10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(3):304-315. doi:10.7150/ijbs.10632 This issue Cite
Research Paper
Molecular Docking and Site-directed Mutagenesis of a Bacillus thuringiensis Chitinase to Improve Chitinolytic, Synergistic Lepidopteran-larvicidal and Nematicidal Activities
1. State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan 430070, Hubei, China
2. Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, Faculty of Life Science, Hubei University, Wuhan 430062, Hubei, China
Received 2014-9-24; Accepted 2014-12-24; Published 2015-1-30
Abstract
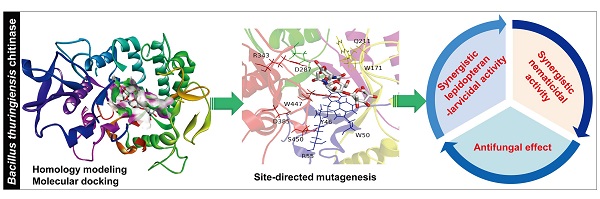
Bacterial chitinases are useful in the biocontrol of agriculturally important pests and fungal pathogens. However, the utility of naturally occurring bacterial chitinases is often limited by their low enzyme activity. In this study, we constructed mutants of a Bacillus thuringiensis chitinase with enhanced activity based on homology modeling, molecular docking, and the site-directed mutagenesis of target residues to modify spatial positions, steric hindrances, or hydrophilicity/hydrophobicity. We first identified a gene from B. thuringiensis YBT-9602 that encodes a chitinase (Chi9602) belonging to glycosyl hydrolase family 18 with conserved substrate-binding and substrate-catalytic motifs. We constructed a structural model of a truncated version of Chi9602 (Chi960235-459) containing the substrate-binding domain using the homologous 1ITX protein of Bacillus circulans as the template. We performed molecular docking analysis of Chi960235-459 using di-N-acetyl-D-glucosamine as the ligand. We then selected 10 residues of interest from the docking area for the site-directed mutagenesis experiments and expression in Escherichia coli. Assays of the chitinolytic activity of the purified chitinases revealed that the three mutants exhibited increased chitinolytic activity. The ChiW50A mutant exhibited a greater than 60 % increase in chitinolytic activity, with similar pH, temperature and metal ion requirements, compared to wild-type Chi9602. Furthermore, ChiW50A exhibited pest-controlling activity and antifungal activity. Remarkable synergistic effects of this mutant with B. thuringiensis spore-crystal preparations against Helicoverpa armigera and Caenorhabditis elegans larvae and obvious activity against several plant-pathogenic fungi were observed.
Keywords: Chitinase, Bacillus thuringiensis, Homology modeling, Molecular docking, Site-directed mutagenesis, Synergistic activity.
INTRODUCTION
Chitinases (E.C.3.2.1.14) comprise a family of extracellular cell-wall hydrolases that hydrolyze the β-1,4-glycosidic bond of chitin to form the monomer N-acetyl-D-glucosamine. Chitinases are found in a wide range of organisms, including bacteria, plants, fungi, insects, and crustaceans [1]. Bacterial chitinases have potential applications in the suppression of plant-pathogenic fungi and nematodes, the control of various pests, and the recycling of chitin to generate carbon and nitrogen sources in ecosystems [2, 3]. Of the various chitinase-producing bacteria, Gram-positive Bacillus species are of special interest due to their ability to form endospores that are resistant to heat and other adverse environments, an advantageous characteristic for agricultural applications [3].
Bacillus thuringiensis is an important insecticidal bacterium that produces insecticidal crystal proteins (ICPs) during sporulation. A variety of B. thuringiensis strains also produce chitinases [1, 4-6]. Because the peritrophic membrane of the insect midgut is composed of a chitinous matrix, the presence of chitinases in B. thuringiensis formulations may increase the uptake of ICPs into epithelial cells and enhance insecticidal activity [7, 8]. Thus, this bacterium is an attractive host for the development of a biocontrol agent with enhanced activity through increasing the expression of chitinases in vivo or through creating a formulation of ICPs and chitinases in vitro. Previous studies have demonstrated that the addition of bacterial chitinases enhances insecticidal effects of B. thuringiensis ICP preparations against pest larvae [9, 10]. Unfortunately, the endogenous chitinase activity of wild-type B. thuringiensis strains is relatively low [1, 6], and improving the chitinolytic activity of B. thuringiensis-derived chitinases would facilitate the development of B. thuringiensis composite formulations [11].
Protein engineering via site-directed mutagenesis is an essential tool for creating a mutant enzyme with altered or novel properties. To improve chitinase activity, site-directed mutagenesis of the Vibrio harveyi chitinase VhChiA, in which amino acid Y435 was replaced with A435, resulted in a mutant enzyme with enhanced binding and catalytic efficiency [12]. The W397F mutant of Vibrio carchariae chitinase A enhanced the hydrolysis of the pNP substrate by 142%, with activity toward colloidal chitin similar to that of the wild-type enzyme [13], while the Y245W mutation at the predicted exterior site of the catalytic cleft enhanced the hydrolysis of crystalline alpha-chitin and colloidal chitin [14]. In Serratia marcescens, the ChiA D313N and ChiB D142N mutants exhibited strongly enhanced transglycosylating activity [15]. These results suggest that structural mutations of active site residues may enhance the hydrolytic activity of chitinases.
B. thuringiensis chitinases structurally consist of a signal peptide, a catalytic domain, a fibronectin type-III-like domain (Fn3D), and a chitin-binding domain [16, 17]. Based on amino acid sequence similarity, these chitinases are grouped into family 18 glycosyl hydrolases [18, 19], which feature a three-dimensional structure in which the catalytic domain is located in a TIM barrel structure composed of eight α-helices and eight β-strands [20, 21]. This conserved molecular structure enables the analysis of amino acid residues critical for enzymatic reactivity using molecular docking and site-directed mutagenesis techniques.
No effort has yet been made to improve the catalytic activity of a B. thuringiensis chitinase through molecular docking and site-directed mutagenesis techniques. In the present study, based on the analysis of the three-dimensional protein structure of the B. thuringiensis chitinase Chi9602 by homology modeling and molecular docking analysis, the impact of 10 amino acid codons on enzymatic activity was analyzed by site-directed mutagenesis and specific chitinolytic activity assays, and three enzyme variants with enhanced activity were identified. The ChiW50A mutation increased activity by more than 60%, and the effects of pH, temperature, and several metal ions on the activity of the ChiW50A mutant and wild-type Chi9602 and their catalytic kinetic constants were compared. The synergistic pest-controlling effects of ChiW50A on B. thuringiensis spore-ICP preparations against Helicoverpa armigera and Caenorhabditis elegans larvae were also examined.
MATERIALS AND METHODS
Bacterial, fungal and C. elegans strains; genes; plasmids; and culture conditions
E. coli DH5α cells (TaKaRa Bio, Inc.) were used to construct various recombinant plasmids. E. coli TOP10 [F-, mcrA Δ (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 (ara-leu)7697galU galK rpsL endA1 nupG] (Invitrogen) was used as the host strain for the expression and purification of target proteins. The fungal strains Sclerotinia sclerotiorum FB014, Physalospora piricola FB016, Fusarium oxysporum FB012, Fulvia fulva FB009, Botrytis cinerea FB007 and Rhizoctonia solani FB010 (Microbial Genetic Stock Center, Wuhan, China) were used as indicators for antifungal assays. B. thuringiensis wild-type strain YBT-1520 (CCTCC No. M94067, China Center for Type Culture Collection), which has lepidopteran-larvicidal activity, and wild-type strain YBT-020 (Microbial Genetic Stock Center, Wuhan, China), which has nematicidal activity, were used to prepare spore-ICP preparations. Neonate larvae of a susceptible strain of H. armigera artificially fed in the laboratory (KeNuo Biotech Co. Ltd, Wuhan, China) and synchronized fourth-stage (L4) larvae of C. elegans wild-type strain N2 (Bristol) were used for toxicity bioassays of the B. thuringiensis spore-ICP preparations.
The chitinase gene chi (GenBank acc. no. KF671757.1) was isolated from B. thuringiensis subsp. tenebrionis wild-type strain YBT-9602 (Microbial Genetic Stock Center, Wuhan, China). The recombinant plasmid pMB332 (Supplementary Material: Fig. S1), which harbors the encoding sequence (codons 35 to 676) of chi and expresses the Chi9602 protein in E. coli, was used as the DNA template to amplify various mutated chi genes by polymerase chain reaction (PCR). The recombinant plasmids pMB581-Y46A, pMB581-W50A, pMB581-R55A, pMB581-W171A, pMB581-E211A, pMB581-D287A, pMB581-R343A, pMB581-D385A, pMB581-W447A, and pMB581-S450A, which harbored single-amino acid mutations of the chi gene (Fig. S1), were constructed to express and purify the mutant Chi9602 proteins.
E. coli strains harboring various recombinant plasmids were grown in Luria-Bertani (LB) medium containing 100 μg/mL ampicillin (Amp) at 37 ºC. B. thuringiensis strains were cultured in 500-mL Erlenmeyer flasks containing 50 mL of LB broth at 210 rpm and 37 ºC, unless specified otherwise. Fungal strains were grown in PDA medium (20% potato infusion 100 mL, dextrose 2 g, biological agar 1.5 g) at 28 °C. The C. elegans N2 strain was cultured using the method previously described by Lewis and Fleming [22].
Homology modeling and molecular docking
The chi sequence was characterized by conducting BLASTN and BLASTP searches of the GenBank nucleotide and amino acid sequence databases at the National Center for Biotechnology Information (NCBI) server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The conserved domain architectures of Chi9602 were analyzed using the NCBI online tool “Conserved Domain Search” (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The signal peptide sequence was predicted by SignalP (http://www.cbs.dtu.dk/services/SignalP/).
For homology modeling of the Chi9602 catalytic domain and partial downstream codons (from codon 35 to 459, in brief, Chi960235-459), homologous proteins were first identified by searching the Protein Data Bank (PDB) database (http://www.rcsb.org/pdb/home/home.do). All matched proteins were aligned based on sequence similarity to Chi960235-459, and the proteins with similarities greater than 30% were considered candidate reference proteins for homology modeling. A structural motif (PDB ID: 1ITX) of the catalytic domain of Bacillus circulans WL-12 chitinase A1 [23] with 61% sequence identity to Chi960235-459 at the corresponding codon regions was then selected as the reference protein.
Homology modeling was performed based on the 1ITX.1.A reference protein model using the “Easy modeler 4.0” tool in the software “MODELLER 9.13”. An optimized Chi960235-459 homology model was constructed from the three generated models by selecting the “DOPE profile” options. The Chi960235-459 homology model was validated using the online tool “Molprobity” (http://molprobity.biochem.duke.edu/index.php) and evaluated by the “Ramachandran Plot Analysis” online tool (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). Subsequently, a stereo image of the Chi960235-459 ribbon model illustrating the TIM-barrel structure and a stereo image illustrating the surface groove structure of the Chi960235-459 model were generated using “PyMol” software (DeLano Scientific LLC).
To perform molecular docking of Chi960235-459 and NAG, a di-[N-Acetyl-D(+)-glucosamine] [(NAG)2] (CID: 446943) molecular ligand was obtained from “PubChem” (http://pubchem.ncbi.nlm.nih.gov/). Protein docking of the Chi960235-459 model with (NAG)2 as the substrate was conducted using AutoDock 4.2 software (The Scripps Research Institute), in which the program Autogrid 4.2 was run at a mesh region with the “Numbers of points in X-, Y-, and Z-dimensions” of 80, 70, and 80, respectively, “Spacing (Å)” of 0.375, and residue E211 set as the center. A total of 20 matched docking conformations were generated using E211 as the flexible residue, and the number of rotatable bonds of (NAG)2 was set as 6/32. An optimal docking configuration of Chi960235-459 and (NAG)2 based on these docking conformations was determined by adjusting the “Binding Energy” to a minimal value of -8.15 kcal/mol. This docking configuration was then plotted as a 3D image using the tool PyMol. The docking area of Chi960235-459 and (NAG)2 featured a substrate-binding, semi-closed groove channel, and 27 amino acid residues were identified as pocket residues in the receptor-ligand groove structure using the Discovery Studio Visualizer tool (Accelrys). From these residues, 10 (Y46, W50, R55, W171, D287, E211, R343, D385, S450, and W447) were selected that were predicted to pack the substrate and form the substrate-binding groove based on their steric distances from (NAG)2 and their hydrophilic/hydrophobic nature.
Plasmid construction and expression and purification of chitinases
The oligonucleotide primers used in this study are listed in Supplementary Material: Table S1. The recombinant plasmids harboring mutated chi genes with either a single mutated codon or dual mutated codons are schematically illustrated in Supplementary Material: Fig. S1. Briefly, the full-length chi was amplified by PCR from the B. thuringiensis YBT-9602 genome with primers Chi-F (Table S1, BglII site underlined) and Chi-R (Table S1, PstI site underlined). The PCR-amplified fragment was sequenced before digestion with BglII and PstI. The digested fragment was then ligated to the BglII/PstI site of the E. coli expression vector pTrcHis B (Invitrogen), yielding the recombinant plasmid pMB332. To obtain a site-directed chi mutant in which the amino acid of interest was replaced with Ala, three separate PCRs were performed using the standard SOE (splicing by overlap extension) method [24]. For example, to construct chiY46A (in which the codon for Tyr46 was replaced with Ala46), the Chi-F (Forward) and Y46A-R (Reverse) primer pair was used to amplify a fragment containing the upstream sequences and the mutation site (from codon 35 to 46), and the second primer pair, Y46A-F and Chi-R, was used to amplify the mutation site and the downstream sequences (from codon 46 to 676). The third primer pair, Chi-F and Chi-R, was used to generate the full-length chi mutant (from codon 35 to 676), with the previously amplified overlapping fragments used as the heteroduplex templates. A similar protocol was used to construct other mutant chi genes with single-codon mutations. To construct the double mutant chiW50A/S450A (W50A and S450A), a similar SOE method was employed, except the previously constructed chiW50A gene was used as the DNA template. All final amplified mutant chi genes were verified by sequencing and ligated into the BglII/PstI sites of pMB332 to yield the recombinant plasmids listed in Supplementary Material: Fig. S1.
Transformed E. coli cells expressing wild-type or mutant Chi9602 were grown in fresh LB broth containing 100 μg/mL Amp and incubated at 37 ºC until the OD600 reached 0.6. Isopropyl-β-D-thiogalactoside (IPTG) was then added at a final concentration of 0.4 mmol/L. The cells were further incubated at 28 ºC for approximately 6 h and harvested by centrifugation (8,000 rpm, 5 min). The cell suspension was treated twice with a French Pressure Cell (Thermo, USA) at 15,000 psi and centrifuged at 13,000 rpm for 10 min. The extract was further purified using the “IMAC” protein purification system following the manufacturer's instructions (Invitrogen).
B. thuringiensis spore-ICP preparation and measurement of ICPs concentration
A seed culture of B. thuringiensis YBT-1520 (or YBT-020) grown at 30 ºC for 10 h was used as an inoculum. The fermentation of YBT-1520 (or YBT-020) was carried out in a 15-L fermentor (with a 7.5-L LB broth loading) by inoculating with 7.5 ml of seed culture. The culture was kept at 30 ºC and with a stirring rate of 400 r/min until ~ 70% spores and crystals released from lytic cells (about 30 h to 32 h). The spore-crystal suspension was harvested, and was then subjected to rapid spray desiccation for preparation of the wettable spore-crystal powder. The ICPs content in the B. thuringiensis spore-ICP preparation was quantified following standard procedures (National Standard of the People's Republic of China, GB/T 19567.2-2004) using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and comparative densitometry of the 130-kDa ICPs bands on SDS-PAGE gels from the prepared powder and that of a standard reference spore-crystal powder (with 8.0% ICPs, KeNuo Biotech Co. Ltd, Wuhan, China).
Analytical assays
The cell density at 600 nm was measured using a UV-VIS spectrophotometer (DU-800 Nucleic Acids/Protein Analyzer, Beckman Coulter). Purified B. thuringiensis wild-type and mutant chitinases were quantified according to Bradford [25] using bovine serum albumin as the standard. The expression of wild-type or mutant chitinases in E. coli was analyzed by SDS-PAGE in 10% polyacrylamide gels as described by Laemmli [26]. Prior to the chitinolytic activity assay, colloidal chitin was prepared from farinose chitin (TaKaRa) following the procedures described by Zhang et al. [27]. The chitinase activity of the wild-type and mutant chitinases was assayed following a previously published method [28], except the modified reaction mixture contained 200 μL of 2% colloidal chitin (pH 6.8), 500 μL of diluted purified Chi (pH 6.5), and Tris-HCl buffer (pH 7.4) to a final volume of 1.0 mL (pH 6.8). The reaction mixture was incubated at 37 ºC for 1 h. After centrifugation, 500 μL of the supernatant fluid was added to 1 mL of dinitrosalicylic acid (DNS) reagent [29] and incubated in boiling water for 10 min. The absorbance of the mixtures was measured at 535 nm, and the reducing sugars in the mixtures were calculated by comparison to a calibration curve constructed using pure NAG (Sigma). One unit of chitinase activity was defined as the amount of enzyme that produced reducing sugars equivalent to 1 μmol of NAG under the above conditions.
Characterization of the purified wild-type and mutant chitinases
The effect of pH on purified chitinase activity was determined in a series of citrate-phosphate-buffered reaction solutions containing 300 µL citrate-phosphate buffer, 200 µL colloidal chitin and 500 µL purified chitinase at pH values ranging from 2.0 to 9.0. The chitinolytic reaction was performed at 37 ºC for 1 h. The enzyme activities at various pH values were determined using the assay described above.
The effect of temperature on the activity of purified chitinase was determined by incubating the reaction solutions at a range of temperatures (25 ºC, 30 ºC, 35 ºC, 37 ºC, 40 ºC, 45 ºC, 50 ºC, 55 ºC, 60 ºC, 70 ºC, and 80 ºC) for 1 h. The chitinase activity was measured using the method described above.
The effect of metal ions, including Fe2+, Ca2+, Zn2+, Pb2+, Mg2+, Mn2+, and Cu2+, at final concentrations of 1 mmol L-1 on chitinase activity was determined by incubating the reaction solutions containing each metal ion at 37 ºC for 1 h. The enzyme activity was then measured following the method described above.
Toxicity bioassay
For toxicity bioassays against H. armigera, 1.00 g B. thuringiensis YBT-1520 spore-ICP powder was added to 10 mL 0.01 M sterile Na2CO3•NaHCO3 buffer (pH 9.6), and stirred vigorously to allow complete dissolution of the crystals, then diluted with sterile double distilled water (ddH2O) to 25.0 mL using a 25 mL volumetric flask to prepare a 40.0 mg/mL spore-ICP powder stock solution. The stock solution was mixed with 0.01 M PBS (pH7.4)-dissolved purified chitinase (Chi9602 or ChiW50A, each at the concentration of 280 μg/mL) at a ratio of 7 : 1 (V/V) to prepare the composite formulations containing spore-ICP powder and chitinase at the final concentration of 35 mg/mL and 35 μg/mL, respectively. YBT-1520 spore-ICP solution at the final concentration of 35 mg/mL was prepared in parallel by adding 1 aliquot ddH2O to 7 aliquots of powder stock solution. Bioassays of spore-ICP preparation and the composite formulations against neonate larvae of H. armigera were performed as previously described [30]. The 50% lethal concentration (LC50) was calculated for each treatment.
For bioassays against C. elegans, 1.00 g B. thuringiensis YBT-020 spore-ICP powder was dissolved with 10 mL sterile 0.01 M Na2CO3•NaHCO3 buffer (pH 9.6), and diluted with sterile ddH2O to 25.0 mL using a 25 mL volumetric flask to prepare a 40.0 mg/mL spore-ICP powder stock solution. The stock solution was further diluted to 2.0 mg/mL, 5.0 mg/mL, 20.0 mg/mL and 30.0 mg/mL solutions with sterile ddH2O. Each 100 μL of above diluted solutions (including the stock solution) was added to different wells of a 96-well cell culture plate, which were loaded with 50 μL 80 μg/mL chitinase (Chi9602 or ChiW50A), 5 μL C. elegans larvae (L4) suspension (containing a total of 40 larvae), 5 μL 10 mM 5-fluoro-2'-deoxy-uridine, and 40 μL E. coli OP50 cell suspension (at OD600 of 0.6). In parallel, each 100 μL of above solutions was added to wells of another plate loading with similar solutions except 50 μL chitinase was substituted with 50 μL sterile ddH2O. The bioassays against C. elegans were performed according to the method described by Dengg and van Mell [31], except that synchronized L4 larvae were used. Lethality was evaluated after three days by probing larvae with a dissecting needle under a stereo microscope. Percentage of lethality was calculated for each treatment. Purified chitinases Chi9602 and ChiW50A at the concentrations of 0-800 μg/mL were used as the controls.
Antifungal assays
Target fungal strains were inoculated onto the central spot of each PDA Petri plate and cultivated for 24-48 h at 28 ºC. A sterile filter paper (diameter, 8 mm) spotted with 50 μL of the purified chitinases at the final concentration of 200 μg/mL (filtered with a 0.45 μm sterile filter membrane) or 50 μL sterile H2O was placed on the surface of each plate. The plates were then incubated at 28 ºC until inhibition of the hyphal extension around the filter papers was observed. The diameters of each inhibition zone were then measured for activity evaluation.
RESULTS
Chi9602 homology modeling
Most available B. thuringiensis genomes contain chitinase-encoding sequences. Based on the sequence of the B. thuringiensis BMB171 chitinase gene [32], a chitinase-encoding gene was amplified from the genome of B. thuringiensis subsp. tenebrionis YBT-9602 by PCR. Nucleotide sequence analysis and nucleotide alignment using the online BLASTN tool suggested that this 2031-bp gene, designated chi (GenBank accession no. KF671757.1), is a chitinase-encoding member of the family 18 glycosyl hydrolases. The chi gene encodes a 676-amino acids protein (Chi9602) with a deduced molecular mass of 74.5 kDa and an isoelectric point of 5.89. Conserved domain architecture analysis of Chi9602 revealed that the protein has a modular structure composed of an N-terminal chitin-catalytic domain (CCD, from amino acids 42 to 452), a fibronectin type-III-like domain (Fn3D, from amino acids 504 to 558), and a C-terminal chitin-binding domain (CBD, from amino acids 582 to 646) (Fig. 1A). The first 34 residues of the N-terminus were predicted to be a signal sequence by SignalP, with a possible cleavage site located between amino acids 34 and 35. Sequence alignments of chi with the chitinase gene of subsp. kurstaki BMB171 (GenBank accession no: CP001903.1), chiA of subsp. kurstaki HD-73 (GenBank accession no: EF581163.2), and chiA of subsp. colmeri 15A3 (GenBank accession no: DQ512474.1) revealed sequence identity in excess of 97%.
Schematic illustration of Chi960235-459 homology modeling and Chi960235-459 and (NAG)2 molecular docking. A, Structural organization of chitinase Chi9602. Abbreviations: AA, amino acid; SS, signal sequence; CCD, chitin-catalytic domain; Fn3D, fibronectin type-III-like domain; CBD, chitin-binding domain. B, Orthogonal view of the Chi960235-459 ribbon model. The colored α-helixes (red) and β-sheet (yellow) constituting the (βα)8 TIM barrel structure are shown. C, Surface view of the spatial structure of Chi960235-459. The substrate-binding semi-closed groove channel is indicated by the rectangular frame. D, Molecular structure of the N-acetyl-D-glucosamine (NAG) dimer. E, The docking model of the Chi960235-459-(NAG)2 complex. The bound red/blue-colored spheroid molecule of (NAG)2 shows the position of the substrate binding groove. F, The docking model of the Chi960235-459-(NAG)2 complex, showing the residues selected for site-directed mutagenesis.
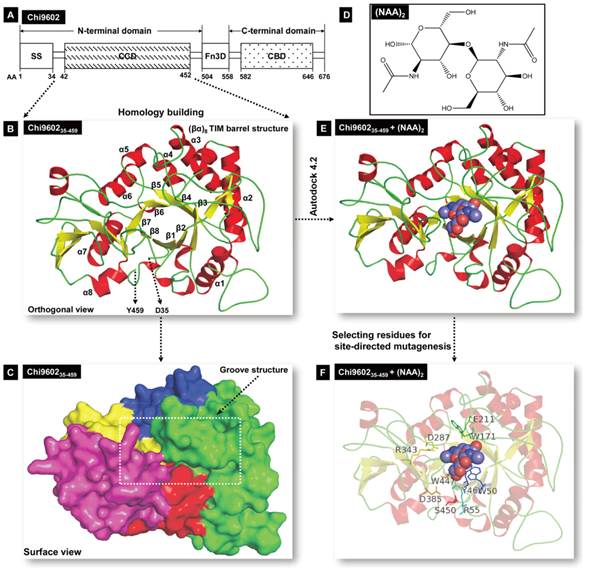
To identify a reference protein for homology modeling of Chi960235-459, the PDB database was searched for homologous proteins. Among the suggested homologous proteins, ten proteins (Supplementary Material: Table S2) with 30% to 70% sequence similarity to Chi960235-459 were considered as possible reference proteins. The sequences of these proteins were aligned with the sequence of Chi960235-459 (Supplementary Material: Fig. S2), which revealed that Chi960235-459 harbored most of the conserved residues of the family 18 chitinases, including the specific chitinase-binding motif “SXGG” and conserved CCD motif “DXDXE”, which are essential for function [23, 33-35]. One of these proteins, Bacillus circulans chitinase A1 (PDB ID: 1ITX), had the highest sequence identity (61%) to Chi960235-459, shared the specific chitinase-binding motif “SVGG” and the conserved CCD motif “DLDWE” with Chi960235-459, and had a structure with a high resolution of 1.10 Å. Thus, 1ITX was selected as the reference protein for Chi960235-459 homology modeling. A three-dimensional structural model of Chi960235-459 was constructed, and the structural coordinates of the optimized Chi960235-457 model were evaluated by Ramachandran plot [36] (Supplementary Material: Fig. S3), which revealed that 93.4% residues (426 of 456 amino acids) were in favored regions and 97.6% residues (445 of 456 amino acids) were in allowed regions, indicating the favorability of this model. A three-dimensional ribbon model of Chi960235-459 was then plotted using PyMol (Fig. 1B), which revealed the classical TIM-barrel structure of the chitinase CCD consisting of 8 α-helices (α1 to α8, Fig. 1B) and 8 β-strands (β1 to β8, Fig. 1B) along with an α-helix-rich terminal domain and an α+β domain, as has been observed in other family 18 chitinases [23, 37, 38]. The barrel fold composed of β-strands formed a long surface groove, with α-helices and coils connecting the strands. A surface view of the Chi960235-459 spatial model (Fig. 1C) clearly showed a channel of this groove that is predicted to be involved in substrate binding and a “bed” structure formed by the base of the TIM barrel, consistent with the structure of Yersinia entomophaga Chi1 [23].
The β4 strand of the TIM-barrel contained the “DLDWE” motif, a characteristic sequence that includes a conserved Glu residue with a crucial role in enzyme function in several other chitinase family 18 members because this Glu residue protonates the oxygen in the scissile glycosidic bond [34, 38-40]. Thus, we refer to this residue, E211 in Chi9602, as a key residue for targeting the orientation range of the residues that interacts with the substrate and modulates enzyme activity during molecular docking.
Molecular docking of Chi9602
Family 18 chitinases hydrolyze C2 N-acetylated β(1-4)-linked substrates, and catalysis occurs until the substrate is hydrolyzed [38, 40]; therefore, we mimicked a dimer fold of NAG (Fig. 1D) to use as the substrate for molecular docking. Flexible docking of Chi960235-459 toward the substrate (NAG)2 was performed to obtain a matched conformation of the Chi960235-459 + (NAG)2 ligand complex (Fig. 1E; Supplementary Material: Fig. S4A). As expected, (NAG)2 matched well to the Chi960235-459 homology model by fully implanting into the groove fold (Fig. 1E). While E211 was set as the core site for activity, a docking area around E211 comprising 27 amino acid residues clearly revealed the coverage of the full groove fold embedding the ligand (NAG)2 (Supplementary Material: Fig. S4A, S4B). To select the candidate residues for mutagenesis to enhance chitinase activity, we considered hydrophilic or hydrophobic residues that could form hydrogen bonds or hydrophobic bonds with (NAG)2, and residues that were oriented toward or surrounding the (NAG)2 with relatively greater steric hindrance. Ten residues, Y46, W50, R55, W171, E211, D287, D385, R343, W447, and S450, predicted to be involved in forming the substrate-binding groove structure and packing the substrate were selected as target residues for site-directed mutagenesis.
Site-directed mutagenesis and activity of mutant chitinases
Site-directed mutagenesis was conducted to mutate all selected target residues to Ala. Ala was chosen because it is an electrically uncharged hydrophobic amino acid with less steric hindrance. Ala is likely appropriate for probing residues that modulate enzyme activity through spatial effects rather than by forming hydrogen bonds or hydrophobic bonds, thereby facilitating the identification of potential activity-enhancing mutations. Using the PCR-SOE strategy, ten mutated Chi9602 variants containing one of the mutated codons (Y46A, W50A, R55A, W171A, E211A, D287A, R343A, D385A, W447A, and S450A) were obtained by substituting each target amino acid codon with a codon encoding Ala. SDS-PAGE analysis demonstrated that wild-type Chi9602 and the mutant Chi9602 variants were expressed as a monomeric protein component with the predicted size (~ 70 kDa) in the cytosol (Supplementary Material: Fig. S5A, S5B, and S5C). These chitinases were then purified from each transformed E. coli strain.
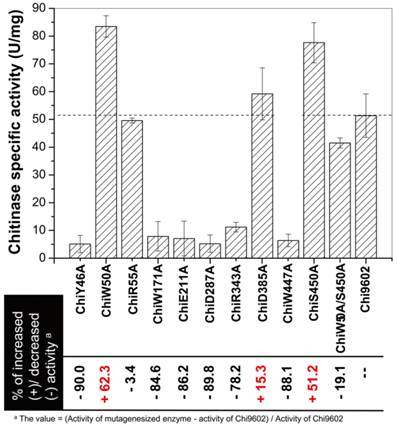
The chitinase activities of purified wild-type and mutant Chi9602 variants were measured under normalized conditions using colloidal chitin as the substrate. As shown in Fig. 2, the mutants ChiR55A and ChiD385A showed almost the same level of activities with Chi9602, and most of other mutants exhibited decreases in enzymatic activity ranging from about 10% to about 90% compared to Chi9602 (51.8 U/mg); however, three mutant enzymes exhibited increased activity: ChiW50A, which contains a mutation in the substrate-binding domain, and ChiD385A and ChiS450, which contain mutations in the catalytic domain, exhibited increases in activity of 62%, 15% and 51%, respectively.
Given the substantially increased activity of ChiW50A and ChiS450A, we constructed the double mutant ChiW50A/S450A. However, this double mutant enzyme did not exhibit synergistically increased activity but rather a decrease in activity of approximately 19%, 50%, and 47% compared to Chi9602, ChiW50A, and ChiS450A, respectively.
Measurement of the specific enzyme activity of the mutant chitinases generated by site-directed mutagenesis.
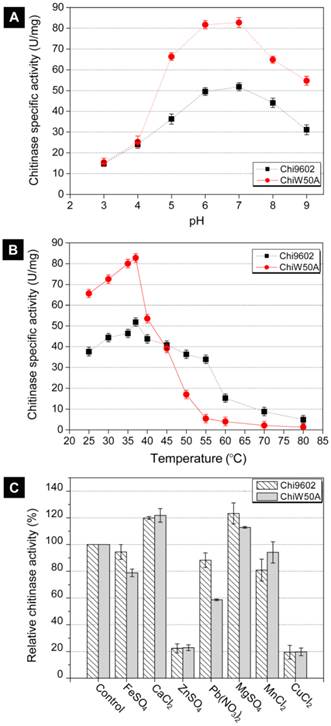
Effect of pH, temperature and metal ions on ChiW50A enzyme activity
Due to its relatively higher activity, the ChiW50A mutant was selected for further studies to determine whether the mutation altered the pH, temperature and metal ion requirements compared to wild-type Chi9602. ChiW50A retained relatively higher activity at pH 5 to 8, with highest activity at pH 7, consistent with Chi9602 (Fig. 3A). Interestingly, as shown in Fig. 3B, ChiW50A exhibited higher activity across the temperature range of 25-37 ºC compared to Chi9602; however, ChiW50A activity rapidly decreased across 40 ºC to 60 ºC and was inactivated at temperatures above 60 ºC. As the temperature increased from 25 ºC to 55 ºC, ChiW50A lost approximately 95% of its activity. Chi9602 exhibited greater thermostability and lost only 10% of its activity at similar temperatures, appearing relatively stable across a temperature range of 25-55 ºC. Fig. 3C shows that Cu2+ and Zn2+ had remarkable inhibitory effects on ChiW50A and Chi9602 activity, whereas Mg2+ and Ca2+ increased enzymatic activity by approximately 15% to 20%. Limited effects of Fe2+ and Mn2+ were observed, but Pb2+ appeared to inhibit the activity of ChiW50A more strongly than Chi9602 activity.
Synergistic lepidopteran-larvicidal and nematicidal activities of ChiW50A
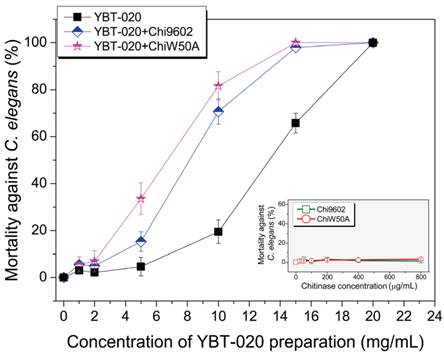
One of most promising applications of chitinases in biocontrol is to enhance the pest-larvicidal activity of B. thuringiensis toxins [8]. To determine if the ChiW50A mutant enhanced lepidopteran-larvicidal activity when added to insecticidal spore-ICP preparations of a lepidopteran-active B. thuringiensis YBT-1520 strain, we performed normalized bioassays with mixtures of the purified chitinases and YBT-1520 spore-ICP preparations against H. armigera larvae. Neither wild-type Chi9602 nor ChiW50A had larvicidal activity; however, both Chi9602 and ChiW50A exhibited synergistically increased activities when added to the YBT-1520 preparations, resulting in decreases in the LC50 of 17.6% and 30.4%, respectively (Table 1). The synergistic nematicidal activities of wild-type Chi9602 and the mutated ChiW50A against C. elegans larvae when added to preparations of a nematicidal B. thuringiensis YBT-020 strain were also evaluated. As shown in Fig. 4, both Chi9602 and ChiW50A mutant possessed no nematicidal activity alone; however, when added to the B. thuringiensis YBT-020 spore-ICP preparation, ChiW50A exhibited a steadily increasing pattern of greater synergistic activity compared to Chi9602 at the YBT-020 concentration range of 1-14 mg/mL. Overall, 100% mortality was observed when both enzymes were added to 15 mg/mL YBT-020 spore-ICP preparations. These results demonstrate that the ChiW50A mutant has greater synergistic lepidopteran-larvicidal and nematicidal activities compared to wild-type Chi9602.
Effects of pH, temperature and metal ions on the chitinase activity of ChiW50A and wild-type Chi9602.
Antifungal assays
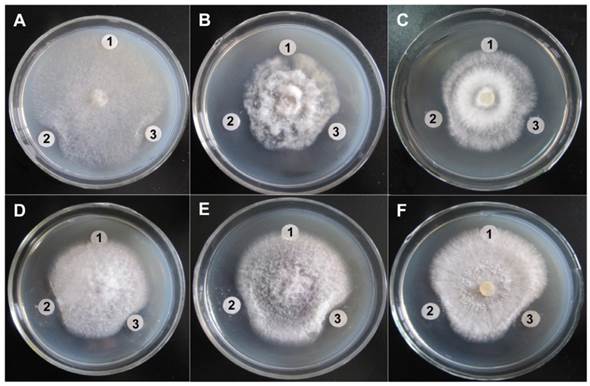
The antifungal activities of purified ChiW50A and Chi9602 were investigated. Both enzymes exhibited obvious inhibitory effects on hyphal growth by the target fungi as visualized by their obvious inhibition zones compared to negative controls (Fig. 5). By comparing the diameters of inhibition zones, ChiW50A showed relatively greater activities against Sclerotinia sclerotiorum, Fusarium oxysporum, Fulvia fulva, and Botrytis cinerea compared to Chi9602, but the difference in activity against Physalospora piricola and Rhizoctonia solani between the two enzyme variants was not remarkable.
Bioassays of the insecticidal mixtures of purified chitinases and YBT-1520 spore-ICP preparations against Helicoverpa armigera.
| Samples a | Linear regression equation b | Linearly dependent coefficient b | LC50 (μg·mL-1) | 95% confidence bounds (μg·mL-1) |
|---|---|---|---|---|
| Chi9602 | - | - | >1000 | - |
| ChiW50A | - | - | >1000 | - |
| YBT-1520 | Y= -5.207+2.064X | 0.9444 | 4.70 | 4.058~5.466 |
| YBT-1520 + Chi9602 | Y= -8.315+3.192X | 0.9653 | 3.88 | 3.384~4.413 |
| YBT-1520 + ChiW50A | Y= -5.963+2.460X | 0.9995 | 3.27 | 2.826~3.719 |
Note: a, Sample initial concentration: Chi9602, 35 μg/mL; ChiW50A, 35 μg/mL; YBT-1520, 35 mg/mL; YBT-1520 + Chi9602/ChiW50A, 35 mg/mL (YBT-1520) and 35 μg/mL (Chi9602 or ChiW50A) at the final concentration. The sample solutions were diluted to 1, 1/2, 1/4, 1/8 and 1/16 diluents with sterile ddH2O for bioassays; b, PROBIT model of lethal-concentration.
Synergistic effect of the ChiW50A mutant on the nematicidal activity of B. thuringiensis YBT-020 spore-ICP preparations against C. elegans larvae. B. thuringiensis YBT-020 spore-ICP powder was prepared as 1.0 mg/mL, 2.5 mg/mL, 10.0 mg/mL, 15.0 mg/mL and 20.0 mg/mL solutions, respectively. Each 20 μg/mL purified ChiW50A or Chi9602 at the final concentration was added in above solution to prepare the composite formulations.
Antifungal activity of the ChiW50A mutant and the Wild-Type Chi9602. A, Sclerotinia sclerotiorum FB014; B, Physalospora piricola FB016; C, Fusarium oxysporum FB012; D, Fulvia fulva FB009; E, Botrytis cinerea FB007; F, Rhizoctonia solani FB010. Labels 1, 2 and 3 in each plate indicate the treatments using sterile H2O (the negative control), ChiW50A and Chi9602 (at the final concentrations of 200 μg/mL), respectively.
DISCUSSION
Chitin is a major structural component in various insect tissues, including the parietal layer, exo- and endocuticle, respiratory tract, peritrophic membrane and intestinal tract. Chitin functions as a mechanically strong supporting scaffold material as well as a permeability barrier lining the trachea and gut epithelium [41]. Chitin is also an integral part of the nematode eggshell and pharyngeal lumen walls [42] and a major cell wall component of many plant-pathogenic fungi [43]. Bacterial chitinases can hydrolyze various chitin-based substrates [10, 11, 44] and exhibit synergistic activity with B. thuringiensis spore-ICP preparations against pest larvae [9, 10]. The biological properties of bacterial chitinases, particularly the chitinase produced by insecticidal B. thuringiensis, indicate their potential for biocontrol of agriculturally important lepidopteran and nematode pests as well as plant-pathogenic fungi.
The molecular docking model permits an intuitive explanation of the binding of the enzyme and ligand molecules, suggesting candidate sites for site-directed mutagenesis [45, 46]. To identify potential active-site residues in chitinases, many previous site-directed mutagenesis studies have substituted substrate-binding residues with hydrophilic or hydrophobic residues that form hydrogen bonds or hydrophobic bonds with the substrate, frequently resulting in decreased enzymatic activity [41-43]. In this study, when E211 was fixed as the central site of the Chi960235-459-(NAG)2 complex model, 27 amino acid residues were identified as the pocket residues forming the substrate-binding groove structure by the Discovery Studio Visualizer tool (Supplementary Material: Fig. S4B). Due to its lack of electric charge and reduced steric hindrance, we chose the uncharged amino acid Ala as the replacement codon in the site-directed mutagenesis experiments. Moreover, we considered both the pocket residues and the substrate-binding residues as the target codons. Based on their hydrophilic/hydrophobic nature as well as their spatial positions and steric hindrance, we selected 10 residues, including substrate-binding residues (e.g., E211) and residues surrounding the substrate-binding residues (e.g., W50). Our results demonstrated that although most of the mutations (Y46A, R55A, W171A, E211A, D287A, R343A, and W447A) decreased activity, as expected, several of the mutations (W50A, D385A, and S450A) increased chitinase activity. Interestingly, all three activity-enhancing mutations involve residues that are relatively far from the substrate-binding fold, indicating that the initial selection of these residues rather than residues close to the center of the fold is reasonable.
Based on their conservation and effects on enzyme activity, these residues can be divided into three groups. Residues in the first group, which include Y46, W171, E211, D287, R343 and W447, are conserved, and their mutagenesis leads to remarkably altered activity. Site-directed mutagenesis of these residues consistently resulted in decreased enzyme activity (Fig. 2). We proposed that these residues are important for interactions with the substrate by forming critical stabilizing interactions or are proximal to the enzyme-ligand binding sites [37, 47]. The Y46 and W447 residues are apparently located below the substrate molecule in the enzyme-substrate pocket fold; therefore, they are likely to support the substrate and stabilize the enzyme-substrate reaction configuration. A similar role was proposed for W171, which is located above the pocket structure and restricts the substrate within the appropriate enzyme-substrate configuration. As indicated above, E211 is a core residue for enzyme activity. The E211 to A211 mutations eliminates the hydrolysis of β-1,4-glycosidic bonds although the enzyme binds to the substrate [39]. D287 and R343 are proposed to be important for substrate delivery into the enzyme core center by forming hydrogen bonds with the substrates. The D287A and R343A mutations hinder the substrate from entering the core center of Chi9602, leading to nearly complete loss of enzyme activity.
The second group of residues includes R55 and D385, which are non-conserved and flank the enzyme-substrate complex fold. As shown in Supplementary Material: Fig. S4B, R55 is apparently located outside the pocket fold that is relatively far from the substrate molecule; therefore, mutation of this residue slightly decreased enzyme activity. However, the mutation of D385 to A385 eliminated negative charge; the residue was closer to the pocket fold, increasing the compactness and stability of the enzyme-substrate complex and thereby increasing enzymatic activity.
The third group of residues includes non-conserved residues that are critical for enzymatic activity, such as W50 and S450. W50 is located in the bottom of the pocket structure, and mutation of W50 to A50 vacates lateral chain space by replacing the larger imidazole group-containing tryptophan residue with the smaller methyl group-containing alanine. The W50A mutation likely enables deeper insertion of the substrate into the groove fold closer to the catalytic center, thereby possibly increasing the affinity between the substrate and enzyme and increasing enzyme activity. The S450A mutation is located in the outer area of the pocket structure and eliminates the hydrophilicity associated with S450, thereby potentially shifting the positions of several adjacent residues (possibly E448 and W447) through the formation of hydrogen bonds. This hydrogen bond formation causes the substrate to move closer to the catalytic active center and increases enzymatic activity.
To determine if the ChiW50A mutant requires similar conditions for enzymatic activity as wild-type Chi9602, we compared the effects of pH, temperature and metal ions on the activity of both enzymes. Our results indicated that the W50A mutation does not radically alter these requirements. This finding suggests that the strategy used in this study is a well-controlled and straight-forward method to modulate only the enzymatic activity of target enzymes without affecting other properties.
We further demonstrated that the ChiW50A chitinase mutant exhibited greater synergistic effects than wild-type Chi9602 in controlling H. armigera larvae in laboratory-based trials. Although various bacterial chitinases exhibited synergistic effects on lepidopteran pests, the relatively low activity of naturally occurring bacterial chitinases has hindered their application in agricultural practices. The results presented here suggest an approach to improve chitinase activity that may be applicable to other lepidopteran-pest biocontrol processes. Moreover, the mutated ChiW50A also exhibited an enhanced synergistic effect on the nematicidal activity of B. thuringiensis spore-ICP preparations; such an effect on nematicidal activity has not been reported previously and should be validated for other nematodes, particularly plant-parasitic nematodes such as the root-knot nematode Meloidogyne incognita, an economically important agricultural pest.
In conclusion, the current study reports a goal-orientated approach to enhance the activity of a B. thuringiensis chitinase through homology modeling, molecular docking and site-directed mutagenesis. One of the resulting mutant chitinases exhibited a greater than 60 % increase in enzymatic activity without radically altered pH, temperature and metal ion requirements and exhibited synergistic effects on the control of lepidopteran and nematode pests and antifungal activity in laboratory trials. Given the simplicity and reliability of the methodology employed here, this strategy is appropriate for the molecular modification of other enzymes.
SUPPLEMENTARY MATERIAL
Tables S1 - S2, Figures S1 - S5.
Acknowledgements
The authors are grateful to Prof. Jibin Zhang for donating fungal strains, Prof. Ming Sun for donating the C. elegans wild-type strain N2, and Dr. Donghai Peng for experimental guidance on the bioassays. This work was supported by grants from the National Basic Research Program of China (973 Program, grant no. 2013CB127504), the National Science Foundation of China (Grant No. 31270158), the Natural Science Foundation of Hubei Province, China (Grant No. 2012FFA055), and the Non-Profit Science and Technology Research Funds of Hubei Province of China (Item no. 2012DBA10001). This work was also supported by the Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University (Grant No. AMLKF201002).
COMPETING INTERESTS
The authors have declared that no competing interest exists.
References
1. Barboza-Corona JE, Contreras JC, Velázquez-Robledo R, Bautista-Justo M, Gómez-Ramírez M, Cruz-Camarillo R. et al. Selection of chitinolytic strains of Bacillus thuringiensis. Biotechnol Lett. 1999;12:1125-1129 doi:10.1023/A:1005626208193
2. Cohen-Kupiec R, Chet I. The molecular biology of chitin digestion. Curr Opin Biotechnol. 1998;9:270-7
3. Neeraja C, Anil K, Purushotham P, Suma K, Sarma P, Moerschbacher BM. et al. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit Rev Biotechnol. 2010;30:231-41 doi:10.3109/07388551.2010.487258 [doi]
4. Gomaa EZ. Chitinase production by Bacillus thuringiensis and Bacillus licheniformis: their potential in antifungal biocontrol. J Microbiol. 2012;50:103-11 doi:10.1007/s12275-012-1343-y [doi]
5. Kuzu SB, Guvenmez HK, Denizci AA. Production of a thermostable and alkaline chitinase by Bacillus thuringiensis subsp. kurstaki strain HBK-51. Biotechnol Res Int. 2012;2012:135498. doi:10.1155/2012/135498 [doi]
6. Rosas-Garcia NM, Fortuna-Gonzalez JM, Barboza-Corona JE. Characterization of the chitinase gene in Bacillus thuringiensis Mexican isolates. Folia Microbiol (Praha). 2013;58:483-90 doi:10.1007/s12223-013-0233-y [doi]
7. Driss F, Rouis S, Azzouz H, Tounsi S, Zouari N, Jaoua S. Integration of a recombinant chitinase into Bacillus thuringiensis parasporal insecticidal crystal. Curr Microbiol. 2011;62:281-8 doi:10.1007/s00284-010-9704-4 [doi]
8. Regev A, Keller M, Strizhov N, Sneh B, Prudovsky E, Chet I. et al. Synergistic activity of a Bacillus thuringiensis delta-endotoxin and a bacterial endochitinase against Spodoptera littoralis larvae. Appl Environ Microbiol. 1996;62:3581-6
9. Ozgen A, Sezen K, Demir I, Demirbag Z, Nalcacioglu R. Molecular characterization of chitinase genes from a local isolate of serratia marcescens and their contribution to the insecticidal activity of Bacillus thuringiensis strains. Curr Microbiol. 2013;67:499-504 doi:10.1007/s00284-013-0395-5
10. Bhattacharya D, Nagpure A, Gupta RK. Bacterial chitinases: properties and potential. Crit Rev Biotechnol. 2007;27:21-8 doi:10.1080/07388550601168223
11. Chavan SB, Deshpande MV. Chitinolytic enzymes: An appraisal as a product of commercial potential. Biotechnol Prog. 2013;29:833-46 doi:10.1002/btpr.1732
12. Sritho N, Suginta W. Role of Tyr-435 of Vibrio harveyi chitinase A in chitin utilization. Appl Biochem Biotechnol. 2012;166:1192-202 doi:10.1007/s12010-011-9504-8
13. Suginta W, Songsiriritthigul C, Kobdaj A, Opassiri R, Svasti J. Mutations of Trp275 and Trp397 altered the binding selectivity of Vibrio carchariae chitinase A. Biochim Biophys Acta. 2007;1770:1151-60 doi:10.1016/j.bbagen.2007.03.012
14. Pantoom S, Songsiriritthigul C, Suginta W. The effects of the surface-exposed residues on the binding and hydrolytic activities of Vibrio carchariae chitinase A. BMC Biochem. 2008;9:2. doi:10.1186/1471-2091-9-2
15. Zakariassen H, Hansen MC, Joranli M, Eijsink VG, Sorlie M. Mutational effects on transglycosylating activity of family 18 chitinases and construction of a hypertransglycosylating mutant. Biochemistry. 2011;50:5693-703 doi:10.1021/bi2002532
16. Barboza-Corona JE, Reyes-Rios DM, Salcedo-Hernandez R, Bideshi DK. Molecular and biochemical characterization of an endochitinase (ChiA-HD73) from Bacillus thuringiensis subsp. kurstaki HD-73. Mol Biotechnol. 2008;39:29-37 doi:10.1007/s12033-007-9025-4
17. Barboza-Corona JE, Nieto-Mazzocco E, Velazquez-Robledo R, Salcedo-Hernandez R, Bautista M, Jimenez B. et al. Cloning, sequencing, and expression of the chitinase gene chiA74 from Bacillus thuringiensis. Appl Environ Microbiol. 2003;69:1023-9
18. Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293( Pt 3):781-8
19. Huang CJ, Guo SH, Chung SC, Lin YJ, Chen CY. Analysis of the involvement of chitin-binding domain of ChiCW in antifungal activity, and engineering a novel chimeric chitinase with high enzyme and antifungal activities. J Microbiol Biotechnol. 2009;19:1169-75 doi:JMB019-10-15
20. Huang QS, Xie XL, Liang G, Gong F, Wang Y, Wei XQ. et al. The GH18 family of chitinases: their domain architectures, functions and evolutions. Glycobiology. 2012;22:23-34 doi:10.1093/glycob/cwr092
21. Ikegami T, Okada T, Hashimoto M, Seino S, Watanabe T, Shirakawa M. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J Biol Chem. 2000;275:13654-61 doi:275/18/13654
22. Lewis JA, Fleming JT. Basic culture methods. In: (ed.) Epstein HF, Shakes DC. Caenorhabditis elegans: Modern biological analysis of an orgainism. San Diego, CA: Academic Press. 1995:3-29
23. Busby JN, Landsberg MJ, Simpson RM, Jones SA, Hankamer B, Hurst MR. et al. Structural analysis of Chi1 Chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga. Journal of molecular biology. 2012;415:359-71 doi:10.1016/j.jmb.2011.11.018
24. Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 2001
25. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-54
26. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-5
27. Zhang H, Liu M, Tian Y, Hu X. Comparative characterization of chitinases from silkworm (Bombyx mori) and bollworm (Helicoverpa armigera). Cell biochemistry and biophysics. 2011;61:267-75 doi:10.1007/s12013-011-9196-2
28. Kang SC, Park S, Lee DG. Purification and characterization of a novel chitinase from the entomopathogenic fungus, Metarhizium anisopliae. Journal of invertebrate pathology. 1999;73:276-81 doi:10.1006/jipa.1999.4843
29. Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426-8
30. Shao Z, Liu Z, Yu Z. Effects of the 20-kilodalton helper protein on Cry1Ac production and spore formation in Bacillus thuringiensis. Appl Environ Microbiol. 2001;67:5362-9 doi:10.1128/aem.67.12.5362-5369.2001
31. Dengg M, van Meel JC. Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. Journal of pharmacological and toxicological methods. 2004;50:209-14 doi:10.1016/j.vascn.2004.04.002
32. He J, Shao X, Zheng H, Li M, Wang J, Zhang Q. et al. Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. Journal of bacteriology. 2010;192:4074-5 doi:10.1128/JB.00562-10
33. Hashimoto M, Honda Y, Nikaidou N, Fukamizo T, Watanabe T. Site-directed mutagenesis of Asp280 suggests substrate-assisted catalysis of chitinase A1 from Bacillus circulans WL-12. J Biosci Bioeng. 2000;89:100-2 doi:S1389-1723(00)90031-8
34. van Aalten DM, Komander D, Synstad B, Gaseidnes S, Peter MG, Eijsink VG. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci U S A. 2001;98:8979-84 doi:10.1073/pnas.151103798
35. Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M. et al. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J Biol Chem. 1993;268:18567-72
36. Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345-64 doi:10.1002/prot.340120407
37. Li H, Greene LH. Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding. PLoS ONE. 2010;5:e8654. doi:10.1371/journal.pone.0008654
38. Papanikolau Y, Prag G, Tavlas G, Vorgias CE, Oppenheim AB, Petratos K. High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis. Biochemistry. 2001;40:11338-43 doi:bi010505h
39. Vaaje-Kolstad G, Houston DR, Rao FV, Peter MG, Synstad B, van Aalten DM. et al. Structure of the D142N mutant of the family 18 chitinase ChiB from Serratia marcescens and its complex with allosamidin. Biochim Biophys Acta. 2004;1696:103-11 doi:S1570963903003017
40. Tews I, Terwisscha van Scheltinga AC, Perrakis A, Wilson KS, Dijkstra BW. Substrate-assisted catalysis unifies two families of chitinolytic enzymes. J Am Chem Soc. 1997;119:7954-9 doi:10.1021/ja970674i
41. Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206:4393-412
42. Zhang Y, Foster JM, Nelson LS, Ma D, Carlow CK. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev Biol. 2005;285:330-9 doi:10.1016/j.ydbio.2005.06.037
43. Gan Z, Yang J, Tao N, Liang L, Mi Q, Li J. et al. Cloning of the gene Lecanicillium psalliotae chitinase Lpchi1 and identification of its potential role in the biocontrol of root-knot nematode Meloidogyne incognita. Appl Microbiol Biotechnol. 2007;76:1309-17 doi:10.1007/s00253-007-1111-9
44. Fukamizo T. Chitinolytic enzymes: catalysis, substrate binding, and their application. Curr Protein Pept Sci. 2000;1:105-24
45. Woodward JD, Wepf R, Sewell BT. Three-dimensional reconstruction of biological macromolecular complexes from in-lens scanning electron micrographs. J Microsc. 2009;234:287-92 doi:10.1111/j.1365-2818.2009.03168.x
46. Lemmon G, Meiler J. Towards ligand docking including explicit interface water molecules. PLoS One. 2013;8:e67536. doi:10.1371/journal.pone.0067536
47. Schueler-Furman O, Baker D. Conserved residue clustering and protein structure prediction. Proteins. 2003;52:225-35 doi:10.1002/prot.10365
Author contact
![]() Corresponding author: Mail address: State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan 430070, and China. Telephone: +86-27-87286952. Fax: +86-27-87280670. E-mail: lilinhzau.edu.cn.
Corresponding author: Mail address: State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan 430070, and China. Telephone: +86-27-87286952. Fax: +86-27-87280670. E-mail: lilinhzau.edu.cn.

 Global reach, higher impact
Global reach, higher impact