate how miRNAs work under normal conditions. Previous reports have shown extensive miRNA expression in the spinal cord of adult mice [19]. Liu et al further demonstrated that approximately 77% of identified rat mature miRNAs were expressed in the spinal cord of adult rats; microarray analysis revealed that 269 of 350 miRNAs were detectable in the adult rat spinal cord, and 36 were expressed with the signal intensities greater than 10,000 [16].
With regard to spatial distribution of miRNAs in spinal cord, both Bcl-2 and miR-9/9* were more abundantly expressed in the dorsal part of the sacral cord, while P53 and miR-124a/125b were more highly expressed in the ventral spinal cord and dorsal root ganglia (DRG). In addition, after the treatment with all-trans-retinoic acid (RA) in spinal bifida rat fetus, the Bcl-2 and miR-9/9* expression dramatically reduced in the dorsal sacral cord and P53 and miR-124a/125b expression dramatically reduced in the ventral spinal cord and DRG. These findings suggest the possible relationship between the expression changes of miR-9/9*, miR-124a, miR-125b and the expression of Bcl-2 and P53 during the spinal cord development [30].
Alterations in miRNA expression following SCI
miRNA expression after SCI
As mentioned above, microarray analysis revealed the expression of nearly 300 kinds of miRNAs changed in the adult rat spinal cord following SCI, and significant change was noted in 97 miRNAs. Of 97 miRNAs, 60 had the signal intensities greater than 500. These miRNAs can be classified into three categories according to their expression patterns: 1) up-regulated miRNAs, 2) down-regulated miRNAs, and 3) miRNAs with bidirectional changes in their expression post-SCI (Table 1). The expression of remaining 37 miRNAs with the signal intensities lower than 500 also significantly reduced after SCI, but they did not exhibit specific patterns. The expression of 172 miRNAs remained unchanged after SCI [16]. Similar results were observed in another microarray study on SCI in rats, in which the expression of 343 miRNAs changed following SCI [31].
Interestingly, another group employed real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) and in situ hybridization to investigate miRNAs, and results showed that the impact of SCI on miRNA expression persisted up to 14 days and expanded both anteriorly and caudally around the injured site; in addition, there was a direct correlation between the number of affected miRNAs and the SCI severity [32].
To investigate the expressional changes in miRNAs following SCI is the first step in understanding the relationship between miRNAs and SCI. miRNAs can be subdivided into hundreds of families with presumably independent origins [33]. It has been known that different members of the same miRNA families can simultaneously target several categories of genes and act together to regulate specific physiological processes [34]. For example, members of the let-7 family are expressed highly in rat spinal cord after chronic constriction injury [35]. A bioinformatics analysis revealed that the potential targets for these miRNAs include genes involved in many pathophysiological processes, such as inflammation, apoptosis and oxidation after SCI [16, 31].
Another study revealed that some mRNAs of inflammatory mediators, such as intercellular adhesion molecule 1 (ICAM-1), interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α), were potential targets of miR-181a, miR-411, miR-99a, miR-34a, miR-30c, miR-384-5p and miR-30b-5p [16], miR-486 [17], which were down-regulated post-SCI, and miR-17 and miR-20 [31], miR-124a [36], which were up-regulated post-SCI, and miR-124a, which was down-regulated at 1 and 7 days post-SCI [36]. On the other hand, some mRNAs of anti-inflammatory mediators, such as TRAF6, annexin A1 and A2, were likely targets of miR-146a [31, 32], miR-223 [31, 36], miR-221 [16] and miR-1 [16, 32], which were up-regulated post-SCI, and miR-1, which was down-regulated at 3 days post-SCI [31, 36]. Furthermore, mRNAs of cytosolic phospholipases A2 (cPLA2) and secretory PLA2 (sPLA2) are likely targets of miR-181a and miR-127, which are down-regulated post-SCI [16]. A recent study demonstrated that SCI-induced PLA2 activation also plays an important role in the pathogenesis of secondary SCI [37]. Some miRNAs that are up-regulated post-SCI (such as miR-1, miR-206, miR-152, and miR-214) may target anti-oxidant genes, including superoxide dismutase 1 (SOD-1) and catalase genes [16].
In recent years, apoptosis has been identified as a vital mechanism of cell death in many neurological diseases and SCI [38-40]. Apoptosis related genes like caspase-3, calpain 1, calpain 2, Bcl-2, c-Myc and apoptosis-inducing factor (AIF) genes become potential targets of miR-138 [31], miR-124 [16, 32], miR-235-3p, miR-137 and miR-30b-3p [16], which are down-regulated post SCI, and miR-1 [31, 32], miR-15b, miR-34 and miR-145 [31], which are up-regulated post SCI. However, anti-apoptosis related genes, including Bcl2-1 and Bcl2-2 genes, are potential targets of miR-21, miR-146a [31, 32], miR-20a [15, 16], miR-145, miR-214, miR-674-5p, miR-15b, miR-17, miR206 and miR-672 [16], which are up-regulated after SCI. In addition, miR-103, miR-107 [31], miR-133a [16], miR-133b [16, 41], which also have influence on the apoptosis, have an up-regulated expression at 4 h post-SCI and a subsequent down-regulated expression at 1 and 7 days post-SCI. It is worth mentioning that let7/miR-98 and miR-133b exhibit both pro-apoptotic and anti-apoptotic effects [31].
microRNAs showing increased or decreased expression following SCI in animal models.
| Expression patterns | miRNA detected at different time points | References | ||||
|---|---|---|---|---|---|---|
| 4 h | 1 d | 4 d | 7 d | After 7 d | ||
| Up-regulated miRNAs | miR-1, miR-15b, miR-20a, miR-20b-5p, miR-21, miR-30a, miR-31, miR-92a, miR-92b, miR-93, miR-98, miR-106b, miR-145, miR-146b, miR-152, miR-199a-3p, miR-203, miR-206, miR-214, miR-221, miR-223, miR-290, miR-333, miR-374, miR-378, miR-672, miR-674-5p, miR-872 | miR-17, miR-146a, miR-124, miR-486 | miR-486 | [15, 16, 32, 51] | ||
| Down-regulated miRNAs | miR-30b-5p, miR-30c, miR-30d, miR-34a, miR-129, miR-137, miR-138, miR-219-2-3p, miR-219-5p, miR-323, miR-325-3p, miR-338, miR-379, miR-384-5p, miR-495, miR-543 | miR-146a, miR-708, miR-125b-3p, miR-126, miR-let-7b | miR-129-1, miR-129-2 | miR-129-3p, miR-342 | miR-199a-3p, | [16, 17, 31, 32, 51, 52, 81] |
The expression of several miRNAs is up-regulated/down-regulated at 4 h, which sustains for several days. In addition, the expression of several miRNAs start to up-regulate/down-regulate at 1 day, 4 days, 7 days or after 7 days.
Relationship of miRNAs with SCI prognosis
A considerable body of researches shows that miRNAs can influence SCI through various pathways, including apoptosis, regeneration, inflammation and demyelination [42]. This has been described in detail in a recent review [10]. In this paper, we focus on the clinical importance of miRNAs in SCI. In available studies, most are carried out in animals, especially in rats. Although some findings are obtained from animal models, they may also offer evidence and indispensable directions to carry out further studies. The prognosis of the SCI, as shown in animal models, can be classified into three types: improvement, deterioration and improvement and thereafter deterioration. In this section, we focus on the outcome of SCI. It has been found that miRNAs and mRNA have no one-to-one relationship: one miRNA can target many mRNAs and a mRNA can also be regulated by many miRNAs [43]. In this case, the expressional changes in miRNAs and the corresponding clinical manifestations may become a good reference for future clinical studies. According to the different expressional changes in miRNAs and the prognosis following SCI, these miRNAs may be classified into three main groups: protective, detrimental and varied.
Anti-inflammatory and anti-apoptotic effects of miR-17-92 cluster (miR-17 and -20), miR-146a, and miR-138 in SCI.
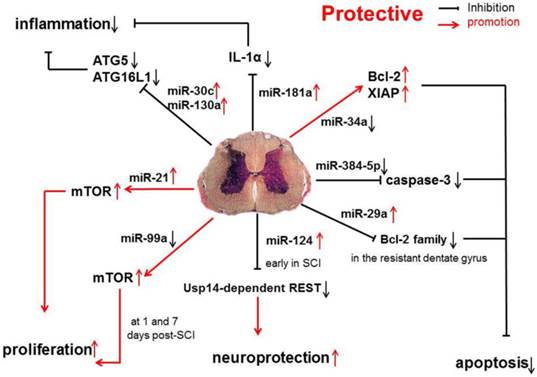
Protective miRNAs in SCI
In one study, the expression of protective miRNAs, which have anti-apoptotic capability, was investigated in SCI, such as miR-17-92 cluster (miR-17 and -20), miR-146a and miR-138 [31] (Figure 1).
Studies have shown that miR-34a can directly inhibit the expression of anti-apoptotic proteins Bcl-2 and XIAP. The increase in caspase-11 following miR-34a over-expression indicates that miR-34a may be related with apoptosis by reducing the expression of anti-apoptotic proteins [44]. However, in SCI, miR-34a expression is significantly down-regulated, which suggests that miR-34a protects against cell death because anti-apoptotic protein expression remains at a high level.
miR-384-5p expression is continuously down-regulated following SCI. Interestingly, cleaved caspase-3 expression is up-regulated by miR-384-5p and down-regulated by an miR-384-5p inhibitor, suggesting that miR-384-5p plays an important role in regulating signaling pathways, such as the apoptotic pathway[45]. Regardless of the identified pathways, it is clear that miR-384-5p down-regulation is protective on SCI.
Evidence has shown that the miR-181a is associated with the modulation of inflammatory pathways, and the up-regulated miR-181a following SCI, may exert anti-inflammatory effects via down-regulating IL-1α. Furthermore, the suppressed expression of several inflammatory factors, such as IL-1β, IL-6 and TNF-α, may also result from the influence of miR-181a mimics [46]. Another study shows that the expression of miR-181 family is down-regulated because of inflammation. Genetic studies also reveal that miR-181 family can inhibit the production of pro-inflammatory cytokines, such as TNF-α, IL-6, IL-1β, IL-8, LIF, and HMGB1, but promote the generation of anti-inflammatory cytokines (IL-10) [47].
Detrimental miRNAs in SCI
Recent evidence shows that miR-30b, miR-30c, miR-223, miR-15b, miR-1, miR-145, miR-17-92 cluster (miR-17 and -20) and hsa-miR-106a may promote the progression of SCI [31] (Figure 2).
A functional screening using novel small RNA expression libraries leads to the identification of hsa-miR-30b and hsa-miR-30c as negative regulators of cell death induced by loss of attachment (anoikis). Importantly, the acquisition of anoikis resistance via these miRNAs is achieved through down-regulating caspase-3 expression. Thus, down-regulation of hsa-miR-30b and hsa-miR-30c expression may result in secondary SCI [48].
Pro-inflammatory and pro-apoptotic effects of miR-30b, miR-30c, miR-223, miR-15b, miR-1, miR-145, miR-17-92 cluster (miR-17 and -20) and hsa-miR-106a in SCI.
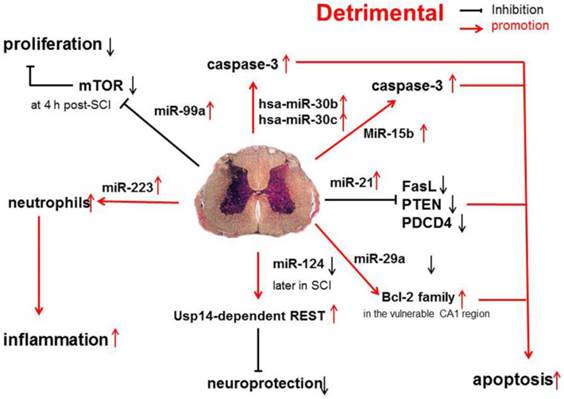
A study on miR-20a shows that miR-20a expression shows an up-regulation at 24 h after SCI, which sustained for 1 week [16]. Ngn1, a key protein that can maintain the cell survival and neurogenesis, may be inhibited by miR-20a, and to block miR-20a expression in SCI animals is effectively to induce neuronal survival and neurogenesis [15].
It has been shown that miR-223 is highly expressed at 12 h after SCI. In situ hybridization and immunohistochemistry showed the signals of miR-223 merged with Gr-1-positive neutrophils. Existing data indicate that miR-223 might regulate neutrophils in the early phase of SCI when there is a cascade of secondary damage to the spinal cord. miR-223 is expressed in the neutrophils that relocate to the epicenter of inflammation after SCI, and inflammatory cytokines are also highly expressed at the same time. Izumi et al found that miR-223 promoted the neutrophil-mediated inflammation following SCI [49].
miR-15b enhances the hypoxia/re-oxygenation-induced apoptosis of cardiomyocytes via the mitochondrial pathway of apoptosis. Inhibition of miR-15b increases the protein expression of Bcl-2 without affecting Bcl-2 mRNA expression, suppresses the release of mitochondrial cytochrome into the cytoplasm, and decreases the caspase-3 activity [50]. However, whether the highly up-regulated miR-15b enhances hypoxia/re-oxygenation-induced apoptosis in SCI remains to be elucidated.
The effects of miR-1, miR-145, miR-17-92 cluster (miR-17 and -20), hsa-miR-106a on SCI were described in a previous study [31].
miRNAs with varied effects in SCI
Some miRNAs may affect several pathways, are expressed in different regions, and have altered expression at different time points after SCI (e.g., early up-regulation at 4 h post-SCI followed by down-regulation at 1 and 7 days post-SCI). Thus, these siRNAs may exert varied effects under distinct conditions, and thus we classified them into the third group.
1) Functioning on different pathways
miR-21 is involved in both proliferation and apoptosis pathways. In SCI, elevated expression of miR-21 and decreased expression of miR-199a-3p correlate with significant changes in the expression of their target genes: mTOR (mammalian target of rapamycin) mRNA expression increases. Western blotting assay confirmed similar change in protein level [45]. An increase in phosphorylated-S6 (a downstream effector of mTOR) in intermediate gray neurons in Ex rats was blocked by rapamycin, suggesting that activity-dependent plasticity in the injured spinal cord is modulated in part through miRNAs that regulate mTOR signaling and may indicate an increase in the regenerative potential of SCI affected neurons [51]. In addition, pro-apoptotic genes for Fas ligand (FasL), phosphatase and tensin homolog (PTEN), and programmed cell death protein 4 (PDCD4) have been shown to be direct targets of miR-21 in many diseases and cell types. In vivo, treatment with antagomir-21 increases the expression of FasL and PTEN, demonstrating that miR-21 down-regulates the expression of these anti-apoptotic genes following SCI [52].
2) Functioning in different regions
Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to the forebrain ischemia. Available data suggest that, by targeting a member of pro-apoptotic Bcl-2 family, to increase miR-29a might emerge as a protective strategy against ischemia-reperfusion injury. Notably, miR-29a significantly increases in the resistant dentate gyrus but decreases in the vulnerable CA1 (cornu ammonis 1) region of the hippocampus, which results in different effects [53].
3) Functioning at different time points
miR-124 protects against focal cerebral ischemia, and ectopic miR-124 expression appears to be an attractive and novel tool in the stroke treatment and may exert neuroprotection via an unknown mechanism involving the Usp14-dependent REST degradation. miR-124 expression increases at initial stage, and decreases later following SCI, indicating that the protection afforded by miR-124 is transient [54].
With an early up-regulation at 4 h post-SCI followed by a down-regulation at 1 and 7 days post-SCI, initial over-expression of miR-99a suppresses the expression of mTOR by directly targeting its 3' un-translated region (3' UTR) in a post-transcriptional manner. In this way, miR-99a inhibits the cell proliferation by suppressing mTOR [55]. At later stages of SCI, a decrease in miR-99a expression will promote the spinal cord recovery by releasing the inhibition of active mTOR pathway [55].
miRNAs of interest in SCI
Following four miRNAs are exampled to illustrate the individual roles of particular miRNAs in different animal models:
miR-21
Abnormal miR-21 expression has been reported in many different human diseases (mostly cancers), and increased miR-21 expression has been found to be associated with the increased cell survival, growth and proliferation and decreased apoptosis [56-59]. Previous studies have shown that miR-21 expression is significantly up-regulated in many CNS diseases, such as traumatic brain injury (TBI), and is related to the TBI-induced pathophysiology, including apoptosis and neuronal plasticity [60].
miR-21 expression initially increases around the lesion 4 days after SCI and then reduces. The transiently elevated miR-21 expression in conjunction with the simultaneous re-expression of nestin (a neural progenitor factor) suggest a protective role of miR-21 in facilitating cell survival and integration of neural progenitor cells at the injury site, which is in accordance with previous findings that miR-21 usually functions as an anti-apoptotic factor by inhibiting the expression of PDCD4 and pro-apoptotic protein PTEN [61, 62].
In another animal model, miR-21 expression also increased significantly at 5 weeks after SCI [63]. miR-21 is expressed mainly by cultured astrocytes [63, 64], and thus to study the role of miR-21 in astrocytes after SCI, transgenic mice with miR-21 over-expression or knockout were used. Results showed that over-expression of miR-21 in astrocytes reduced the hypertrophic response to SCI. On the other hand, inhibition of miR-21 in astrocytes led to the increased axon density within the lesion site [63].
Together, these findings demonstrate a novel role of miR-21 in SCI recovery. Notably, a short-term exercise after SCI leads to a significant increase in miR-21 expression, leading to reduced structural and functional deficit after initial injury, which will be discussed later [42].
miR-486
Similar to miR-21, miR-486 expression increases at 7 days post-SCI in a murine contusion model. Studies have demonstrated that infusion of miR-486 into the spinal cord of healthy mice decreases the motor function and increases the neuronal death. Conversely, knocking down miR-486 in a mouse SCI model improves the hind limb functional recovery [17]. The major target of miR-486 in the motor neuron is NeuroD6, which is an important protein for neuronal differentiation and the oxidative stress response [65]. NeuroD6 induces the expression of glutathione peroxidase 3 (GPx3) and thioredoxin-like 1 (TXNL1) in the lesioned spinal cord, which effectively scavenges excessive reactive oxygen species (ROS) and attenuates inflammation in SCI [17].
Based on these findings, a promising therapeutic strategy for SCI might be the inhibition of miR-486 by up-regulating NeuroD6 expression, which significantly decreases the apoptosis and improves the SCI-induced functional deficits.
miR-133b
A SCI model of adult zebrafish shows the extreme up-regulation of miR-133b expression in regenerating brainstem neurons after spinal cord transection [66]. Antisense morpholino (MO) induced inhibition of miR-133b expression attenuates the locomotor recovery and reduces the axonal regeneration. miR-133b targets RhoA, the expression of which is significantly up-regulated following SCI [67], and inhibition of RhoA facilitates the corticospinal tract repair and exerts neuroprotection by decreasing tissue damage after SCI [68, 69].
Studies have shown that miR-133b serves as an important determinant in the spinal cord regeneration in the adult zebrafish by reducing RhoA protein expression [66], making miR-133b and RhoA attractive therapeutic targets in SCI.
Exercise-related miRNAs
As a non-invasive therapy for SCI, exercise is beneficial for maintaining the muscle mass of paralyzed hindlimbs [70], the stabilization of rhythmic firing patterns of spinal motoneurons [71] and the improvement of functional recovery [72].
Evidence has demonstrated that a short-term exercise leads to a significant increase in miR-21 expression and a decrease in miR-15b expression. A long-term exercise has no effects on the miRNAs expression [42]. miR-21 functions as an anti-apoptotic factor in SCI by inhibiting the expression of pro-apoptotic proteins PTEN and PDCD4 [61]. Similar mechanisms are also found in the miR-15 and miR-16, which directly influences the Bcl-2 expression, facilitating the caspases-3, -7 and -9 dependent apoptosis [42]. Thus, non-invasive exercise may influence multiple miRNAs that target genes regulating caspase expression, resulting in decreased apoptosis.
miRNAs as diagnostic tools for SCI
Studies have demonstrated the possibility of scanning cerebrospinal fluid (CSF) and blood for identification of miRNA biomarkers of diseases and injuries. Circulating miRNAs may serve as biomarkers when their expression changes. For instance, the expression of multiple miRNAs has been found to change in some brain regions and blood in rats with ischemic stroke [73]. A study also shows that plasmic miR-124 increases in rats after brain injury [74]. Further work is needed to examine the usefulness of these miRNAs as SCI biomarkers.
A recent study inspires us to make a right diagnosis: the distribution characteristics of miRNA-positive cells and miRNAs can be detected in related regions. For example, miR-9 expression is up-regulated in miR-9-positive cells of the ventral horn and dorsal horn of the spinal cord after injury [75].
Moreover, the changes in the expression of miRNAs may become promising biomarkers for predicting the severity of SCI. With the different compressing power worked on spinal cord, the expression of miR-9*, miR-219 and miR-384-5p is up-regulated by different degree in the serum following the SCI [76]. In this way, we can judge the severity of SCI by testing the expression of miRNAs. However, further study should be done in clinical trials.
Therapeutic implications of miRNAs in SCI
miRNA and mimics as new therapeutic reagents
The possibility of using miRNAs as therapeutic reagents has drawn high attention, because of their capability to modulate gene expression though tuning their expression [10]. While RhoA (a protein with highly up-regulated expression following SCI) has been known as a therapeutic reagent for SCI, miR-133 may down-regulate the RhoA expression [66]. miRNAs with the ability of molecular suppression may become novel reagents for the neuranagenesis after SCI.
Studies on miRNA mimics for SCI patients are lacking, but these molecules might be effective on SCI. As mentioned above, miRNA and its mimics have the advantage of small size, and administration of a miRNA mimetic may increase the endogenous miRNA and repress the expression of detrimental genes. Besides, with current knowledge of miRNA biogenesis, modified RNAs can be clinically delivered as synthetic, pre-processed miRNA oligonucleotides [77].
Anti-miRNA molecules as new drugs to target miRNAs
Several studies have described the potentials of certain kinds of anti-miRNA molecules targeting specific pathologic miRNAs in vitro and in vivo [77, 78]. For example, a study showed si-miR-20a treated rats had significantly decreased size of lesions and compromised blood clotting [15], which shows that to inhibit miR-20a expression may prevent inflammation-mediated secondary injury and also simulate neurogenesis. Although anti-miRNA molecules have been effectively delivered to many tissues, including the CNS [79], and whether these molecules at high doses and administered in different ways are still effective in vivo are required to be confirmed in future studies [80].
Up-regulating or down-regulating the expression of major target genes may exert similar effects as inhibition of miRNAs [15].
Conclusions
Existing studies demonstrate that SCI can induce extended miRNA dys-regulation that is parallel to the changes in mRNA expression and affects the key processes of SCI, including inflammation and apoptosis. Collectively, available findings suggest that targeting miRNA dys-regulation is an attractive therapeutic strategy for SCI.
In this review, we discuss the changes in miRNAs following SCI and the physiologic and pathologic effects of these alterations. The miRNAs in adult rats can be classified into three categories according to their alterations in the temporal expression following SCI: 1) miRNAs with up-regulated expression, 2) miRNAs with down-regulated expression, and 3) those with an early up-regulation at 4 h and a subsequent down-regulation at 1 and 7 days post-SCI. Moreover, miRNAs appear to exert protective, detrimental and varied effects on SCI. However, current understanding of the mechanisms underlying the pathogenesis of SCI is still limited, and additional miRNAs remain to be discovered. More studies are required to investigate the functions and targets of SCI-related miRNAs.
microRNAs showing varied changes in their expression following SCI in animal models.
| Expression patterns | miRNA detected at different time points | References |
|---|---|---|
| miRNAs with early up-regulation at 4 h post-SCI followed by down-regulation at 1 day post-SCI | miR-99a, miR-100, miR-103, miR-107, miR-124, miR-127, miR-128, miR-154, miR-181a, miR-434, miR-487b, miR-124a | [16, 32, 36] |
| miRNAs with early up-regulation at 4 h post-SCI followed by down-regulation at 7 days post-SCI | miR-133a, miR-133b, miR-45 | [16, 41] |
Several miRNAs show varied changes in their expression and the varied time can be the 1 day or the 7 days after SCI.
Injury models:
15. Adult female SD rats (200-230 g) underwent a T10 contusive spinal cord injury using an NYU impactor (10 g, 12.5 mm)
76. Male C57BL/6 mice, approximately 8 weeks of age, SCI was made at the level of T11-12 by compressing the cord laterally from both sides for 10 s with a number 5 forceps (FONTAX, Lausanne, Switzerland) modified with a spacer so that a 0.5 mm space remained at maximal closure
31. Adult, female Wistar rats weighing approximately 200 g, Animals were anesthetized with intraperitoneal sodium pentobarbital at 40 mg/kg (Dolethal, Vetoquinol, Lure Cedex, France). Contusion was performed at vertebral thoracic level 8 (T8) using an IH Spinal Cord Impactor from Precision System and Instrumentation, LLC, Va, which induced an impact of 200 kilodynes
16. Adult female ICR mice weighing 30 g (6 weeks of age, The contusion injuries were delivered to the T11 spinal cord following a L1 laminectomy
14. Adult female ICR mice weighing 30 g (*6 weeks of age), remove the neural spines of the T9 and T10 thoracic vertebrae and to carry out a laminectomy to expose the outer surface of the spinal dura mater
46.Adult, female Sprague-Dawley rats (225-250 g), T10, gentle aspiration
61. Adult zebrafish (Danio rerio, body length > 2.5 cm, age > 6 months), thoracic level, complete transection
50.- Sprague Dawley rats, T10, Contusion, mod. AllenWeight Drop (8 g, 40 mm)
79.- Rats, lumbo-sacral segments, Ischemic-Reperfusion
Acknowledgements
The authors thank Dr. Dong Zhenhua, Jiang Jianhao and Zhao Kai for their reviewing the manuscript. This work was supported by the National Natural Science Foundation of China (No. 81401014 M.H.), the Young Scientists Awards Foundation of Shandong Province (No. BS2013YY049).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. TheScientificWorldJournal. 2007;7:155-66 doi:10.1100/tsw.2007.201
2. Kosik KS. The neuronal microRNA system. Nature reviews Neuroscience. 2006;7:911-20 doi:10.1038/nrn2037
3. Shafi G, Aliya N, Munshi A. MicroRNA signatures in neurological disorders. The Canadian journal of neurological sciences Le journal canadien des sciences neurologiques. 2010;37:177-85
4. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5 doi:10.1038/nature02871
5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
7. Bareyre FM, Schwab ME. Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends in neurosciences. 2003;26:555-63 doi:10.1016/j.tins.2003.08.004
8. Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Annals of neurology. 2003;53:454-68 doi:10.1002/ana.10472
9. Nesic O, Svrakic NM, Xu GY, McAdoo D, Westlund KN, Hulsebosch CE. et al. DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. Journal of neuroscience research. 2002;68:406-23 doi:10.1002/jnr.10171
10. Nieto-Diaz M, Esteban FJ, Reigada D, Munoz-Galdeano T, Yunta M, Caballero-Lopez M. et al. MicroRNA dysregulation in spinal cord injury: causes, consequences and therapeutics. Frontiers in cellular neuroscience. 2014;8:53. doi:10.3389/fncel.2014.00053
11. Xu J, Kim GM, Chen S, Yan P, Ahmed SH, Ku G. et al. iNOS and nitrotyrosine expression after spinal cord injury. Journal of neurotrauma. 2001;18:523-32 doi:10.1089/089771501300227323
12. Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R. et al. Cerebellar neurodegeneration in the absence of microRNAs. The Journal of experimental medicine. 2007;204:1553-8 doi:10.1084/jem.20070823
13. Han H, Xia Y, Wang S, Zhao B, Sun Z, Yuan L. Synergistic effects of galectin-1 and reactive astrocytes on functional recovery after contusive spinal cord injury. Archives of orthopaedic and trauma surgery. 2011;131:829-39 doi:10.1007/s00402-010-1233-x
14. Myers SA, DeVries WH, Andres KR, Gruenthal MJ, Benton RL, Hoying JB. et al. CD47 knockout mice exhibit improved recovery from spinal cord injury. Neurobiology of disease. 2011;42:21-34 doi:10.1016/j.nbd.2010.12.010
15. Jee MK, Jung JS, Im YB, Jung SJ, Kang SK. Silencing of miR20a is crucial for Ngn1-mediated neuroprotection in injured spinal cord. Hum Gene Ther. 2012;23:508-20 doi:10.1089/hum.2011.121
16. Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Experimental neurology. 2009;219:424-9 doi:10.1016/j.expneurol.2009.06.015
17. Jee MK, Jung JS, Choi JI, Jang JA, Kang KS, Im YB. et al. MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain. 2012;135:1237-52 doi:10.1093/brain/aws047
18. Wei H, Wang C, Zhang C, Li P, Wang F, Zhang Z. Comparative profiling of microRNA expression between neural stem cells and motor neurons in embryonic spinal cord in rat. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2010;28:545-51 doi:10.1016/j.ijdevneu.2010.04.007
19. Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B. et al. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14:432-44 doi:10.1261/rna.783108
20. Hohjoh H, Fukushima T. Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene. 2007;391:39-44 doi:10.1016/j.gene.2006.11.018
21. Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP. et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817-21 doi:10.1126/science.1121158
22. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-14 doi:10.1016/j.cell.2007.04.040
23. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Developmental cell. 2006;11:441-50 doi:10.1016/j.devcel.2006.09.009
24. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ. et al. Dicer is essential for mouse development. Nature genetics. 2003;35:215-7 doi:10.1038/ng1253
25. Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E. et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5614-9 doi:10.1073/pnas.0801689105
26. Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Developmental biology. 2010;340:459-67 doi:10.1016/j.ydbio.2010.01.039
27. Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:2800-12 doi:10.1002/dvdy.22109
28. Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochimica et biophysica acta. 2008;1779:471-8 doi:10.1016/j.bbagrm.2007.12.006
29. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437-41 doi:10.1126/science.1102513
30. Zhao JJ, Sun DG, Wang J, Liu SR, Zhang CY, Zhu MX. et al. Retinoic acid downregulates microRNAs to induce abnormal development of spinal cord in spina bifida rat model. Child's nervous system: ChNS: official journal of the International Society for Pediatric Neurosurgery. 2008;24:485-92 doi:10.1007/s00381-007-0520-5
31. Yunta M, Nieto-Diaz M, Esteban FJ, Caballero-Lopez M, Navarro-Ruiz R, Reigada D. et al. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One. 2012;7:e34534. doi:10.1371/journal.pone.0034534
32. Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146-60 doi:10.1016/j.neuroscience.2011.03.063
33. Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. Journal of experimental zoology Part B, Molecular and developmental evolution. 2006;306:575-88 doi:10.1002/jez.b.21118
34. Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Current opinion in cell biology. 2008;20:214-21 doi:10.1016/j.ceb.2008.01.006
35. Brandenburger T, Castoldi M, Brendel M, Grievink H, Schlosser L, Werdehausen R. et al. Expression of spinal cord microRNAs in a rat model of chronic neuropathic pain. Neuroscience letters. 2012;506:281-6 doi:10.1016/j.neulet.2011.11.023
36. Nakanishi K, Nakasa T, Tanaka N, Ishikawa M, Yamada K, Yamasaki K. et al. Responses of microRNAs 124a and 223 following spinal cord injury in mice. Spinal cord. 2010;48:192-6 doi:10.1038/sc.2009.89
37. Liu NK, Zhang YP, Titsworth WL, Jiang X, Han S, Lu PH. et al. A novel role of phospholipase A2 in mediating spinal cord secondary injury. Annals of neurology. 2006;59:606-19 doi:10.1002/ana.20798
38. Beattie MS, Farooqui AA, Bresnahan JC. Review of current evidence for apoptosis after spinal cord injury. Journal of neurotrauma. 2000;17:915-25
39. Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nature medicine. 1997;3:73-6
40. Liu XZ, Xu XM, Hu R, Du C, Zhang SX, McDonald JW. et al. Neuronal and glial apoptosis after traumatic spinal cord injury. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17:5395-406
41. Yu YM, Gibbs KM, Davila J, Campbell N, Sung S, Todorova TI. et al. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur J Neurosci. 2011;33:1587-97 doi:10.1111/j.1460-9568.2011.07643.x
42. Liu G, Keeler BE, Zhukareva V, Houle JD. Cycling exercise affects the expression of apoptosis-associated microRNAs after spinal cord injury in rats. Experimental neurology. 2010;226:200-6 doi:10.1016/j.expneurol.2010.08.032
43. Jin L, Wu Z, Xu W, Hu X, Zhang J, Xue Z. et al. Identifying gene expression profile of spinal cord injury in rat by bioinformatics strategy. Molecular biology reports. 2014;41:3169-77 doi:10.1007/s11033-014-3176-8
44. Truettner JS, Motti D, Dietrich WD. MicroRNA overexpression increases cortical neuronal vulnerability to injury. Brain research. 2013;1533:122-30 doi:10.1016/j.brainres.2013.08.011
45. Bao Y, Lin C, Ren J, Liu J. MicroRNA-384-5p regulates ischemia-induced cardioprotection by targeting phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta (PI3K p110delta). Apoptosis: an international journal on programmed cell death. 2013;18:260-70 doi:10.1007/s10495-013-0802-1
46. Xie W, Li M, Xu N, Lv Q, Huang N, He J. et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PloS one. 2013;8:e58639. doi:10.1371/journal.pone.0058639
47. Hutchison ER, Kawamoto EM, Taub DD, Lal A, Abdelmohsen K, Zhang Y. et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia. 2013;61:1018-28 doi:10.1002/glia.22483
48. Moreno-Mateos MA, Barragan V, Torres B, Rodriguez-Mateo C, Mendez-Vidal C, Berezikov E. et al. Novel small RNA expression libraries uncover hsa-miR-30b and hsa-miR-30c as important factors in anoikis resistance. Rna. 2013 doi:10.1261/rna.039461.113
49. Izumi B, Nakasa T, Tanaka N, Nakanishi K, Kamei N, Yamamoto R. et al. MicroRNA-223 expression in neutrophils in the early phase of secondary damage after spinal cord injury. Neuroscience letters. 2011;492:114-8 doi:10.1016/j.neulet.2011.01.068
50. Liu L, Zhang G, Liang Z, Liu X, Li T, Fan J. et al. MicroRNA-15b enhances hypoxia/reoxygenation-induced apoptosis of cardiomyocytes via a mitochondrial apoptotic pathway. Apoptosis: an international journal on programmed cell death. 2013 doi:10.1007/s10495-013-0899-2
51. Liu G, Detloff MR, Miller KN, Santi L, Houle JD. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Experimental neurology. 2012;233:447-56 doi:10.1016/j.expneurol.2011.11.018
52. Hu JZ, Huang JH, Zeng L, Wang G, Cao M, Lu HB. Anti-apoptotic effect of microRNA-21 after contusion spinal cord injury in rats. Journal of neurotrauma. 2013;30:1349-60 doi:10.1089/neu.2012.2748
53. Ouyang YB, Xu L, Lu Y, Sun X, Yue S, Xiong XX. et al. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013;61:1784-94 doi:10.1002/glia.22556
54. Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B. et al. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta neuropathologica. 2013;126:251-65 doi:10.1007/s00401-013-1142-5
55. Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao Y. et al. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Medical oncology. 2013;30:411. doi:10.1007/s12032-012-0411-9
56. Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113-29 doi:10.1053/j.gastro.2006.02.057
57. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-803 doi:10.1038/sj.onc.1210083
58. Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H. et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circulation research. 2007;100:1579-88 doi:10.1161/CIRCRESAHA.106.141986
59. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS. et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and cellular biology. 2008;28:5369-80 doi:10.1128/MCB.00479-08
60. Redell JB, Zhao J, Dash PK. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J Neurosci Res. 2011;89:212-21 doi:10.1002/jnr.22539
61. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer research. 2005;65:6029-33 doi:10.1158/0008-5472.CAN-05-0137
62. Sayed D, He M, Hong C, Gao S, Rane S, Yang Z. et al. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. The Journal of biological chemistry. 2010;285:20281-90 doi:10.1074/jbc.M110.109207
63. Bhalala OG, Pan L, Sahni V, McGuire TL, Gruner K, Tourtellotte WG. et al. microRNA-21 regulates astrocytic response following spinal cord injury. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:17935-47 doi:10.1523/JNEUROSCI.3860-12.2012
64. Sahni V, Mukhopadhyay A, Tysseling V, Hebert A, Birch D, McGuire TL. et al. BMPR1a and BMPR1b signaling exert opposing effects on gliosis after spinal cord injury. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:1839-55 doi:10.1523/JNEUROSCI.4459-09.2010
65. Uittenbogaard M, Baxter KK, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN neuro. 2010;2:e00034. doi:10.1042/AN20100005
66. Bin H, Yujie Z, Yuyang L, Dongmei S, Yingxin Z, Dean J. et al. Impact of metabolic syndrome on clinical outcomes after drug-eluting stent implantation in patients with coronary artery disease. Angiology. 2011;62:440-6 doi:10.1177/0003319711398473
67. Conrad S, Schluesener HJ, Trautmann K, Joannin N, Meyermann R, Schwab JM. Prolonged lesional expression of RhoA and RhoB following spinal cord injury. The Journal of comparative neurology. 2005;487:166-75 doi:10.1002/cne.20561
68. Dergham P, Ellezam B, Essagian C, Avedissian H, Lubell WD, McKerracher L. Rho signaling pathway targeted to promote spinal cord repair. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22:6570-7 doi:20026637
69. Holtje M, Djalali S, Hofmann F, Munster-Wandowski A, Hendrix S, Boato F. et al. A 29-amino acid fragment of Clostridium botulinum C3 protein enhances neuronal outgrowth, connectivity, and reinnervation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23:1115-26 doi:10.1096/fj.08-116855
70. Houle JD, Morris K, Skinner RD, Garcia-Rill E, Peterson CA. Effects of fetal spinal cord tissue transplants and cycling exercise on the soleus muscle in spinalized rats. Muscle & nerve. 1999;22:846-56
71. Beaumont E, Houle JD, Peterson CA, Gardiner PF. Passive exercise and fetal spinal cord transplant both help to restore motoneuronal properties after spinal cord transection in rats. Muscle & nerve. 2004;29:234-42 doi:10.1002/mus.10539
72. van den Brand R, Heutschi J, Barraud Q, DiGiovanna J, Bartholdi K, Huerlimann M. et al. Restoring voluntary control of locomotion after paralyzing spinal cord injury. Science. 2012;336:1182-5 doi:10.1126/science.1217416
73. Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke; a journal of cerebral circulation. 2008;39:959-66 doi:10.1161/STROKEAHA.107.500736
74. Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK. et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clinical chemistry. 2009;55:1977-83 doi:10.1373/clinchem.2009.131797
75. Zhou F, Guan Y, Chen Y, Zhang C, Yu L, Gao H. et al. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. International journal of clinical and experimental pathology. 2013;6:1826-38
76. Hachisuka S, Kamei N, Ujigo S, Miyaki S, Yasunaga Y, Ochi M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal cord. 2014 doi:10.1038/sc.2014.86
77. Medina PP, Slack FJ. Inhibiting microRNA function in vivo. Nature methods. 2009;6:37-8 doi:10.1038/nmeth0109-37
78. Mirnezami AH, Pickard K, Zhang L, Primrose JN, Packham G. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. European journal of surgical oncology: the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2009;35:339-47 doi:10.1016/j.ejso.2008.06.006
79. Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T. et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic acids research. 2007;35:2885-92 doi:10.1093/nar/gkm024
80. Broderick JA, Zamore PD. MicroRNA therapeutics. Gene therapy. 2011;18:1104-10 doi:10.1038/gt.2011.50
81. Hu JR, Lv GH, Yin BL. Altered microRNA expression in the ischemic-reperfusion spinal cord with atorvastatin therapy. Journal of pharmacological sciences. 2013;121:343-6
Author contact
![]() Corresponding author: Zhe-Yu Chen, Department of Neurobiology, Shandong Provincial Key Laboratory of Mental Disorders, School of Medicine, Shandong University, Jinan, Shandong 250012, PR China; E-mail: zheyuchenedu.cn.
Corresponding author: Zhe-Yu Chen, Department of Neurobiology, Shandong Provincial Key Laboratory of Mental Disorders, School of Medicine, Shandong University, Jinan, Shandong 250012, PR China; E-mail: zheyuchenedu.cn.

 Global reach, higher impact
Global reach, higher impact