10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(9):1072-1083. doi:10.7150/ijbs.9719 This issue Cite
Review
Caspases: A Molecular Switch Node in the Crosstalk between Autophagy and Apoptosis
1. Department of Neurosurgery, Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China;
2. Department of Cardiology, Zhejiang Provincial People's Hospital, Hangzhou, China;
3. Clinical Research Center, Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China.
4. Department of Neurological Surgery, Weill Cornell Medical College, New York, New York, USA.
*These authors provided equal contribution to this work.
Received 2014-5-22; Accepted 2014-8-20; Published 2014-9-13
Abstract
Autophagy and apoptosis are two important catabolic processes contributing to the maintenance of cellular and tissue homeostasis. Autophagy controls the turnover of protein aggregates and damaged organelles within cells, while apoptosis is the principal mechanism by which unwanted cells are dismantled and eliminated from organisms. Despite marked differences between these two pathways, they are highly interconnected in determining the fate of cells. Intriguingly, caspases, the primary drivers of apoptotic cell death, play a critical role in mediating the complex crosstalk between autophagy and apoptosis. Pro-apoptotic signals can converge to activate caspases to execute apoptotic cell death. In addition, activated caspases can degrade autophagy proteins (i.e., Beclin-1, Atg5, and Atg7) to shut down the autophagic response. Moreover, caspases can convert pro-autophagic proteins into pro-apoptotic proteints to trigger apoptotic cell death instead. It is clear that caspases are important in both apoptosis and autophagy, thus a detailed deciphering of the role of caspases in these two processes is still required to clarify the functional relationship between them. In this article, we provide a current overview of caspases in its interplay between autophagy and apoptosis. We emphasized that defining the role of caspases in autophagy-apoptosis crosstalk will provide a framework for more precise manipulation of these two processes during cell death.
Keywords: autophagy, apoptosis, caspases, Atg proteins, crosstalk, cell death.
Introduction
Macroautophagy (hereafter refer to as autophagy) is an intracellular catabolic process by which portions of cytoplasmic components are delivered to autolysosomes for degradation [1]. Apoptosis is a well-studied form of programmed cell death that displays typical morphological characteristics, such as chromatin condensation, nuclear fragmentation, and membrane blebbing [2, 3]. Both autophagy and apoptosis are highly orchestrated cascades that play a vital role in embryonic development and tissue homeostasis [4, 5]. Dysregulation of them have been associated with a number of pathologies, such as cancer, autoimmune diseases, and neurodegenerative diseases [6]. It's known that autophagy extensively communicates with apoptosis during cell fate decision in a myriad of physiological and pathological conditions [7, 8]. In low-stress situations, cells initiate autophagy as a pro-survival mechanism to combat apoptotic cell death. However, as stress increases towards a point of no return where cells are doomed to die, cells block autophagy and begin initiating apoptotic cascades [9]. Thus, a better knowledge of those interactions between autophagy and apoptosis is of great importance, in order to optimally manipulate these two crucial pathways for therapeutic purposes.
An overview of the cellular and molecular events during autophagy.
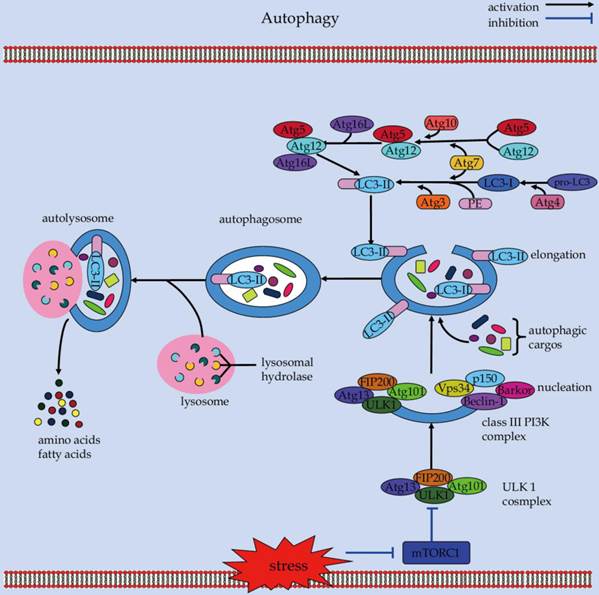
Caspases, a family of cysteinyl aspartate-requiring proteases, play a central role in the transduction of apoptotic signals [10]. Apoptotic caspases can be divided into two general categories: the initiator caspases consisting of caspase-2, -8, -9 and -10; and the effector caspases consisting of caspases-3, -6 and -7 [11, 12]. Caspases normally appear as inactive zymogenic precursors. They can be activated to mediate apoptosis in response to diverse pro-apoptotic stimuli from outside and inside of the cells [13]. Emerging evidences demonstrate that caspases can also influence the non-apoptotic signaling events, in particular, the “self-eating” autophagy [14, 15]. Further research shows that caspases can inhibit autophagy by cleaving and destroying the pro-autophagic activity of autophagy-related (Atg) proteins, such as Atg3 and Beclin-1 [16, 17]. Whereas, caspase-9 can facilitate autophagosome formation by promoting the Atg7-dependent conversion process of microtubule-associated protein 1 light chain 3 (LC3) [18]. On the other hand, the cytoprotective autophagy counter-balances apoptosis by continuous sequestration of active caspase-8 into autophagosomes for its subsequent degradation in Bax-/- Hct116 colon carcinoma cells [19]. However, autophagosomal membrane also serves as a platform for intracellular death-inducing signaling complex (DISC)-mediated caspase-8 activation and apoptosis [20]. Therefore, elucidating these unexpected functions of caspases in linking the autophagic and apoptotic signaling pathways helps decipher the molecular basis underlying autophagy-apoptosis crosstalk, which can drive therapeutic development by optimally manipulating these two important processes.
In this review article, we focus on the biological effects and underlying mechanisms of caspases in directing the conversation between autophagy and apoptosis in the mammalian systems (unless otherwise stated). In particular, we emphasize that further insights into the role of caspases in mediating autophagy-apoptosis crosstalk will guide future therapeutic strategies during cell death control.
The autophagic process
Autophagy is a catabolic process which involves the turnover of cytosolic protein aggregates and damaged organelles in the lysosomes via double-membrane autophagosomes (Figure 1) [21]. The autophagic process consists of initiation, elongation, maturation, fusion, and degradation [22]. It begins with the nucleation and expansion of an isolated membrane called the phagophore, which can sequester portions of cytoplasmic materials to form an autophagosome [23]. Subsequently, the autophagosome fuses with a lysosome to form an autolysosome, where the captured cargos are degraded by hydrolytic enzymes for recycling [24].
The autophagy machinery is tightly controlled by a set of Atg proteins that can assemble into several different complexes to produce autophagosomes [25, 26]. Among these core multimolecular complexes, the ULK1 complex, made of ULK1, Atg13, Atg101, and FIP200 (focal adhesion kinase family-interacting protein of 200 kD), plays a critical role in the initiation of autophagy [27]. Under nutrient-rich conditions, the serine/threonine kinase mammalian Target of rapamycin (mTOR) acts in concert with other proteins of the mTOR complex 1 (mTORC1) to suppress autophagy through direct interaction with the ULK1 complex [27]. In contrast, in response to stress conditions such as starvation, mTOR is inhibited and dissociates from the ULK1 complex [28]. This can result in the activation of ULK1 and ULK1-catalyzed phosphorylation of Atg13, FIP200, and ULK1 itself, which is essential for initiating autophagy [29]. Also, AMP activated protein kinase (AMPK) can activate ULK1 via a direct phosphorylation mechanism [30]. AMPK interacts with ULK1 and phosphorylates it on Ser 317 and Ser 777, which can lead to activation of ULK1 kinase and autophagy induction in response to glucose starvation [29]. Downstream of the ULK1 complex, the class III phosphatidylinositol 3-kinase (PI3K) complex exerts an essential role in the nucleation and assembly of the initial phagophore membrane [31]. Beclin-1 functions as a platform by binding several cofactors such as barkor (Beclin-1-associated autophagy-related key regulator), p150, and UVRAG (UV irradiation resistance-associated gene) to assemble the class III PI3K complex during autophagosome formation [32-36]. Of note, the phosphorylated ULK1 can promote the class III PI3K complex-mediated phagophore nucleation [37], possible through phosphorylation of key molecules such as AMBRA (activating molecule in Beclin-1-regulated autophagy protein 1) [38], and Beclin-1 [39]. The subsequent elongation process requires the involvement of two ubiquitin-like conjugation reactions: the Atg5-Atg12 conjugation and LC3-PE (phosphatidylethanolamine) conjugation [40]. Analogous to ubiquitination, Atg12 is conjugated to Atg5 to form Atg12-Atg5 conjugate, with the help of the E1-like enzyme Atg7 and the E2-like enzyme Atg10. Similarly, the conjugation of LC3 to PE is mediated by the E1-like enzyme Atg7 and the E2-like enzyme Atg3. The Atg12-Atg5 then conjugates with Atg16L (Atg16-like protein) to form an approximately 800-kDa protein complex containing Atg12-Atg5-Atg16L [41]. This complex serves as a platform for stimulating the LC3-PE conjugation reaction [42, 43], which is essential for the elongation of the pre-autophagosomal membranes [44, 45]. Noteworthily, although molecules such as Beclin-1 interacting proteins (i.e., UVRAG and Rubicon) have been proven to be involved in the maturation of autophagosomes [46-48], the role of Atg proteins in the maturation and fusion step of autophagosomes, however, remains unclear and requires further characterization.
The apoptotic cascades
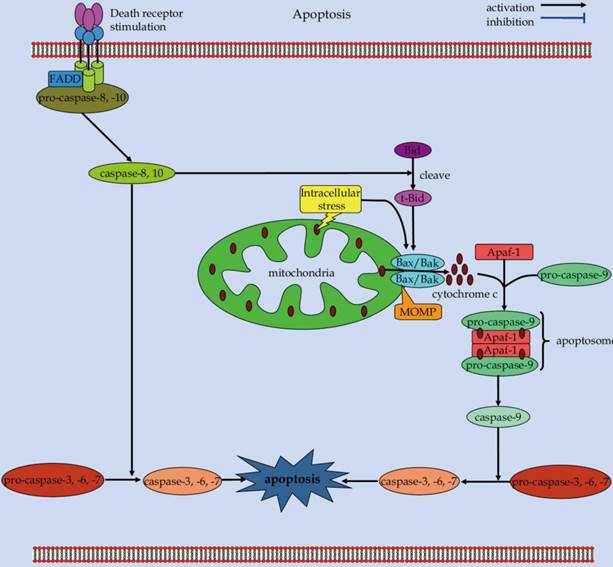
Apoptosis is a self-destructive process with carefully choreographed steps (Figure 2) [49]. During the apoptotic process, a series of intracellular events come into play to decommission the unwanted and dangerous cells [50, 51]. Apoptosis is crucial for embryonic development, organogenesis, as well as tissue homeostasis [3, 52]. Aberrant regulation of apoptosis can lead to either pathological cell accumulation or inappropriate cell loss, which eventually results in the pathogenesis of various human disorders, including cancer, infection, neurodegeneration, and autoimmune diseases [5, 50, 53].
Caspases, both the initiators and the effectors, play a major role in execution of apoptotic cascades [54]. Mechanically, two major apoptotic signaling pathways, including the extrinsic and intrinsic signals, converge onto these caspases to initiate cell death [3, 55]. The extrinsic apoptotic cascade is triggered by ligation of cell-surface death receptors of the tumor necrosis factor (TNF) receptor superfamily, such as CD95/Fas or TNF-related apoptosis-inducing ligand (TRAIL) receptors [56, 57]. Upon ligand binding, the receptors become oligomerized forms, recruiting Fas-associated death domain (FADD) and procaspase-8 to form the death-inducing signaling complex (DISC) [58]. Recruitment of procaspase-8 to the DISC facilitates its self-cleavage into active caspase-8 enzyme that later cleaves effector caspases [59]. These effector caspases can further cleave a number of cytosolic and nuclear substrates to execute apoptotic cascades that leads to morphological and biochemical features of apoptosis [60]. Alternatively, caspase-8 can propagate apoptotic death signals through proteolysis of the Bcl-2 homology-3 (BH3)-only protein Bid. The truncated Bid (tBid) translocates to the outer mitochondrial membrane to orchestrate a process that mediating mitochondrial outer membranes permeabilization (MOMP) [61, 62]. MOMP is a crucial event during apoptosis [63]. As an outcome of this process, pores are formed in the outer mitochondrial membrane, membrane integrity is compromised, and apoptogenic proteins (i.e., cytochrome c) within the intermembrane space are released into the cytosol to induce apoptotic cell death.
The intrinsic pathway of apoptosis is triggered by a wide range of intracellular death signals, such as DNA damage, cytotoxic stress, and growth factor withdrawal [64]. In response to these apoptotic stimuli, the integrity of the outer mitochondrial membrane is lost, and the permeabilization of outer mitochondrial membranes is culminated [65]. This will thereby permit release of many pro-apoptotic proteins that are normally confined to the intermembrane space, in particular, cytochrome c, into the cytoplasm to promote caspase activation [66, 67]. In the cytoplasm, the apoptogenic factor cytochrome c associates with apoptotic protease-activating factor 1 (Apaf-1) and initiator caspase-9 to form a multi-protein complex known as apoptosome [65, 68]. The apoptosome serves as a molecular platform for caspase-9 processing and activation, which can subsequently activate downstream executioner caspases to orchestrate apoptotic cell death [69].
Scheme of the extrinsic and intrinsic apoptotic pathways.
Caspases: key orchestrators of autophagy-apoptosis crosstalk
Autophagy and apoptosis have been proven to extensively cross talk with each other in cell fate decision [70-73]. With series of key findings, molecular mechanisms underlying the complex counter-regulation between autophagy and apoptosis are emerging. Of much interest, caspases, a family of cysteine proteases that are instrumental to the execution of apoptosis, serve as important players in directing these conversations between autophagy and apoptosis. Multiple pro-apoptotic signals, both inside and outside the cells, can converge to activate caspases to initiate apoptosis. In addition, activated caspases can degrade essential autophagy proteins (i.e., Beclin-1, Atg5, and Atg7) to shut down the autophagic response instead [74-76]. Moreover, after been cleaved by caspases, some pro-autophagic proteins can be even converted into pro-apoptotic ones to trigger apoptotic cell death. On the other hand, autophagy can influence apoptotic cascades by regulating the amount and activity of caspases. And autophagic elimination of caspases, such as caspase-8, can hamper their involvement in the apoptotic pathways [19].
Initiator caspases (Figure 3)
Caspase-8 is a well-known initiator caspase implicated in the death receptor-triggered apoptosis [77]. As aforementioned above, in the context of extrinsic apoptosis, caspase-8 is activated by dimerization and autoproteolysis inside the DISC, and subsequently released into the cytoplasm to activate effector caspases for efficient apoptosis execution [57, 78-81].
Apart from its pro-apoptotic properties, caspase-8 plays an important role in the modulation of autophagy. Data from in vitro studies demonstrated that caspase-8 can rescue T cells from hyperactive autophagy [15, 82]. In response to energetic demands for rapid clonal expansion, the autophagic responses inside T cells are stimulated [82]. The autophagic signaling can induce an interaction among Atg5-Atg12-Atg16L, FADD, caspase-8, and receptor interacting protein kinase 1 (RIPK1), which promotes the activation of caspase-8 in mitogenically stimulated live T cells [83]. Activated caspase-8 can directly cleave the serine/threonine kinase RIPK1 as part of a negative feedback loop to limit autophagic signaling [82, 84]. In fact, RIPK1 also has emerged as a key upstream regulator which controls of apoptosis and necroptosis [85-87]. Besides, RIPK1 is required for MAPK8 activation and induction of protective autophagy to blunt apoptosis in TRAIL-treated cancer cells [88]. Whereas, RIPK1 can be cleaved by the active caspase-8 during apoptotic cascades [84]. These evidences indicate that the caspase-8/RIPK1 is involved in the crosstalk between autophagy, apoptosis and necroptosis, which is of interest for future investigation. Atg3 is a critical regulatory component for autophagosome biogenesis. It is conceived as a new substrate of caspase-8 during receptor-mediated cell death [16]. By targeting the evolutionary conserved LETD sequence (Atg3 amino acids 166-169) of this protein, caspase-8 mediates the cleavage of Atg3. As a result, the pro-autophagic activity of Atg3 is abolished after it has been cleaved, which is a critical event in autophagy inactivation during death receptor-triggered apoptosis [16].
Scenarios for initiator caspases-mediated crosstalk between apoptosis and autophagy.
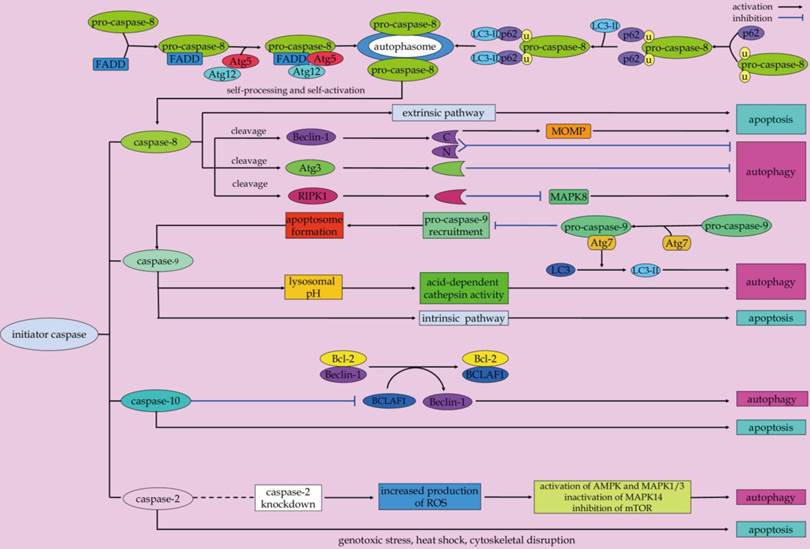
On the other hand, autophagy contributes to the regulation of caspase-8-mediated apoptosis under the condition of proteasome inhibition [82, 89]. In response to proteasome inhibition, the autophagy flux can increase as a compensatory mechanism for protein degradation [90, 91]. Moreover, it has been shown that autophagosomes serve as a platform for intracellular DISC formation for caspase-8 self-processing, which is responsible for initiation of the caspase cascades and execution of apoptosis [20]. The association of FADD with the Atg12-Atg5 complex facilitates caspase-8 recruitment to the autophagosomal membrane to form a proper higher-order oligomer structure for caspase-8 activation [20]. Also, the adapter protein p62 can facilitate the recruitment of self-associated caspase-8 to autophagosomal membranes via its interaction with LC3-II [20]. This event contributes to the formation of proper oligomer structures for caspase-8 self-processing and self-activation [20, 92, 93]. It is important to note that the functional domains of p62, including UBA (ubiquitin-associated) which binds ubiquitinated proteins, PB1 (Phox and Bem1p) that mediates dimerization/oligomerization, and LC3 interaction region, are required for p62 in mediating caspase-8 activation [94-97]. In particular, the UBA domain at the C-terminus of p62 is an important domain for binding and aggregation of polyubiquitinated caspase-8 for its final activation [98, 99]. As a consequence, the enhancement of caspase-8 self-aggregation and activation due to p62 upregulation in the setting of autophagy inhibition can sensitize human colon carcinoma cells to the BH3 mimetic agent ABT-263-induced apoptosis [97]. Consequently, blockage of autophagolysosomal degradation pathway can result in an increase in the association of aggregated caspase-8 with p62 and LC3, which contributes to caspase-8 oligomerization, activation, and apoptotic signal transduction in the mammalian degenerin homolog G430F mutant-triggered cells [93]. Conversely, the autophagic degradation of caspase-8 precursor or a subunit of the active caspase-8 enzyme can hinder caspase-8-induced apoptosis [19]. In Bax-/- Hct116 colon carcinoma cells that are resistant to TRAIL-mediated apoptosis, TRAIL signaling mediates a protective autophagic response. This keeps the caspase-8-dependent apoptotic response at bay, probably through continuous elimination of the caspase-8 precursor and/or the active caspase-8 large subunit [19]. And autophagy inhibition due to Beclin-1 RNA interference allows for a significant increase in caspase-8 enzymatic activity, resulting in downstream caspase processing and apoptosis activation [19]. Summarizing it all together, caspase-8 functions as a switch point in polarization between autophagy and apoptosis during cell-fate determination.
Caspase-9 is another essential initiator caspase that is involved in the intrinsic apoptosis pathway [100, 101]. Once activated, caspase-9 is able to process downstream effector caspases, including caspase-3, -6, and -7, all of which can target key structural and regulatory proteins to bring about apoptotic cell death [102]. Intriguingly, Jeong and colleagues demonstrated the involvement of caspase-9 in promoting autophagy-mediated cell survival of breast cancer MCF-7 cells [103]. Inhibition of caspase-9 can negatively regulate lysosomal pH and acid-dependent cathepsin activities that causes blockage of cytoprotective autophagy and augment of cell death following the exposure of the non-steroidal anti-inflammatory drug FR122047 [103]. In addition, Han and coworkers described that caspase-9 acts as a novel co-regulator of autophagy and apoptosis through its mutually regulation with Atg7 [18]. Caspase-9 can interact with Atg7 via the C-terminal region of the large subunit caspase-9, which facilitates the Atg7-dependent formation of autophagosomal LC3-II and autophagic function. The interaction of caspase-9 with Atg7 can potentially interfere with the recruitment and processing of caspase-9 in the apoptosome. The interference can ultimately inhibit caspase-9 activation and apoptotic signal transduction [18]. Although the cross-regulation between Atg7 and caspase-9 is a possible mechanism that determines the participation of caspase-9 either in autophagy or apoptosis, additional molecular scenarios should be still considered. For instance, a phosphorylation mechanism may serve as a potential determinant in the involvement of caspase-9 in either apoptotic or autophagic cascades [18, 104, 105]. However, the exact molecular mechanisms that forces caspase-9 into either autophagic or apoptotic mode should be addressed.
Caspase-2, one of the most conserved members of the caspase family, has been proven to be a crucial mediator of apoptotic cascades in a context-dependent manner. Examples include genotoxic stress [106], heat shock [107, 108], and cytoskeletal disruption [109]. In addition to its role in apoptosis, evidence suggests that caspase-2 is involved in the negative regulation of autophagy [110]. Loss of caspase-2 has been shown to lead to an enhanced autophagy in mouse embryonic fibroblast. Increased production of reactive oxygen species and oxidative stress in cells lacking caspase-2 contribute to upregulation of AMPK, activation of MAPK1/3, downregulation of mTOR induction, and inactivation of MAPK14, which play a role autophagy induction [110]. Additionally, when the neurons cultured from young adult mice cannot undergo apoptosis due to lack of caspase-2, the pro-survival autophagy can be induced at an early stage in response to rotenone-mediated mitochondrial oxidative stress [111]. These evidences suggested that caspase-2 can be a molecular link between autophagy and apoptosis.
Caspase-10, the closest homolog of caspase-8, functions as an initiator caspase of extrinsic apoptotic signaling pathway [112, 113]. Of interest, Lamy et al. discovered that caspase-10 also plays an important role in maintaining the proper autophagic activities in myeloma cells [114]. Caspase-10 can cleave and inactivate Bcl-2-associated transcription factor 1 (BCLAF1), a potent autophagy inducer that can induce autophagy by displacing Beclin-1 from Bcl-2 complex, thus preventing myeloma cells from uncontrolled autophagy [114]. The enhanced association between the caspase-like protein cFLIPL and caspase-10 in myeloma cells promotes caspase-10-mediated BCLAF1 cleavage, thus limits BCLAF1-induced autophagic cell death of myeloma cells [114].
Effector caspases (Figure 4)
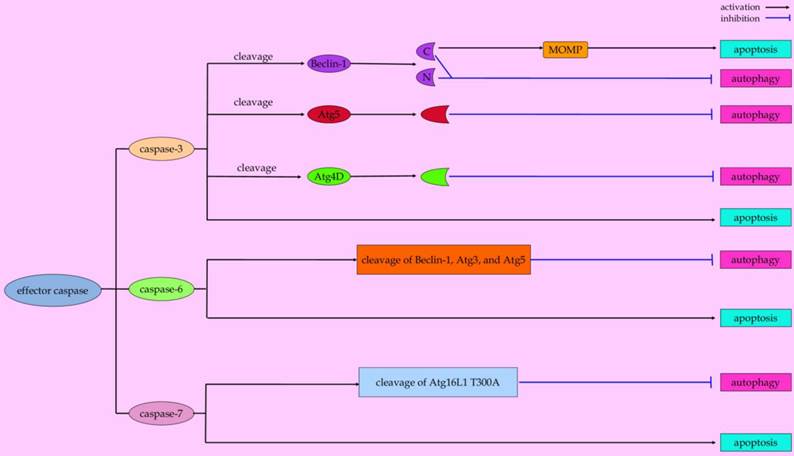
Caspase-3 is a predominant player in the execution of apoptotic cell death. However, recent studies indicated that capase-3 plays a role in autophagic processes [115]. In human apoptotic endothelial cells under conditions of nutrient shortage, activated caspase-3 favors the extracellular export of autophagic vacuoles via rerouting autophagic vacuoles toward the cell membrane [116]. The externalization of large autophagic vacuoles may contribute to apoptotic decrease in volume, a geometric determinant for cell dismantling into apoptotic bodies [117]. These evidences represent caspase-3 as a potential molecular switch in mediating crosstalk between the autophagic and apoptotic programs [116]. Zhu and coworkers reported that caspase-3 can cleave autophagy-associated protein Beclin-1 at positions 124 and 149 during staurosporine-induced apoptosis, thus abrogating the pro-autophagic effect of Beclin-1 in HeLa cells [118]. Moreover, the cleavage of Beclin-1 disrupts its interaction with Bcl-2, which leads to the exposure of the BH3 domain of Beclin-1 to other anti-apoptotic Bcl-2 family members [119-121]. The competitive binding of truncated Beclin-1 with anti-apoptotic Bcl-2 family members allows pro-apoptotic BH3-only molecules released from the Bcl-2/Bcl-xL complex to initiate intrinsic apoptotic cascades [121, 122]. Also, Atg4D, one member of the Atg4 family in mammalian cells that contributes to starvation-induced autophagy, can be cleaved by caspase-3 at the DEVD63K motif [123]. More importantly, cleavage of Atg4D by caspase-3 enables it to stimulate the priming and delipidation of the LC3 paralogue, GABARAP-L1 (γ-aminobutyric acid receptor-associated protein (GABARAP)-like 1), indicating that caspase-3 promotes Atg4D-coordinated autophagy [123]. It is noteworthy that caspase-cleaved ΔN63 Atg4D can induce cell death when overexpressed in human cells [123]. The cleaved Atg4D-mediated cell toxicity is associated with its recruitment to mitochondria to induction of apoptosis [123]. Unfortunately, whether Atg4D can cause the release of proapoptotic mitochondrial factors by targeting Bax or Bak is still unknown.
Other effector caspases, including caspase-6 and caspase-7, not only play a role in the execution phase of apoptosis [124, 125], but also affect the autophagic pathway. When melanoma cell lines undergo arginine deprivation, TRAIL-initiated caspase-6 activation contributes to disruption of pro-survival autophagy by cleavage of two key autophagic proteins (Beclin-1 and Atg5) [74]. And evidence from an in vitro study demonstrated that proteins such as Atg3 and p62 can be cleaved by caspase-6, suggesting its potential role in the regulation of autophagy [126]. By using knock-in mice, the effects of the Atg16L1 T300A (a common threonine to alanine coding variant at position 300 in Atg16L) polymorphism on the autophagy pathway were studied by Lassen et al. They found that Atg16L is a substrate for caspase-7, while the Atg16L1 T300A is more susceptible to caspase-7-mediated cleavage compared with the wild-type protein, which is associated with the reduced antibacterial autophagy in cells of Atg16L T300A mice [76]. Nevertheless, other mechanisms underlying these two caspases in linking autophagy and apoptosis are warranted to be further elucidated.
Paradigms of effector caspases-mediated conversation between autophagy and apoptosis.
The calpain system
Calpains represent a well-conserved family of calcium-sensitive cysteine proteases localized to the cytosol and mitochondria [127]. They exert important roles in a number of fundamental physiological processes, including the execution of apoptosis [128-130]. Evidence demonstrates calpains are involved in the apoptotic process at numerous steps. For instance, calpain is capable of processing and activation of several caspases, such as caspase-7 and caspase-12, which can sensitize cells to Ca2+-dependent apoptosis [131-133]. Calpain is also able to cleave pro-apoptotic Bcl-2 family proteins including Bid and Bax, which in turn mediates cytochrome c release and initiates the apoptotic execution [134-136].
Not surprisingly, similar to the caspase family proteases, the calpain plays a role in linking autophagy to apoptosis [137]. In sphingosine-1-phosphate phosphohydrolase 1 (SPP1)-depleted cells, doxorubicin switches protective autophagy to apoptosis via calpain-mediated Atg5 cleavage [138]. The truncated Atg5 cleaved by calpain translocates from the cytosol to mitochondria and associates with the anti-apoptotic Bcl-xL to trigger cytochrome c release and caspase activation, and triggers apoptotic cell death [139]. Taken together, these experimental data suggest the potential importance of calpain in the crosstalk between autophagy and apoptosis and the need for further investigation of the cellular functions of these cysteine proteases in the switch between autophagy and apoptosis.
Caspase-mediated autophagy-apoptosis crosstalk in other model systems
The role of caspase in the cross-regulation between apoptosis and autophagy has also been surveyed in other model systems, in particular, the Drosophila system. As an effector caspase, death caspase-1 (Dcp-1) plays a role in maintaining the balance between autophagy and apoptosis during Drosophila melanogaster oogenesis [140-142]. DeVorkin et al., found that Dcp-1 is a positive regulator of starvation-induced autophagic flux during Drosophila midoogenesis [143]. Dcp-1 promotes autophagy by negatively regulating the levels of mitochondrial adenine nucleotide translocase stress-sensitive B and adenosine triphosphate, suggesting a novel mechanism of caspase-mediated regulation of autophagy [143]. By performing a large-scale in vivo genetic screen, Kim and coworkers found that full-length Dcp-1 can induce autophagy, whereas the truncated active Dcp-1 can result in apoptosis instead [144]. It is also hypothesized that the levels of Dcp-1 activity determine the sensitivity thresholds of autophagic and apoptotic responses [142]. Low levels of Dcp-1 activity due to starvation initially induce a pro-survival autophagy for Drosophila melanogaster cells [145]. Whereas, higher levels of Dcp-1 activity in response to prolonged starvation signals will result in the cleavage and activation of another effector caspase, drICE [146], which triggers apoptotic death of these cells. Nevertheless, the molecular details of Dcp-1 in governing the autophagy-apoptosis switch are warranted to be elucidated [145].
Concluding Remarks and Perspectives
Autophagy and apoptosis are two evolutionarily conserved processes that play a crucial role in determining cell fate. Despite of the marked differences between these two catabolic processes, autophagy and apoptosis are intimately connected with each other. In the majority of cases, autophagy and apoptosis tend to be mutually inhibitory. Caspases have a cardinal role in apoptotic cell death and plays a critical role in directing autophagy-apoptosis crosstalk (Table 1). Processed caspases can shut off the autophagic response by degradation of Atg proteins (i.e., Beclin-1, Atg5, and Atg7). In some special cases, the pro-autophagic proteins can be even converted into pro-apoptotic ones to mediate apoptotic cell death after having been cleaved by caspases. On the other hand, autophagy can influence the apoptotic cascades by regulating the amount and activity of caspases. Undoubtedly, these key findings have renewed our knowledge of the unique roles of caspases in directing autophagy-apoptosis conversation.
However, molecular mechanisms underlying caspase-mediated autophagy-apoptosis crosstalk are still incomplete and fragmented. Current evidence emphasized that direct protein-protein interactions between caspases and autophagy-associated proteins enable the multiple layers of communications between autophagy and apoptosis during cell fate decision [9, 147]. For instance, most cases merely illustrate a functional outcome for the given interacting proteins (i.e., cleavage of Beclin-1 by caspase-3 abolishs its pro-autophagic function) in the in vitro settings. However, these binary protein interactions of specific caspases and Atg proteins exhibit clear directionality that does suggest the effects of these given caspases on autophagic and apoptotic processes in an in vitro assay under specific conditions. It remains a pending conundrum whether such binary interactions actually reflect the nature of the crosstalk between autophagy and apoptosis in an in vivo circumstance. Thus, further research is necessary to evaluate that the functions of the protein-protein interactions between caspases and Atg proteins in the autophagy-apoptosis crosstalk in animal models and human clinical models. This will provide a more global and integrative understanding of how they converge together to determine autophagic and apoptotic response in the physiological and pathological settings.
In addition, there is a substantial body of literatures implying that caspases-mediated cleavage of Atg proteins underlies the apoptotic pathway to block the autophagic flux. In contrast, less is known at the mechanistic level about how Atg-dependent autophagic mechanism controls apoptosis by regulating the amount and activity of caspase. Therefore, it is interesting to assess whether autophagy is capable of modulating the apoptotic process by selectively degrading other caspases beyond caspase-8.
Moreover, the interrelationship between autophagy and apoptosis are highly context-dependent and cell-specific [8], which places the caspase network in a more complicated situation in driving the switch between autophagy and apoptosis. Thus, effective techniques such as proteomics studies and transgenic technology (i.e., transgenic mice) are required to further reveal the potential substrates of caspases in the autophagic process. More potent and specific “druggable” inhibitors are anticipated for the experimental or therapeutic inhibition of autophagy. More reliable and meaningful assays for measuring autophagic flux are needed as well. Together, these chemical and proteomics technologies help manipulate and analyze the caspase interactome to probe the mutual relationships between autophagic and apoptostic processes.
Summary of current evidences concerning the molecular basis of caspase-mediated autophagy-apoptosis crosstalk.
| Model | Specific caspases | Associated molecular mechanisms | Main findings | References |
|---|---|---|---|---|
| OT1 and OT-II splenocytes; FADD-/- MEFs; caspse8-/- T cells | caspase-8 | RIPK1 cleavage | Anti-autophagic effect | Bell et al., [82] |
| Jurkat T cells; U937 cells; HEK293T cells | caspase-8 | Atg3 cleavage (Atg3 amino acids 166-169) | Anti-autophagic effect | Oral et al., [16] |
| Atg5+/+ and Atg5-/- MEF cells; Atg3+/+ and Atg3-/- MEF cells; FADD+/+ and FADD-/- MEF cells; KG-1 cells | caspase-8 | Association with FADD:Atg12-Atg5 complex | Pro-apoptotic effect | Young et al., [20] |
| H460 cells; HEK293T cells; Bax+/- HCT116 cells; HT29 cells; JHH6 cells; MG63 cells; U2OS cells | caspase-8 | Association with p62 | Pro-apoptotic effect | Jin et al., [92] |
| HEK293T cells; bax-/-bak-/- BMK cells | caspase-8 | Association with p62 | Pro-apoptotic effect | Pan et al., [93] |
| HCT116 Bax-/-cells; caspase-8-deficient, FADD-deficient, and parental Jurkat cells | caspase-8 | Association with p62 | Pro-apoptotic effect | Huang et al., [97] |
| Bax-/- HCT116 cells; Jurkat cells | caspase-8 | Anti-apoptotic effect | Hou et al., [19] | |
| MCF-7 breast cancer cells | caspase-9 | Pro-autophagic effect | Jeong et al., [103] | |
| Tumor cell lines, including Hct116, HeLa, MB-MDA-231, and RKO, VP-16-resistant MDA-MB-231 cells | caspase-9 | Atg7-caspase-9 complex | Pro-autophagic effect Anti-apoptotic effect | Han et al., [18] |
| Myeloma cell lines, lymphoma cell lines, and the human stromal cell line HS-5 | caspase-10 | Cleavage of BCLAF1 | Anti-autophagic effect | Lamy et al., [114] |
| Neurons, astrocytes, osteoclasts, and embryonic fibroblasts cultured from caspase-2 knockout mice | caspase-2 | Anti-autophagic effect | Tiwari et al., [110, 111] | |
| HeLa cells | caspase-3 | cleavage of Beclin-1 at positions 124 and 149 | Anti-autophagic effect Pro-apoptotic effect | Zhu et al., [118] |
| HeLa cells; HEK293 cells | caspase-3 | cleavage of Atg4D at the DEVD63K motif | Pro-autophagic effect Pro-apoptotic effect | Betin et al., [123] |
| Melanoma cell lines A375 and A2058 | caspase-6 | cleavage of Beclin-1 and Atg5 | Anti-autophagic effect | You et al., [74] |
| Atg16L1 T300A mice | caspase-7 | cleavage of ATG16L1 T300A | Anti-autophagic effect | Lassen et al., [76] |
Abbreviations
AMBRA: activating molecule in Beclin1-regulated autophagy protein 1; AMPK: AMP activated protein kinase; Apaf-1: apoptotic protease-activating factor 1; Atg: autophagy-related; Atg16L: Atg16-like protein; Atg16L1 T300A: a common threonine to alanine coding variant at position 300 in Atg16L; barkor: Beclin-1-associated autophagy-related key regulator; BCLAF1: Bcl-2-associated transcription factor 1; BH3, BCL-2 homology-3; Dcp-1: death caspase-1; DISC: death-inducing signaling complex; FADD: Fas-associated death domain; FIP200: focal adhesion kinase family-interacting protein of 200 kD; GABARAP: γ-aminobutyric acid receptor-associated protein; LC3: the protein microtubule-associated protein 1 light chain 3; MOMP: mitochondrial outer membranes permeabilization; mTOR: mammalian Target of rapamycin; PE: phosphatidylethanolamine; PI3K: phosphatidylinositol 3-kinase; RIPK1: receptor interacting protein kinase 1; SPP1: sphingosine-1-phosphate phosphohydrolase 1; TNF: tumor necrosis factor; TRAIL: TNF-related apoptosis-inducing ligand; UBA: ubiquitin-associated; UVRAG: UV irradiation resistance-associated gene.
Acknowledgements
This study was supported by National Natural Science Foundation of China (grant. 81171096, 81371433, 81271273, and 81371369), Research Fund for the Doctoral Program of Higher Education of China (20120101120030), Zhejiang Provincial Medical Science and Technology Planning Project (grant. 2012RCA030 and 2013KYA088), Zhejiang Provincial Education Planning Project (Y201226287) and Zhejiang Provincial Natural Science Foundation of China (grant Y13H090007 and LY13H090002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nature cell biology. 2013;15:713-20 doi:10.1038/ncb2788
2. Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770-6 doi:10.1038/35037710
3. Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nature reviews Molecular cell biology. 2008;9:231-41 doi:10.1038/nrm2312
4. Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nature cell biology. 2010;12:823-30 doi:10.1038/ncb0910-823
5. Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456-62
6. Caroppi P, Sinibaldi F, Fiorucci L, Santucci R. Apoptosis and human diseases: mitochondrion damage and lethal role of released cytochrome C as proapoptotic protein. Current medicinal chemistry. 2009;16:4058-65
7. Moretti L, Cha YI, Niermann KJ, Lu B. Switch between apoptosis and autophagy: radiation-induced endoplasmic reticulum stress? Cell cycle. 2007;6:793-8
8. Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nature reviews Molecular cell biology. 2014;15:81-94 doi:10.1038/nrm3735
9. Rubinstein AD, Kimchi A. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. Journal of cell science. 2012;125:5259-68 doi:10.1242/jcs.115865
10. Nunez G, Benedict MA, Hu Y, Inohara N. Caspases: the proteases of the apoptotic pathway. Oncogene. 1998;17:3237-45 doi:10.1038/sj.onc.1202581
11. Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nature reviews Molecular cell biology. 2004;5:897-907 doi:10.1038/nrm1496
12. Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nature reviews Molecular cell biology. 2010;11:621-32 doi:10.1038/nrm2952
13. Parrish AB, Freel CD, Kornbluth S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harbor perspectives in biology. 2013 5. doi:10.1101/cshperspect.a008672
14. Stupack DG. Caspase-8 as a therapeutic target in cancer. Cancer letters. 2013;332:133-40 doi:10.1016/j.canlet.2010.07.022
15. Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S. et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500-2 doi:10.1126/science.1096645
16. Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S. et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis: an international journal on programmed cell death. 2012;17:810-20 doi:10.1007/s10495-012-0735-0
17. Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717-9 doi:10.1038/onc.2009.519
18. Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H. A Complex between Atg7 and Caspase-9: A NOVEL MECHANISM OF CROSS-REGULATION BETWEEN AUTOPHAGY AND APOPTOSIS. The Journal of biological chemistry. 2014;289:6485-97 doi:10.1074/jbc.M113.536854
19. Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891-900 doi:10.4161/auto.6.7.13038
20. Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J. et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. The Journal of biological chemistry. 2012;287:12455-68 doi:10.1074/jbc.M111.309104
21. Nixon RA. The role of autophagy in neurodegenerative disease. Nature medicine. 2013;19:983-97 doi:10.1038/nm.3232
22. Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nature reviews Neurology. 2012;8:108-17 doi:10.1038/nrneurol.2011.200
23. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-75 doi:10.1038/nature06639
24. Vinod V, Padmakrishnan CJ, Vijayan B, Gopala S. 'How can I halt thee?' The puzzles involved in autophagic inhibition. Pharmacological research: the official journal of the Italian Pharmacological Society. 2014;82C:1-8 doi:10.1016/j.phrs.2014.03.005
25. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental cell. 2004;6:463-77
26. Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxidants & redox signaling. 2011;14:2201-14 doi:10.1089/ars.2010.3482
27. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67-93 doi:10.1146/annurev-genet-102808-114910
28. Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124-37 doi:10.4161/auto.23323
29. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132-41 doi:10.1038/ncb2152
30. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W. et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456-61 doi:10.1126/science.1196371
31. Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM. et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2003-8 doi:10.1073/pnas.1112848109
32. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19211-6 doi:10.1073/pnas.0810452105
33. Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. The Journal of biological chemistry. 1997;272:2477-85
34. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y. et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature cell biology. 2007;9:1142-51 doi:10.1038/ncb1634
35. Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46-52
36. Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534-6
37. Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes to cells: devoted to molecular & cellular mechanisms. 2007;12:209-18 doi:10.1111/j.1365-2443.2007.01050.x
38. Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M. et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. The Journal of cell biology. 2010;191:155-68 doi:10.1083/jcb.201002100
39. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J. et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nature cell biology. 2013;15:741-50 doi:10.1038/ncb2757
40. Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO reports. 2008;9:859-64 doi:10.1038/embor.2008.163
41. Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424-5 doi:10.4161/auto.22931
42. Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO reports. 2013;14:206-11 doi:10.1038/embor.2012.208
43. Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. The Journal of biological chemistry. 2007;282:37298-302 doi:10.1074/jbc.C700195200
44. Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C. et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. The EMBO journal. 2012;31:4304-17 doi:10.1038/emboj.2012.278
45. Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469-81 doi:10.1016/j.cell.2013.12.022
46. Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA. et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature cell biology. 2008;10:776-87 doi:10.1038/ncb1740
47. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature cell biology. 2009;11:385-96 doi:10.1038/ncb1846
48. Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT. et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nature cell biology. 2009;11:468-76 doi:10.1038/ncb1854
49. Mattson MP. Apoptosis in neurodegenerative disorders. Nature reviews Molecular cell biology. 2000;1:120-9 doi:10.1038/35040009
50. Reed JC. Apoptosis-based therapies. Nature reviews Drug discovery. 2002;1:111-21 doi:10.1038/nrd726
51. Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742-58 doi:10.1016/j.cell.2011.10.033
52. White E. Life, death, and the pursuit of apoptosis. Genes & development. 1996;10:1-15
53. Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802-9 doi:10.1038/35037739
54. Boyce M, Degterev A, Yuan J. Caspases: an ancient cellular sword of Damocles. Cell death and differentiation. 2004;11:29-37 doi:10.1038/sj.cdd.4401339
55. Strasser A, O'Connor L, Dixit VM. Apoptosis signaling. Annual review of biochemistry. 2000;69:217-45 doi:10.1146/annurev.biochem.69.1.217
56. Sayers TJ. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer immunology, immunotherapy: CII. 2011;60:1173-80 doi:10.1007/s00262-011-1008-4
57. Kaufmann T, Strasser A, Jost PJ. Fas death receptor signalling: roles of Bid and XIAP. Cell death and differentiation. 2012;19:42-50 doi:10.1038/cdd.2011.121
58. Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798-811 doi:10.1038/sj.onc.1209608
59. Fulda S, Debatin KM. Exploiting death receptor signaling pathways for tumor therapy. Biochimica et biophysica acta. 2004;1705:27-41 doi:10.1016/j.bbcan.2004.09.003
60. Mahalingam D, Szegezdi E, Keane M, de Jong S, Samali A. TRAIL receptor signalling and modulation: Are we on the right TRAIL? Cancer treatment reviews. 2009;35:280-8 doi:10.1016/j.ctrv.2008.11.006
61. Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: Orchestrators of apoptosis. Biochimica et biophysica acta. 2011;1813:508-20 doi:10.1016/j.bbamcr.2010.11.024
62. Wang Y, Tjandra N. Structural insights of tBid, the caspase-8-activated Bid, and its BH3 domain. The Journal of biological chemistry. 2013;288:35840-51 doi:10.1074/jbc.M113.503680
63. Bender T, Martinou JC. Where killers meet--permeabilization of the outer mitochondrial membrane during apoptosis. Cold Spring Harbor perspectives in biology. 2013;5:a011106. doi:10.1101/cshperspect.a011106
64. Franklin JL. Redox regulation of the intrinsic pathway in neuronal apoptosis. Antioxidants & redox signaling. 2011;14:1437-48 doi:10.1089/ars.2010.3596
65. Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harbor perspectives in biology. 2013 5. doi:10.1101/cshperspect.a008706
66. Pradelli LA, Beneteau M, Ricci JE. Mitochondrial control of caspase-dependent and -independent cell death. Cellular and molecular life sciences: CMLS. 2010;67:1589-97 doi:10.1007/s00018-010-0285-y
67. Harris MH, Thompson CB. The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell death and differentiation. 2000;7:1182-91 doi:10.1038/sj.cdd.4400781
68. Zou H, Yang R, Hao J, Wang J, Sun C, Fesik SW. et al. Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. The Journal of biological chemistry. 2003;278:8091-8 doi:10.1074/jbc.M204783200
69. Zentgraf H, Scheer U, Franke WW. Characterization and localization of the RNA synthesized in mature avian erythrocytes. Experimental cell research. 1975;96:81-95
70. Yang Y, Xing D, Zhou F, Chen Q. Mitochondrial autophagy protects against heat shock-induced apoptosis through reducing cytosolic cytochrome c release and downstream caspase-3 activation. Biochemical and biophysical research communications. 2010;395:190-5 doi:10.1016/j.bbrc.2010.03.155
71. Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5:329-38
72. Kim R, Emi M, Tanabe K. Role of mitochondria as the gardens of cell death. Cancer chemotherapy and pharmacology. 2006;57:545-53 doi:10.1007/s00280-005-0111-7
73. Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L. et al. Chikungunya virus-induced autophagy delays caspase-dependent cell death. The Journal of experimental medicine. 2012;209:1029-47 doi:10.1084/jem.20110996
74. You M, Savaraj N, Kuo MT, Wangpaichitr M, Varona-Santos J, Wu C. et al. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Molecular and cellular biochemistry. 2013;374:181-90 doi:10.1007/s11010-012-1518-1
75. Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer letters. 2009;274:95-100 doi:10.1016/j.canlet.2008.09.004
76. Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM. et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7741-6 doi:10.1073/pnas.1407001111
77. Fulda S. Caspase-8 in cancer biology and therapy. Cancer letters. 2009;281:128-33 doi:10.1016/j.canlet.2008.11.023
78. Zhao Y, Sui X, Ren H. From procaspase-8 to caspase-8: revisiting structural functions of caspase-8. Journal of cellular physiology. 2010;225:316-20 doi:10.1002/jcp.22276
79. Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491-501
80. Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C. et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. The Journal of biological chemistry. 1999;274:1156-63
81. Ferreira KS, Kreutz C, Macnelly S, Neubert K, Haber A, Bogyo M. et al. Caspase-3 feeds back on caspase-8, Bid and XIAP in type I Fas signaling in primary mouse hepatocytes. Apoptosis: an international journal on programmed cell death. 2012;17:503-15 doi:10.1007/s10495-011-0691-0
82. Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA. et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16677-82 doi:10.1073/pnas.0808597105
83. Bell BD, Walsh CM. Coordinate regulation of autophagy and apoptosis in T cells by death effectors: FADD or foundation. Autophagy. 2009;5:238-40
84. Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes & development. 1999;13:2514-26
85. Orozco S, Yatim N, Werner MR, Tran H, Gunja SY, Tait SW. et al. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell death and differentiation. 2014 doi:10.1038/cdd.2014.76
86. Duprez L, Bertrand MJ, Vanden Berghe T, Dondelinger Y, Festjens N, Vandenabeele P. Intermediate domain of receptor-interacting protein kinase 1 (RIPK1) determines switch between necroptosis and RIPK1 kinase-dependent apoptosis. The Journal of biological chemistry. 2012;287:14863-72 doi:10.1074/jbc.M111.288670
87. Kaiser WJ, Daley-Bauer LP, Thapa RJ, Mandal P, Berger SB, Huang C. et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7753-8 doi:10.1073/pnas.1401857111
88. He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G. et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8:1811-21 doi:10.4161/auto.22145
89. Laussmann MA, Passante E, Dussmann H, Rauen JA, Wurstle ML, Delgado ME. et al. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell death and differentiation. 2011;18:1584-97 doi:10.1038/cdd.2011.27
90. Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB. et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859-63 doi:10.1038/nature05853
91. Zhu K, Dunner K Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451-62 doi:10.1038/onc.2009.343
92. Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR. et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721-35 doi:10.1016/j.cell.2009.03.015
93. Pan JA, Fan Y, Gandhirajan RK, Madesh M, Zong WX. Hyperactivation of the mammalian degenerin MDEG promotes caspase-8 activation and apoptosis. The Journal of biological chemistry. 2013;288:2952-63 doi:10.1074/jbc.M112.441063
94. Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Molecular and cellular biology. 2004;24:8055-68 doi:10.1128/MCB.24.18.8055-8068.2004
95. Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Molecular cell. 2006;23:631-40 doi:10.1016/j.molcel.2006.08.002
96. Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends in biochemical sciences. 2007;32:95-100 doi:10.1016/j.tibs.2006.12.002
97. Huang S, Okamoto K, Yu C, Sinicrope FA. p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. The Journal of biological chemistry. 2013;288:33654-66 doi:10.1074/jbc.M113.518134
98. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131-45 doi:10.1074/jbc.M702824200
99. Zhang YB, Zhao W, Zeng RX. Autophagic degradation of caspase-8 protects U87MG cells against H2O2-induced oxidative stress. Asian Pacific journal of cancer prevention: APJCP. 2013;14:4095-9
100. Wurstle ML, Laussmann MA, Rehm M. The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Experimental cell research. 2012;318:1213-20 doi:10.1016/j.yexcr.2012.02.013
101. Reubold TF, Eschenburg S. A molecular view on signal transduction by the apoptosome. Cellular signalling. 2012;24:1420-5 doi:10.1016/j.cellsig.2012.03.007
102. Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. Journal of cell science. 2010;123:3209-14 doi:10.1242/jcs.073643
103. Jeong HS, Choi HY, Lee ER, Kim JH, Jeon K, Lee HJ. et al. Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochimica et biophysica acta. 2011;1813:80-90 doi:10.1016/j.bbamcr.2010.09.016
104. Allan LA, Clarke PR. Apoptosis and autophagy: Regulation of caspase-9 by phosphorylation. The FEBS journal. 2009;276:6063-73 doi:10.1111/j.1742-4658.2009.07330.x
105. Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E. et al. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318-21
106. Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843-6 doi:10.1126/science.1095432
107. Tu S, McStay GP, Boucher LM, Mak T, Beere HM, Green DR. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nature cell biology. 2006;8:72-7 doi:10.1038/ncb1340
108. Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD. Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Molecular biology of the cell. 2006;17:2150-7 doi:10.1091/mbc.E05-12-1107
109. Ho LH, Read SH, Dorstyn L, Lambrusco L, Kumar S. Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene. 2008;27:3393-404 doi:10.1038/sj.onc.1211005
110. Tiwari M, Sharma LK, Vanegas D, Callaway DA, Bai Y, Lechleiter JD. et al. A nonapoptotic role for CASP2/caspase 2: modulation of autophagy. Autophagy. 2014;10:1054-70 doi:10.4161/auto.28528
111. Tiwari M, Lopez-Cruzan M, Morgan WW, Herman B. Loss of caspase-2-dependent apoptosis induces autophagy after mitochondrial oxidative stress in primary cultures of young adult cortical neurons. The Journal of biological chemistry. 2011;286:8493-506 doi:10.1074/jbc.M110.163824
112. Fischer U, Stroh C, Schulze-Osthoff K. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene. 2006;25:152-9 doi:10.1038/sj.onc.1209015
113. Milhas D, Cuvillier O, Therville N, Clave P, Thomsen M, Levade T. et al. Caspase-10 triggers Bid cleavage and caspase cascade activation in FasL-induced apoptosis. The Journal of biological chemistry. 2005;280:19836-42 doi:10.1074/jbc.M414358200
114. Lamy L, Ngo VN, Emre NC, Shaffer AL 3rd, Yang Y, Tian E. et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer cell. 2013;23:435-49 doi:10.1016/j.ccr.2013.02.017
115. Sadasivan S, Waghray A, Larner SF, Dunn WA Jr, Hayes RL, Wang KK. Amino acid starvation induced autophagic cell death in PC-12 cells: evidence for activation of caspase-3 but not calpain-1. Apoptosis: an international journal on programmed cell death. 2006;11:1573-82 doi:10.1007/s10495-006-7690-6
116. Sirois I, Groleau J, Pallet N, Brassard N, Hamelin K, Londono I. et al. Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy. 2012;8:927-37 doi:10.4161/auto.19768
117. Nunez R, Sancho-Martinez SM, Novoa JM, Lopez-Hernandez FJ. Apoptotic volume decrease as a geometric determinant for cell dismantling into apoptotic bodies. Cell death and differentiation. 2010;17:1665-71 doi:10.1038/cdd.2010.96
118. Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X. et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein & cell. 2010;1:468-77 doi:10.1007/s13238-010-0048-4
119. Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. The Journal of biological chemistry. 2007;282:13123-32 doi:10.1074/jbc.M700492200
120. Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S. et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561-8
121. Ku B, Woo JS, Liang C, Lee KH, Jung JU, Oh BH. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy. 2008;4:519-20
122. Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nature reviews Molecular cell biology. 2014;15:49-63 doi:10.1038/nrm3722
123. Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. Journal of cell science. 2009;122:2554-66 doi:10.1242/jcs.046250
124. Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. The Journal of biological chemistry. 2001;276:7320-6 doi:10.1074/jbc.M008363200
125. Eguchi R, Tone S, Suzuki A, Fujimori Y, Nakano T, Kaji K. et al. Possible involvement of caspase-6 and -7 but not caspase-3 in the regulation of hypoxia-induced apoptosis in tube-forming endothelial cells. Experimental cell research. 2009;315:327-35 doi:10.1016/j.yexcr.2008.10.041
126. Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6:1042-56
127. Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiological reviews. 2003;83:731-801 doi:10.1152/physrev.00029.2002
128. Kar P, Samanta K, Shaikh S, Chowdhury A, Chakraborti T, Chakraborti S. Mitochondrial calpain system: an overview. Archives of biochemistry and biophysics. 2010;495:1-7 doi:10.1016/j.abb.2009.12.020
129. Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nature reviews Cancer. 2011;11:364-74 doi:10.1038/nrc3050
130. Sharma AK, Rohrer B. Calcium-induced calpain mediates apoptosis via caspase-3 in a mouse photoreceptor cell line. The Journal of biological chemistry. 2004;279:35564-72 doi:10.1074/jbc.M401037200
131. Gafni J, Cong X, Chen SF, Gibson BW, Ellerby LM. Calpain-1 cleaves and activates caspase-7. The Journal of biological chemistry. 2009;284:25441-9 doi:10.1074/jbc.M109.038174
132. Tan Y, Dourdin N, Wu C, De Veyra T, Elce JS, Greer PA. Ubiquitous calpains promote caspase-12 and JNK activation during endoplasmic reticulum stress-induced apoptosis. The Journal of biological chemistry. 2006;281:16016-24 doi:10.1074/jbc.M601299200
133. Martinez JA, Zhang Z, Svetlov SI, Hayes RL, Wang KK, Larner SF. Calpain and caspase processing of caspase-12 contribute to the ER stress-induced cell death pathway in differentiated PC12 cells. Apoptosis: an international journal on programmed cell death. 2010;15:1480-93 doi:10.1007/s10495-010-0526-4
134. Wood DE, Thomas A, Devi LA, Berman Y, Beavis RC, Reed JC. et al. Bax cleavage is mediated by calpain during drug-induced apoptosis. Oncogene. 1998;17:1069-78 doi:10.1038/sj.onc.1202034
135. Gao G, Dou QP. N-terminal cleavage of bax by calpain generates a potent proapoptotic 18-kDa fragment that promotes bcl-2-independent cytochrome C release and apoptotic cell death. Journal of cellular biochemistry. 2000;80:53-72
136. Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. The Journal of biological chemistry. 2001;276:30724-8 doi:10.1074/jbc.M103701200
137. Shi M, Zhang T, Sun L, Luo Y, Liu DH, Xie ST. et al. Calpain, Atg5 and Bak play important roles in the crosstalk between apoptosis and autophagy induced by influx of extracellular calcium. Apoptosis: an international journal on programmed cell death. 2013;18:435-51 doi:10.1007/s10495-012-0786-2
138. Lepine S, Allegood JC, Edmonds Y, Milstien S, Spiegel S. Autophagy induced by deficiency of sphingosine-1-phosphate phosphohydrolase 1 is switched to apoptosis by calpain-mediated autophagy-related gene 5 (Atg5) cleavage. The Journal of biological chemistry. 2011;286:44380-90 doi:10.1074/jbc.M111.257519
139. Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature cell biology. 2006;8:1124-32 doi:10.1038/ncb1482
140. Song Z, Guan B, Bergman A, Nicholson DW, Thornberry NA, Peterson EP. et al. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Molecular and cellular biology. 2000;20:2907-14
141. Lee G, Wang Z, Sehgal R, Chen CH, Kikuno K, Hay B. et al. Drosophila caspases involved in developmentally regulated programmed cell death of peptidergic neurons during early metamorphosis. The Journal of comparative neurology. 2011;519:34-48 doi:10.1002/cne.22498
142. Hou YC, Hannigan AM, Gorski SM. An executioner caspase regulates autophagy. Autophagy. 2009;5:530-3
143. DeVorkin L, Go NE, Hou YC, Moradian A, Morin GB, Gorski SM. The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB. The Journal of cell biology. 2014;205:477-92 doi:10.1083/jcb.201303144
144. Kim YI, Ryu T, Lee J, Heo YS, Ahnn J, Lee SJ. et al. A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC cell biology. 2010;11:9. doi:10.1186/1471-2121-11-9
145. Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. The Journal of cell biology. 2008;182:1127-39 doi:10.1083/jcb.200712091
146. Fraser AG, McCarthy NJ, Evan GI. drICE is an essential caspase required for apoptotic activity in Drosophila cells. The EMBO journal. 1997;16:6192-9 doi:10.1093/emboj/16.20.6192
147. Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell death and differentiation. 2009;16:966-75 doi:10.1038/cdd.2009.33
Author contact
![]() Corresponding author: Dr. Yuan Hong Or Dr. Jianmin Zhang, Department of Neurosurgery, Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang 310009, China; Tel: +86-571-87784785; Fax: +86-571-87784755; E-mail: hy0904cn.
Corresponding author: Dr. Yuan Hong Or Dr. Jianmin Zhang, Department of Neurosurgery, Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang 310009, China; Tel: +86-571-87784785; Fax: +86-571-87784755; E-mail: hy0904cn.

 Global reach, higher impact
Global reach, higher impact