10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(4):456-471. doi:10.7150/ijbs.11060 This issue Cite
Research Paper
Hints for Metal-Preference Protein Sequence Determinants: Different Metal Binding Features of the Five Tetrahymena thermophila Metallothioneins
1. Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, 08028-Barcelona, Spain;
2. Departament de Química, Facultat de Ciències, Universitat Autònoma de Barcelona, 08193-Cerdanyola del Vallès (Barcelona), Spain;
3. Departamento de Microbiología-III, Facultad de Biología, Universidad Complutense, 28040-Madrid, Spain.
Received 2014-11-14; Accepted 2015-1-21; Published 2015-3-18
Abstract
The metal binding preference of metallothioneins (MTs) groups them in two extreme subsets, the Zn/Cd- and the Cu-thioneins. Ciliates harbor the largest MT gene/protein family reported so far, including 5 paralogs that exhibit relatively low sequence similarity, excepting MTT2 and MTT4. In Tetrahymena thermophila, three MTs (MTT1, MTT3 and MTT5) were considered Cd-thioneins and two (MTT2 and MTT4) Cu-thioneins, according to gene expression inducibility and phylogenetic analysis. In this study, the metal-binding abilities of the five MTT proteins were characterized, to obtain information about the folding and stability of their cognate- and non-cognate metal complexes, and to characterize the T. thermophila MT system at protein level. Hence, the five MTTs were recombinantly synthesized as Zn2+-, Cd2+- or Cu+-complexes, which were analyzed by electrospray mass spectrometry (ESI-MS), circular dichroism (CD), and UV-vis spectrophotometry. Among the Cd-thioneins, MTT1 and MTT5 were optimal for Cd2+ coordination, yielding unique Cd17- and Cd8- complexes, respectively. When binding Zn2+, they rendered a mixture of Zn-species. Only MTT5 was capable to coordinate Cu+, although yielding heteronuclear Zn-, Cu-species or highly unstable Cu-homometallic species. MTT3 exhibited poor binding abilities both for Cd2+ and for Cu+, and although not optimally, it yielded the best result when coordinating Zn2+. The two Cu-thioneins, MTT2 and MTT4 isoforms formed homometallic Cu-complexes (major Cu20-MTT) upon synthesis in Cu-supplemented hosts. Contrarily, they were unable to fold into stable Cd-complexes, while Zn-MTT species were only recovered for MTT4 (major Zn10-MTT4). Thus, the metal binding preferences of the five T. thermophila MTs correlate well with their previous classification as Cd- and Cu-thioneins, and globally, they can be classified from Zn/Cd- to Cu-thioneins according to the gradation: MTT1>MTT5>MTT3>MTT4>MTT2. The main mechanisms underlying the evolution and specialization of the MTT metal binding preferences may have been internal tandem duplications, presence of doublet and triplet Cys patterns in Zn/Cd-thioneins, and optimization of site specific amino acid determinants (Lys for Zn/Cd- and Asn for Cu-coordination).
Keywords: Metallothionein, Functional Differentiation, Metal specificity, Zinc, Copper, Tetrahymena thermophila.
Introduction
The massive explosion of Genome and Proteome projects in the last decades demonstrated the wide existence of gene/protein families, instead of single-copy elements, in all types of genomes along the tree of life. A broadly accepted Molecular Evolution principle considers gene duplication events and subsequent specialization of paralogs as the optimal scenario for the acquisition of novel and differentiated functions, from the unicellular Eukaryote organisms and first Metazoa (1,2) up to the Chordates/Vertebrates (3). Consequently, the characterization of the protein structure/function relationships in any polymorphic gene/protein system, and precisely the features of the specialized paralogous forms, should shed light to determine the evolutionary determinants that had caused the differentiation of the initially identical duplicates. Unfortunately, in many gene/protein families this basis for paralogous differentiation cannot be analyzed because even the function of every family member is unknown.
Metallothioneins (MTs) are small, ubiquitous, proteins exhibiting an extraordinary Cys content (ca. 30 %), which allows them the coordination of heavy-metal ions through the corresponding metal-thiolate bonds (4,5). They are polymorphic in practically all the organisms (plants and animals) studied up to now. It is supposed that the diversification of MT isoforms had its origin in successive gene duplication events (6,7) occurred independently in different taxa, where they constitute different homology groups. In each case, the MT function may have evolved to serve different molecular metal-related functions, such as essential metal ion homeostasis (Zn2+ or Cu+), the defense in front of toxic metal ions (i.e. Cd2+, Pb2+ or Hg2+), the scavenging of free radicals and ROS, and a wide range of cell stresses (8,9). Therefore, MTs are a very useful model to study function (in this case, metal-binding) differentiation and specificity. MT isoforms in a given organism exhibit either equivalent or opposite preferences for divalent (Zn2+ and Cd2+) vs. monovalent (Cu+) metal ion coordination (5,10), independently of the degree of their similarity at protein sequence level. At present, there is no clear clue about the molecular determinants of this specificity, a question that is framed in the more global subject of protein/metal interaction specificity in living systems (11,12). Sequence/function relationship evolution is best investigated in gene/protein families that simultaneously include highly differentiated members. In the case of MTs, this assumes the coexistence in the same organism of MTs optimized for Zn/Cd-binding (Zn/Cd-thioneins) and for Cu-binding (Cu-thioneins). Significantly, our recent thorough analysis of the MT system in pulmonate gastropod Molluscs (the Helix pomatia and Cantareus aspersus snails), which consists of highly similar MT paralogs with extreme opposite metal ion binding specialization (Cd vs. Cu), revealed that this “metal specificity” lies in their protein sequence attributes and not in other possible factors, such as gene expression inducibility, metal availability, or cell environment (13). Precisely, the specific constraints imposed by the coordination geometry of each metal ion should be in accordance with the number and disposition of ligands (i.e. thiolate groups or alternative amino acid side chains) in the MT polypeptide sequences. As a consequence, the MT protein synthesis and folding about their cognate metal ions results in a unique, energetically optimized complex, while when taking place about non-cognate metal ions, a mixture of species is produced, none of them representing an energy well conformation, but principally reflecting the amount of metal ions available in its molecular environment (14).
The first studies of function and structure in metallothioneins took for granted that the most primitive eukaryotic MTs might have been extremely short peptides of Cu-thionein character, represented nowadays by the fungal N. crassa and A. bisporus MTs, which evolved to produce all the ß-like domains of MTs in higher Eukaryotes, including Vertebrates (15). Since then, this hypothesis has been superseded by multiple experimental evidence, among which the molecular characterization of the Tetrahymena (Ciliophora, Protozoa) MT system in several species of the genus offers a most striking example. In fact, Tetrahymena MTs are among the longer MTs reported (up to 191 amino acids) and include MTs classified both as Cd-thioneins (Family 7a in the Kägi's classification (16)), and as Cu-thioneins (Family 7b) (17,18), while the evolutionary origin of Ciliates has been proposed for around 109 years ago, thus, notably before the emergence of fungi and other major eukaryotic lineages (19). These features triggered a more extensive study of the MT system in different Tetrahymena species in terms of molecular evolution and differentiation (T. thermophila (20,21,22,23,24), T. pigmentosa (20,25,26,27,28), T. pyriformis (25,29,30,31), T. rostrata (32), T. tropicalis lahorensis (33,34,35) and lately T. hegewischi, T. malaccensis and T. mobilis (36)), all of them exhibiting a high degree of polymorphism. At this point, it is worth remembering that the classification of a given MT peptide as Zn/Cd- or Cu-thionein can be performed according to three different criteria, that logically converge in their results: gene expression inducibility, protein sequence similarity, and protein metal-binding behavior (37). The wealth of information gathered from the above mentioned literature refers almost exclusively to the first two criteria. Hence, on the one hand, all the reported Tetrahymena MTs have been so far classified according to the type of metal ion that provokes or enhances the expression of its gene, and the promoter response to different metals and stresses has been deeply characterized, also in view of biotechnological applications (38,39,40,41,42). On the other hand, the origin, relationships and evolution of the corresponding protein sequences has been the object of deep and thorough analyses that have revealed close internal relationships in the Cd- and Cu-thioneins clades, as well as an interesting modular organization of the MT Cd-thionein sequences showing their more than probable origin from tandem duplications of primeval amino acid stretches (17,18). However, it is striking that studies on the third criterion, i.e. metal binding behavior or metal preference, are almost absent. Hence, only the metal ion binding features of the T. pyriformis MT1 isoform were shown in full concordance with its Cd-thionein character (43), and a partial attempt to compare the T. thermophila MTT1 and MTT2 isoforms has been recently published (44).
Thus, to fill the gap of protein functional studies on Tetrahymena MTs, we present here the full characterization of the Zn-, Cd- and Cu-binding abilities of the five T. thermophila MT isoforms (named MTT1 to MTT5 (17), cf. Figure 1 for polypeptide features). The MTT1, MTT3 and MTT5 Cd-thioneins exhibit Cys patterns typical of MTs (XCCX, CXC, XXCXX), and also some atypical Cys arrangements, such as CCC, CXCC, and CXCXC, while the MTT2 and MTT4 isoforms only enclose typical CXC motifs. Comprehensive interpretation of our results, obtained from the spectrometric and spectroscopic analyses of the recombinantly synthesized, as well as in vitro-reconstituted, metal-MTT complexes confirm that the MTT1 and MTT5 isoforms are optimized for divalent metal binding, MTT2 and MTT4 forms behave as clear Cu-thioneins and MTT3 shows an undefined behavior. However, clear differences can be defined among the coordination abilities of the five isoforms. This allows some relationships between the metal preference traits and the amino acid composition of the Tetrahymena MTs to be proposed, which will contribute to the understanding of the factors determining metal preference in proteins. Finally, the correspondence of a modular sequence structure, as proposed for the Cd-isoforms, and the metal clusters formed, is examined. Overall, it remains clear that from the first steps of the eukaryotic world, two complementary forces have driven the evolution of metallothioneins: a qualitative one, for metal specificity; and a quantitative one, to enlarge the metal binding capacity of a basic peptide fragment. This resulted in protein lengthening by internal tandem repeats (as the case of Tetrahymena, or the recently reported fungal MTs (cf. C. neoformans Cu-thioneins (45,46)), or in entire gene duplication events, as is reported for S. cerevisiae Cup1 (47).
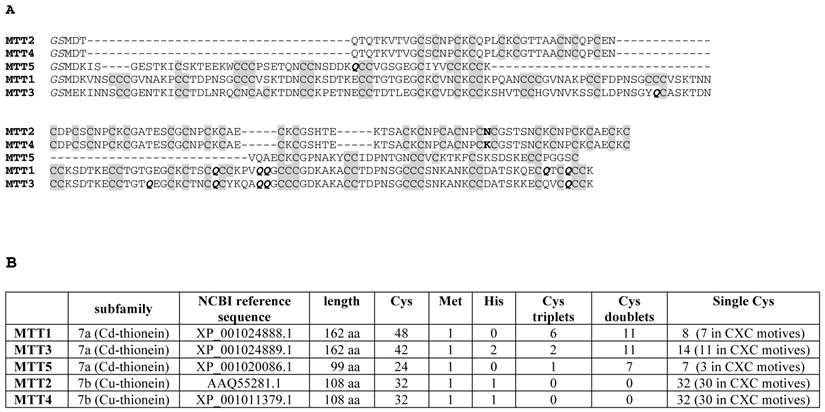
(A) Multiple sequence alignment (Clustal Omega) of the five Tetrahymena thermophila MT isoforms. The Cys residues are in grey. The unique amino acid substitution between MTT2 and MTT4 is marked in bold. The Glu (Q) residues encoded by mutated codons are marked in bold italics. The initial GS residues (in italics) result from the recombinant synthesis rationale. (B) Comparison of the main sequence features of the five Tetrahymena thermophila MT isoforms.
Materials and Methods
Construction of MTT cDNAs and E.coli expression vectors
The cDNAs corresponding to the five T. thermophila MT isoforms were obtained by mRNA retrotranscription, from cultures previously treated with Cd2+ (27 μM), Zn2+ (870 μM) or Cu2+ (80 μM) for 1h, and subcloned in PCR2.1-TOPO-TA vectors (Invitrogen), as previously reported (17). Since in Tetrahymena nuclear genes, the TAA and TAG triplets encode a glutamine instead of being stop codons (as in the Universal Gene Code) (48), the cDNAs of the MTT1, MTT3 and MTT5 isoforms had to be site-directed-mutated before cloning in the bacterial expression plasmid (pGEX-4T1). MTT2 and MTT4 cDNAs include no TAA or TAG codons, thus they could be directly subcloned. Two different site-directed-mutagenesis methods were used, owing to the different location of the bases to be mutated inside the cDNA length; hence the MTT1 and MTT5 cDNAs were mutated through Megaprimer PCR reactions (49) and the QuickChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies) was used for the MTT3 cDNA mutagenesis. In all cases the T position of the TAA and TAG codons was changed to C, the CAA and CAG codons encoding Gln in the Universal Genetic Code.
In the MTT1 cDNA, four TAA (encoding Gln110, Gln116, Gln117 and Gln159) and one TAG (encoding Gln156) triplets were present. The first PCR amplified a MTT1 cDNA fragment which included the five target codons, by using as primer oligonucleotides: 5'AAATGTACAAGTTGCCAATGCTGCAAACCTGTTCAACAAGGATGTTGTTGTG-3' (forward) and 5'-GGAACTCGAGTCATTTACAACATTGACAAGTCTGACACTCTTGCTTTGA-3' (reverse). An XhoI restriction site (underlined) was added to the reverse primer for cloning purposes. 30-cycle amplification reactions were performed with a thermo-resistant Taq DNA polymerase (Expand High Fidelity PCR System, Roche) under the conditions: 2 min at 94 oC (initial denaturation), 15 s at 94oC (denaturation), 30 s at 57oC (annealing) and 30 s at 72oC (elongation). The second PCR reaction was required to amplify the whole cDNA sequence of MTT1, using a new oligonucleotide 5'-GGGGAGGATCCATGGATAAAGTTAATAGC-3' (forward) and the product of the first PCR (reverse) as primers. Now, the BamHI restriction site (underlined) was added to the forward primer for cloning purposes. The 30-cycle amplification reactions were performed with the same Taq DNA polymerase as before, under the conditions: 2 min at 94oC (initial denaturation), 15 s at 94oC (denaturation), 30 s at 52oC (annealing) and 30 s at 72oC (elongation).
The MTT5 cDNA included only one TAA (encoding Gln36) that had to be mutated. Here, the first PCR amplified a MTT5 cDNA fragment using as primers: 5'-GCCGGGGATCCATGGATAAAATTTCTGGTGA-3' (forward BamHI site underlined) and 5'-TCTCCTGAACCGACACAACATTGTTTATCATCAGAATTGCAGCAA-3' (reverse). The 30-cycle amplification reactions were performed with the same Taq DNA polymerase as for MTT1, under the following conditions: 2 min at 94oC (initial denaturation), 15 s at 94oC (denaturation), 30 s at 57oC (annealing) and 30 s at 72oC (elongation). The second PCR was performed using the product of the previous PCR as forward megaprimer and the oligonucleotide 5'-AAAAGCTCGAGTCAGCAACTACCTCCAGGGC-3' (XhoI restriction site underlined) as reverse primer. The procedure and reagents in the second PCR were the same as for the first reaction.
As mentioned before, the MTT3 cDNA was mutagenized by using the QuickChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies), because the location of the involved codons (one TAG, encoding Glu81) and four TAA (encoding Gln102, Gln111, Gln117, Gln118 and Gln159)) made it impossible to use the megaprimer strategy. Four oligonucleotides were required to introduce the desired mutations: ol-1: (to mutate the T nucleotide in the 159-TAA triplet) 5'-ACTTCAAAGAAAGAGTGTCAGGTATGTCAATGTTGTAAATGA-3';
ol-2: (to mutate the T nucleotide in the 111-, 117- and 118-TAA triplet) 5'-CACTAATTGTCAATGCTACAAACAAGCTCAACAAGGATGTTGTTG-3';
ol-3: (to mutate the T nucleotide in the 102-TAA triplet) 5'-CTAAAGAATGTTGTACTGGCACTCAAGAAGGATG-3'; and ol-4: (to mutate the T nucleotide in the 81-TAG triplet) 5'-TTAGATCCAAATAGTGGATATCAGTGTGCAAGTAAAACTG-3'. The 30-cycle amplification reactions were performed following the kit instructions: 20 s at 95oC (initial denaturation), 30 s at 55oC (annealing) and 30 s at 65oC (elongation). Finally, an additional PCR reaction added the suitable restriction sites for cloning into the expression vector (BamHI in forward and XhoI in reverse, underlined), to the fully mutated cDNA product. The designed primers were: 5'-GGGAAGGATCCATGGAAAAAATTAATAAC-3' (forward) and 5'-GGGGACTCGAGTCATTTACAACATTGACA-3' (reverse) and the PCR conditions were the same as before.
The MTT2 and MTT4 cDNA sequences were directly amplified using the following oligonucleotides as primers: 5'- GGGGAGGATCCATGGACACTCA-3' (forward) and 5'-GAAACTCGAGTCAGCATTTGCATT-3' (reverse) for MTT2; and 5'-GGGGAGGATCCATGGACACCCA-3' (forward) and 5'-GGGGACTCGAGTCAGCATTTGC-3' (reverse) for MTT4. These primers introduced the 5' BamHI and 3' XhoI restriction sites required for subsequent subcloning. The PCR conditions and reagents were the same as before.
In all cases, the final PCR products were analyzed by 1% agarose gel electrophoresis and the expected bands were excised and purified (Genelute™ Gel Extraction Kit, Sigma Aldrich) to be subcloned into the BamHI/XhoI sites of the pGEX-4T1 E.coli expression vector (GE Healthcare) by ligation using the DNA Ligation Kit 2.1 (Takara Bio Inc.). The recombinant vectors were transformed into E. coli MachI strains. All the mutated MTT cDNAs were sequenced before expression, using the Big Dye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems). The recombinant clones were then transformed into BL21 E. coli protease deficient cells for GST-MTT fusion protein synthesis.
Synthesis and purification of recombinant and in vitro-constituted metal-MTT complexes
5-l Luria-Bertani (LB) cultures of the transformed BL21 E. coli strains were the source of recombinant metal-MTT complexes. Gene induction was switched on with 100 µM (final concentration) of isopropyl β-D-thiogalactopyranoside (IPTG) 30 min before the addition of the suitable metal supplement (300 µM ZnCl2, 300 µM CdCl2 or 500 µM CuSO4, final concentrations) to allow the synthesis of the corresponding metal complex. The cultures grew for 3 h, and in the case of Cu-supplementation, cultures were aerated to obtain either a normal oxygenation (1-l of LB media in a 2-l Erlenmeyer flask at 250 rpm) or a low oxygenation (1.5-l of LB media in a 2-l Erlenmeyer flask at 150 rpm), since this condition highly determines the level of intracellular copper in the host cells, as described in (50). It is worth noting that to prevent oxidation of the metal-MTT complexes, argon was bubbled in all the subsequent steps of the purification protocol. The 2.5-h cultures were centrifuged and the recovered cell mass was resuspended in ice-cold PBS (1.4 M NaCl, 27 mM KCl, 101 mM Na2HPO4, 18 mM KH2PO4)-0.5% v/v β-mercaptoethanol, and disrupted by sonication. The total protein extract was obtained in the supernatant of a 12,000 xg, 30 min centrifugation, which was then incubated with Glutathione-Sepharose 4B (GE Healthcare) beads at gentle agitation for 1 h at room temperature, for GST-MTT purification by batch affinity chromatography. After three washes in PBS, the GST-MTT proteins were digested with thrombin (10 u per mg of fusion protein, overnight at 17 °C) to separate the metal-MTT complexes from the GST fragment, which remains bound to the gel matrix. The recovered solution was concentrated using Centriprep 3 kDa cut-off Microcons (Amicon) and finally fractionated through a Superdex-75 FPLC column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 7.0, and run at 0.8 ml min-1. Aliquots of the protein-containing fractions were identified by their absorbance at 254 and 280 nm, and later analyzed in 15% SDS-PAGE gels stained with Coomassie Blue. MTT-containing samples were pooled and stored at -80 °C until further use. Due to the pGEX recombinant expression system specificities, the five synthesized MTT isoforms contained two additional residues (Gly-Ser) as their N-termini, but these amino acids have been shown not to alter the MT metal-binding features (51). Further details about the synthesis and purification procedures can be found in our previous publications (51, 52).
The so-called “in vitro complexes”, to differentiate them from the “in vivo” recombinantly synthesized complexes, were prepared via metal replacement by adding the corresponding metal ions (Cd2+ or Cu+) to the recombinant Zn-MTT samples. These reactions were performed at pH 7.0 following the procedures previously reported for mammalian MTs (52, 53). Characterization of the in vitro complexes was performed by UV-Vis and CD spectroscopies, as well as ESI-MS analysis, as explained below for the recombinant complexes. All assays were carried out in an Ar atmosphere, and the pH remained constant throughout all the experiments, without the addition of any extra buffers.
Spectroscopic characterization of the metal-MTT complexes
The S, Zn, Cd and Cu content of all the metal-MTT preparations was analyzed by Inductively Coupled Plasma Atomic Emission Spectroscopy (ICP-AES), using a Polyscan 61E (Thermo Jarrell Ash) spectrometer, measuring S at 182.040 nm, Zn at 213.856 nm, Cd at 228.802 nm, and Cu at 324.803 nm. Samples were routinely treated as reported in (54). Alternatively their incubation in 1 M HCl at 65 °C for 15 min prior to analyses allowed the elimination of labile sulfide ions (55). Protein concentrations were calculated from the ICP-AES sulfur measurement, assuming that all S atoms were contributed by the MTT peptides. CD spectra were recorded in a Jasco spectropolarimeter (Model J-715) interfaced to a computer (J700 software), where a 25 °C temperature was maintained constant by a Peltier PTC-351S equipment. Electronic absorptions measurements were performed on an HP-8453 Diode array UV-visible spectrophotometer. 1-cm capped quartz cuvettes were used to record all the spectra, which were corrected for the dilution effects and processed using the GRAMS 32 program.
ElectroSpray ionization mass spectrometry (ESI-MS) analyses of the metal-MTT complexes
Electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) was performed on a Micro TOF-Q instrument (Bruker) interfaced with a Series 1200 HPLC Agilent pump, equipped with an autosampler, all of which controlled by the Compass Software. The ESI-L Low Concentration Tuning Mix (Agilent Technologies) was used for equipment calibration. For the analysis of Zn- and Cd-MTT complexes, samples were run under the following conditions: 20 µl of protein solution injected through a PEEK (polyether heteroketone) tubing (1.5 m x 0.18 mm i.d.) at 40 µl min-1; capillary counter-electrode voltage 5 kV; desolvation temperature 90-110 °C; dry gas 6 l min-1; spectra collection range 800-2500 m/z. The carrier buffer was a 5:95 mixture of acetonitrile:ammonium acetate (15 mM, pH 7.0). Instead, the Cu-MTT samples were analyzed as follows: 20 µl of protein solution injected at 40 µl min-1; capillary counter-electrode voltage 3.5 kV; lens counter-electrode voltage 4 kV; dry temperature 80 °C; dry gas 6 l min-1. Here, the carrier was a 10:90 mixture of acetonitrile:ammonium acetate, 15 mM, pH 7.0. Acidic-MS conditions, which causes the demetalation of the peptides loaded with divalent metal ions, but keeps the Cu+ ions bound to the protein, were used to generate the apo-MTT forms and to analyze the Cu-containing MTT samples. For it, 20 µl of the preparation were injected under the same conditions described previously, but using a 5:95 mixture of acetonitrile:formic acid, pH 2.4, as liquid carrier. For all the ESI-MS results, the error associated with the mass measurements was always inferior to 0.1%. Masses for the holo-species were calculated according the rationale previously described (56).
Results and Discussion
MTT1 to MTT5 peptide identity and classification
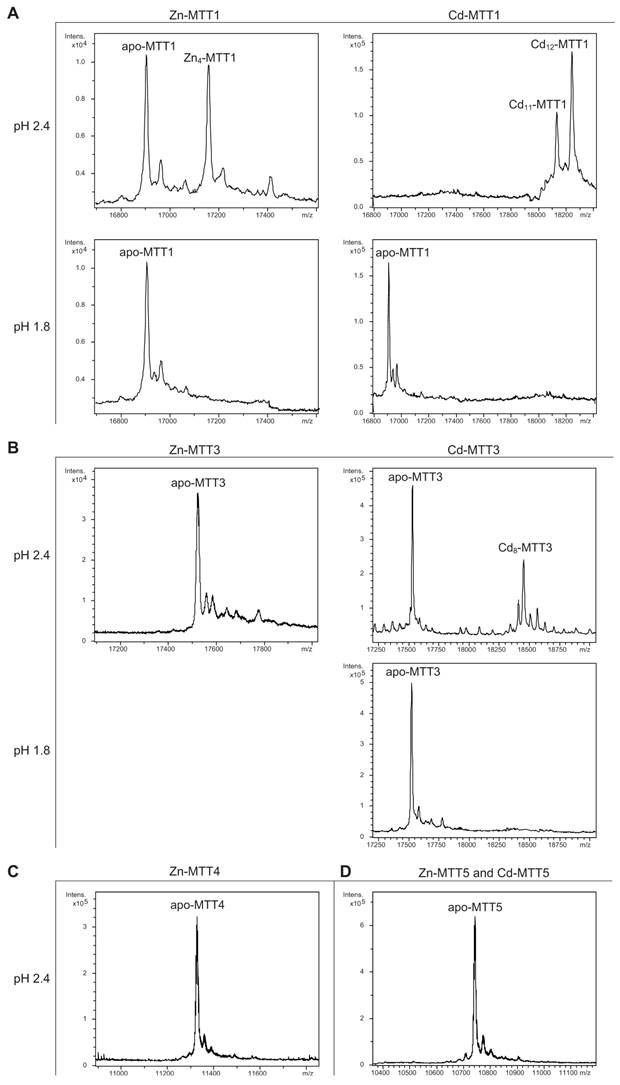
The MTT1 to MTT5 cDNAs constructed by site-directed mutagenesis according to the standard genetic code were confirmed by DNA sequencing. In total, nine TAA and two TAG triplets (coding for Gln in Tetrahymena and Stop in the standard genetic code) had been replaced by CAA and CAG codons: five in MTT1, five in MTT3, and one in MTT5 (protein positions indicated in Figure 1). SDS-PAGE analyses of total protein extracts from BL21 cells transformed with each one of the pGEX-MTT plasmids revealed the presence of bands corresponding to the expected GST-MTT sizes (data not shown). Homogeneous metal-MTT complex preparations were obtained from 5-l E. coli cultures at final concentrations varying in the 10-4 M range, as detailed in Table 1. Firstly, Zn-MTT and Cd-MTT aliquots were acidified to pH 2.4 to verify the molecular weight of the corresponding apo-forms, since this acid pH conventionally results in demetalation of the complexes formed by MTs and divalent metal ions. Some unusual results were already obtained at this stage. Since it was impossible to recover the corresponding Zn- or Cd-complexes for MTT2, no apo-MTT2 could be characterized. Nevertheless, the coherent results obtained for the Cu-MTT2 species (which will be analyzed in a following section) led to assuming the correct identity and integrity of the MTT2 peptide. For MTT4 and MTT5, the molecular masses of the acidified samples were in accordance with the expected values calculated from their respective amino acid sequences (Figure 1 and Table 1). Strikingly, the MTT1 and MTT3 isoforms, those first classified as Cd-thioneins according to gene induction criteria, yielded both Zn- and Cd-complexes that were extremely resistant to demetalation (Figure 2 and Table 1). Hence, the Zn-MTT1 preparation acidified to pH 2.4 yielded a mixture of almost equimolar apo-MTT1 and Zn4-MTT1 forms, while in the acidified Cd-preparations, a major Cd12-MTT1 and minor Cd11-MTT1 were detected. In contrast, the Zn-MTT3 complexes exhibited the usual complete demetalation at pH 2.4, yielding an apo-form with the expected molecular weight, and only the Cd-MTT3 preparation was reluctant to yield the corresponding apo-form, yielding Cd8-MTT3 complexes instead (Figure 2). Since the Cys content of MTT1 (48 Cys/162 aa) is considerably higher than that of MTT3 (42 Cys/162 aa) it is sensible to hypothesize that the resistance to acid demetalation exhibited by the Cd-MTT1 in relation to the Cd-MTT3 complexes may be related to the capacity of the former to fold into a more compact cluster, which would be stabilized by a higher number of Cd-thiolate bonds, the Cd content of both, Cd-MTT1 and Cd-MTT3, being roughly equivalent (cf. Table 2). Also the fact that, for both isoforms, the Zn species are more prone to demetalation than the Cd species is concordant with the higher strength of Cd-thiolate than Zn-thiolate bonds. These results suggest that the Cd-MTT1 and Cd-MTT3 complexes include highly stable Cd-SCys cores, which are formed by coordination of 4, or multiples of 4, Cd2+, and that the bound Cd2+ ions are only released under harsh acidification conditions. Therefore, the incubation of the Zn-MTT1, Cd-MTT1, and Cd-MTT3 preparations with increasing strength of formic acid (final pH of 1.82) yielded the expected apo-MTT1 and apo-MTT3 polypeptides, as shown in both cases by the single ESI-MS peak corresponding to the expected molecular size (Figure 2).
Deconvoluted ESI-MS spectra of the demetalated Zn- and Cd-MTT complexes, recorded at acidic pH. The spectra correspond to the demetalated preparations in Zn- and/or Cd-enriched cultures of (A) MTT1, (B) MTT3, (C) MTT4, and (D) MTT5. For those isoforms that were resistant to demetalation, the ESI-MS was run at pH 2.4 and pH 1.8.
General features of the recombinant MTT syntheses. Total protein yield of the recombinant metal-MTT preparations. The molecular weight of the acidified (pH 2.4) Zn-, and Cd- MTT preparations compared to the expected theoretical MW of the respective apo-MTT polypeptides.
| [MT]a (mg per L of culture) | Molecular Weight (Da) | |||||
|---|---|---|---|---|---|---|
| Isoform | Zn supplemented in culture | Cd supplemented in culture | Cu supplemented in culture | Acidified (pH 2.4) Zn- and Cd-complexesb | Acidified (pH 1.8) | Theoretical value for the apoforms |
| MTT1 | 1.21-1.62 | 2.55 | 0.54 | 16903.0 + Zn4-MTT1 Cd12- > Cd11-MTT1 | 16903.0 16903.0 | 16901.5 |
| MTT2 c | --- | --- | 0.32-3.15 | --- | --- | 11316.0 |
| MTT3 | 1.90-2.27 | 6.31-7.36 | 0.11 | 17530.0 Cd8-MTT3 | 17530.0 17530.0 | 17529.9 |
| MTT4 | 0.49-1.47 | 0.21 | 0.32-1.14 | 11328.0 | --- | 11330.1 |
| MTT5 | 1.67 | 1.72-2.36 | 0.86-2.40 | 10739.6 | --- | 10741.4 |
a The values were calculated from the sulphur content in normal ICP-AES measurements.
b Acidification of complexes with divalent metal ions commonly renders the demetalated polypeptides, as observed for MTT3, MTT4 and MTT5. The cases of MTT1 and MTT3 are fully commented in the text.
c MTT2 failed to yield Zn- and Cd-complexes.
Summary of the metal-to-protein-stoichiometries found in the recombinant metal-MTT preparations.
| MT Isoform | Zn supplemented in culture | Cd supplemented in culture | Cu supplemented in culture (normal aeration) | Cu supplemented in culture (low aeration) |
|---|---|---|---|---|
| MTT1 | Zn17- Zn16-, Zn18- | Cd17- Cd12- | --- | --- |
| MTT3 | Zn12- Zn11-, Zn13- Zn10-, Zn14- | Cd15S-, Cd16S-,Cd18- several Cdx- and CdyS- | Znx(Cu8, Cu4, Cu12) | --- |
| MTT5 | Zn6-, Zn5- Zn7-, Zn4- Zn8-, Zn3- | Cd8- Cd9- | M12- M9-, M8- Znx(Cu8, Cu9, Cu12) | (Cu8, Cu4) |
| MTT2 | --- | --- | Cu20- Cu16- Zn5Cu12- | Cu20- Cu23- Cu21-, Cu22- |
| MTT4 | Zn10- Zn11-, Zn9- Zn12-, Zn8- | --- | M16-, M13- Znx(Cu8, Cu4, Cu12) | Cu20- Cu23-, Cu24- Cu21-, Cu22- |
Major species are highlighted in bold. (---) means that neither protein nor metal complexes were recovered.
Zn-, Cd- and Cu-binding abilities of the Cd-MTT isoforms (family 7a): MTT1, MTT3 and MTT5
The metal binding abilities of the T. thermophila MTs previously described as Cd-thioneins (i.e. family 7a, including MTT1, MTT3 and MTT5) (17) were studied using ESI-MS and spectroscopic characterization of their corresponding recombinant Zn2+, Cd2+ and Cu+-complexes (Figure 3 and 4, respectively).
MTT1 could only be recovered from Zn2+- and Cd2+-supplemented cultures, this pointing to a complete inability of the protein for folding in vivo into stable Cu-complexes. MTT1 yielded a major Zn17-MTT1, together with minor Zn18-, Zn16- and other much minor complexes of lower and higher stoichiometry, when synthesized in the presence of Zn2+ (Table 2, Figure 3). Conversely, an almost unique peak was detected as the result of the synthesis by Cd2+-enriched bacteria, which corresponded to Cd17-MTT1, accompanied only by a very minor Cd12-complex, in total coincidence with the major Zn17- stoichiometry found for the Zn-MTT1 preparation (Table 2, Figure 3). Interestingly, although both syntheses yielded major M17 complexes, their CD fingerprints are quite different, and reflect the nature of the samples. Zn-MTT1 shows a practically featureless CD envelope, mainly contributed by the protein, and where the absorptions expected at ca. 240 nm for the Zn(SCys)4 chromophores are not perceptible, it is probably as a consequence of the mixture of coexisting species. Conversely, Cd-MTT1 gives rise to a very intense CD spectrum with maxima at 245(+) and 260(-) that can be attributed to the major Cd17-MTT1 species. This fingerprint could be contributed by a Gaussian band centered at the characteristic wavelength of the Cd(SCys)4 chromophores, 250 nm, and an exciton coupling at the same wavelength. The presence of two types of Cd-thiolate entities could be hypothesized, so that perhaps the exciton coupling signal arises from the Cd12 “robust cluster”, while the remaining Cd-SCys units forming the Cd17-MTT1 complex just generate a Gaussian band in the spectrum. Finally, it is worth noting that the results reported here are highly consistent with the stoichiometric data recently reported after apo-MTT1 metal reconstitution experiments, which showed the formation of Cd16-MTT1 (44), and the theoretical Cd17 maximum capacity, estimated from the available coordinating Cys residues of the polypeptide (18).
Deconvoluted ESI-MS spectra of the recombinant preparations of MTT1, MTT3 and MTT5. The metal-MTT complexes were synthesized in recombinant cultures supplemented with Zn, Cd, or Cu, and in the case of Cu-enriched media, the synthesis was carried out under regular and low aeration conditions. (---) denotes that no metal-MTT complexes could be purified from the corresponding cultures.
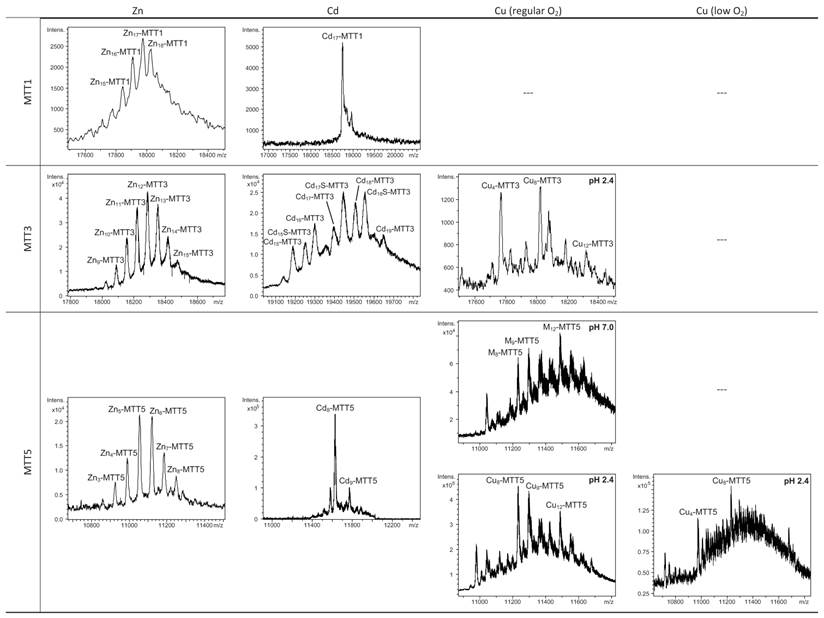
MTT3 could be recovered from Zn2+, Cd2+ and also Cu2+-supplemented cultures, but the latter only if they had been grown under normal aeration (normal cell Cu content), so that high Cu may be assumed to preclude the folding into stable complexes. Synthesis of MTT3 in Zn-supplemented E. coli cells yielded a mixture of species ranging from major Zn12-MTT3 complexes to minor Zn9- to Zn15-MTT3 species (Table 2, Figure 3), and this sample showed a CD spectrum such as that expected for an apo-MT, i.e. a silent to CD above 250 nm (Figure 4), once again reflecting the mixture of species in the sample. Although this multiplicity of Zn species resembled the behavior of MTT1, MTT3, unlike the former isoform, also yielded a mixture of complexes when synthesized under Cd supplementation, which, most significantly, included sulfide-containing species as major components (Table 2, Figure 3). Hence, Cd16S-, Cd15S-, and Cd18-MTT3 were predominant, but Cd13S-, Cd14S-, Cd15-, Cd16-, Cd17- and Cd19-MTT3 were also clearly identifiable. The presence of sulfide-containing species was confirmed by the corresponding ICP measurements, in which the S content proved to be significantly different depending on whether or not the sample had been subjected to acid treatment prior to analysis (data not shown). Additionally, the recombinant Cd-MTT3 sample exhibited a CD profile very similar to that of the Cd-MTT1 preparation, but the latter including the typical absorption of the Cd-S2- binding motifs absorbing at ca. 280 nm(-) (Figure 4). The synthesis in Cu2+-supplemented media also yielded poor results, consisting of heterometallic complexes (ICP-AES results of almost equimolar Zn:Cu content) where only Cu8- and Cu4- and minor Cu12- cores were stable enough to resist ESI-MS analysis conditions. Furthermore, these complexes were invariably CD silent at the metal-to-protein transition wavelength range (cf. Table 2, Figures 3 and 4). Therefore, MTT3 had a very atypical behavior, since it yielded mixtures of species with the three assayed metal ions.
Circular dichroism spectra of the recombinant preparations of MTT1, MTT3 and MTT5. The metal-MTT complexes were synthesized in recombinant cultures supplemented with Zn, Cd, or Cu, and in the case of Cu-enriched media, the synthesis was carried out under regular (solid line) and low aeration (dashed line) conditions.
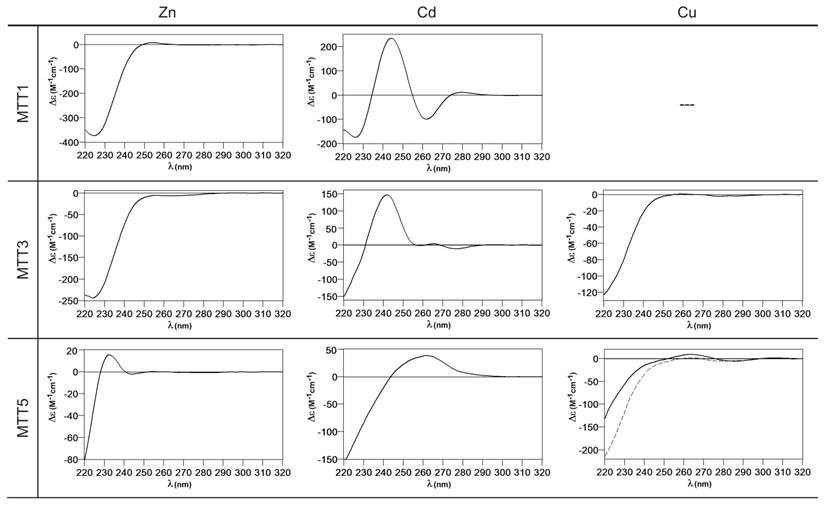
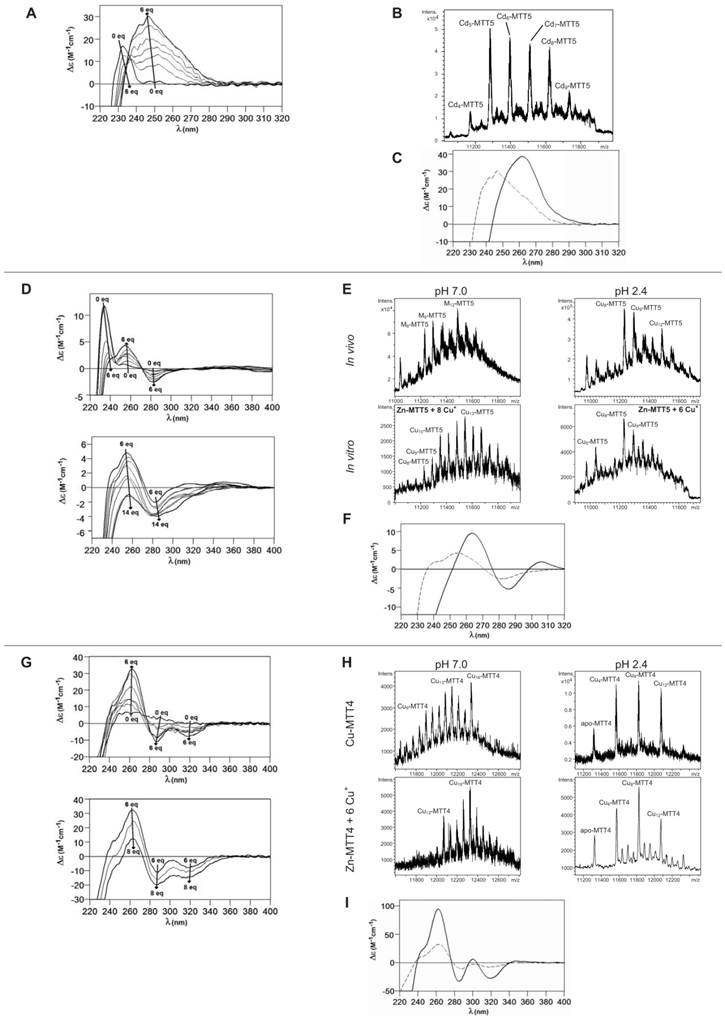
Finally, MTT5 results were significantly interesting, because it was the unique isoform that yielded stable complexes with the three metal ions analyzed, although the better results for Zn2+ and Cd2+ than for Cu+ confirmed their classification as a 7a-subfamily MT. In view of this special behavior, the in vitro Zn/Cd and Zn/Cu replacement reactions were studied for this isoform. The recombinant synthesis of MTT5 in Zn-enriched bacteria yielded two major Zn species (Zn6- and Zn5-MTT5, as revealed by ESI-MS (Table 2, Figure 3), together with minor Zn7-, Zn8- and Zn4-, Zn3-MTT5. The CD spectrum of this preparation exhibited a low intensity Gaussian band centered at 240(+) nm, in correspondence with the typical signals of the Zn-thiolate chromophores. Following a behavior similar to MTT1, MTT5 yielded an almost unique Cd-complex when synthesized in the presence of Cd2+, here Cd8-MTT5, and only very minor Cd9-MTT5 species accompanied it (Table 2, Figure 3). However, this Cd8-MTT5 complex exhibited a CD fingerprint less intense, and different in shape, to those of Cd-MTT1 and Cd-MTT3, with a wide Gaussian band ranging from 240 to 280 nm indicative of the different folding of this Cd8 complex (Figure 4). For MTT5, the Zn/Cd exchange reaction was followed in detail by CD and UV-Vis spectrophotometry and ESI-MS at discrete steps of the Cd2+ addition to the Zn-MTT5 preparation (Figure 5). This reaction demonstrated the progressive incorporation of Cd2+ ions into MTT5 (Figure 5A), but this caused the generation of a considerable mixture of Cdx-MTT5 species, (x = from 3 to 9), even for 10 Cd2+ ions added (Figure 5B). Hence, it is clear that the composition of the in vivo preparations (i.e. an almost unique Cd8-MTT5 species, Figure 3) could not be reproduced by the Zn2+/Cd2+ replacement, which was also highly evident by the comparison of the CD spectra of the respective samples (Figure 5C).
As commented before, the biosynthesis of Cu-loaded MTT5 proved to be feasible in both normal- and low-aerated Cu-supplemented cultures. Under normal Cu conditions, a mixture of Mx-MTT5 (major peaks being, in decreasing order, M12- > M9- > M8-MTT5, M=Zn or Cu) was detected by ESI-MS at neutral pH. Since ICP-AES analyses of this sample showed a ratio of 1.6 Zn:11.2 Cu, and acid ESI-MS of the sample revealed a major content of Cu9- and Cu8-MTT5, followed by Cu12-MTT5 cores (Table 2, Figure 3), it was reasonable to conclude that some of the recombinant complexes were indeed heterometallic Zn,Cu-MTT5 species. Conversely, cultures grown at high intracellular copper concentrations (i.e. low culture aeration) led to the formation of homonuclear Cu-MTT5 species, which, however, showed an extremely high instability, and only Cu8- and Cu4-cores were clearly identified among a myriad of peaks in the corresponding acid ESI-MS analyses (Table 2, Figure 3). The Zn/Cu replacement studies on Zn-MTT5 demonstrated the successive incorporation of Cu+ into the protein (Figure 5D) and revealed that a mixture of heterometallic species, similar to that yielded in vivo when this peptide was synthesized in regular cell copper concentrations, was reached at the interval of 6-to-8 Cu+ eq added (Figure 5E), despite the fact that the CD fingerprint of this sample was not reproduced at this stage of the Cu+ addition (Figure 5F). The addition of further Cu+ ions led to the detection of apo-MTT5, for 12 eq added if the sample was analyzed at neutral ESI-MS conditions, and already at 8 eq added if the sample was subjected to acid (pH 2.4) ESI-MS. This was consistent with a high instability of these complexes, which, logically, was more apparent under the harsh acid ESI-MS conditions.
Metal-binding ability comparison between the three MTT isoforms classically classified as Tetrahymena Cd-thioneins (MTT1, MTT3 and MTT5) is not straightforward, because, unlike the Cu-thionein MTT isoforms, they differ either in size and/or in Cys content and patterns (cf. Figure 1). However, several of the Zn/Cd- vs. Cu-thionein classification criteria coincide in pointing to MTT1 as the isoform with a more pronounced Zn/Cd-thionein character, because, according to these (5,10): (i) MTT1 is unable to yield stable Cu-complexes in any of the conditions assayed for Cu-supplemented cultures; (ii) the Zn-MTT1 preparation is a mixture of multiple species, exhibiting an almost silent CD spectrum, and (iii) in contrast with the two preceding points, an almost unique Cd17-MTT1 species, with very particular CD features, is the result of MTT1 folding upon Cd2+ ions. Unlike this clearly defined MTT1 behavior, MTT3 and MTT5 somehow present contradictory results. Both isoforms yield several complexes when synthesized under Zn2+ surplus, this suggesting a non-optimized polypeptide composition for Zn2+ coordination. If considering Cd2+, results clearly indicate the patent ability of MTT5 to fold into a unique, well folded complex, while MTT3 yields a poor mixture of species, the most abundant of which being sulfide-containing complexes, a feature typical of Cu-thioneins (5,10). However, the synthesis of MTT3 in Cu-supplemented media was only successful under regular intracellular Cu concentrations, and it only yielded heterometallic species with a high Zn2+ content; while MTT5, yielded stable Cu-species also at high Cu concentrations (low aeration of the cultures), with a markedly minimum Zn2+ content. All these consideration led us to suggest that MTT5 may be considered as a second-best Zn/Cd-thionein, while MTT3 would in fact behave as a MT peptide with patent deficiencies whatever the metal ion considered.
Zn-, Cd- and Cu-binding abilities of the Cu-MTT isoforms (family 7b): MTT2 and MTT4
According to their gene expression profile, the MTT2 and MTT4 isoforms were previously classified as Cu-thioneins (i.e. family 7b MTs) (17). Following the same approach described above for family 7a MTTs, we studied here the features of their Zn2+-, Cd2+- and Cu+-complexes, in order to corroborate if the copper responsiveness of their genes was coincident with the metal binding abilities of the encoded peptides and, furthermore, to evaluate if there was any differential behavior between these two T. thermophila MT isoforms. First, their divalent metal ion binding abilities were studied. Very significantly, and even after repeated attempts, no MTT2 complexes could be recovered from the Zn- and Cd-supplemented bacterial cells, this indicating the incapacity of MTT2 to fold into stable Zn- or Cd-complexes in an intracellular environment. It is worth commenting that we have commonly encountered this situation the other way round, i.e. when attempting to synthesize non-strict Cu-thioneins in copper-enriched host cells (57,58), but never in the case of divalent metal ion supplementation. Therefore, it is the first time that we report a Cu-thionein unable to bind Zn2+ or Cd2+ in vivo. Conversely, and for both MTT2 and MTT4, the two types of Cu-supplemented cultures (i.e., low aeration -meaning high cell Cu content-; and regular aeration -meaning normal cell Cu content-), respectively, yielded stable heterometallic and homometallic Cu-containing complexes. Hence, at normal aeration, a major M20-MTT2 complex (M=Zn or Cu) coexisted with minor M16- and M17-MTT2 species, as revealed by ESI-MS at neutral pH (Table 2, Figure 6). Since the major peaks detected at acid ESI-MS were Cu20-, Cu16- and Cu12-MTT2, it is reasonable to deduce that the species present in this sample were homometallic Cu20- and Cu16-MTT2 complexes, together with heterometallic Zn5Cu12-MTT2, which fits with the ICP-AES-quantification of the total metal in these preparations (3.0 Zn:15.1 Cu per MTT2).
Characterization of in vitro prepared metal-MTT5 and metal-MTT4 complexes. (A) Circular dichroism -CD- spectra recorded after the addition of up to 6 Cd2+ eq to Zn-MTT5 at pH 7.0. (B) Deconvoluted ESI-MS spectrum recorded after the addition of 10 Cd2+ eq to Zn-MTT5. (C) CD spectra corresponding to the Cd-MTT5 preparation (solid line) and that recorded after the addition of 10 Cd2+ eq to Zn-MTT5. (D) CD spectra recorded after the addition of up to 14 Cu+ eq to Zn-MTT5 at pH 7.0. Comparison of (E) the deconvoluted ESI-MS and (F) the CD spectra of the recombinant Cu-MTT5 preparation (solid line) and those recorded at several stages of the titration of Zn-MTT5 with Cu+ (dashed line for the addition of 6 Cu+ eq). (G) CD spectra recorded after the addition of up to 8 Cu+ eq to Zn-MTT4 at pH 7.0. Comparison of (H) the deconvoluted ESI-MS and (I) the CD spectra of the recombinant Cu-MTT4 preparation (solid line) and those recorded after the addition of 6 Cu+ eq to Zn-MTT4 (dashed).
Deconvoluted ESI-MS spectra of the recombinant preparations of MTT2 and MTT4. The metal-MTT complexes were synthesized in recombinant cultures supplemented with Zn, Cd, or Cu, and in the case of Cu-enriched media, the synthesis was carried out under regular and low aeration conditions. ESI-MS was run at pH 7.0 and pH 2.4. (M=Zn or Cu). (---) denotes that no metal-MTT complexes could be purified from the corresponding cultures.
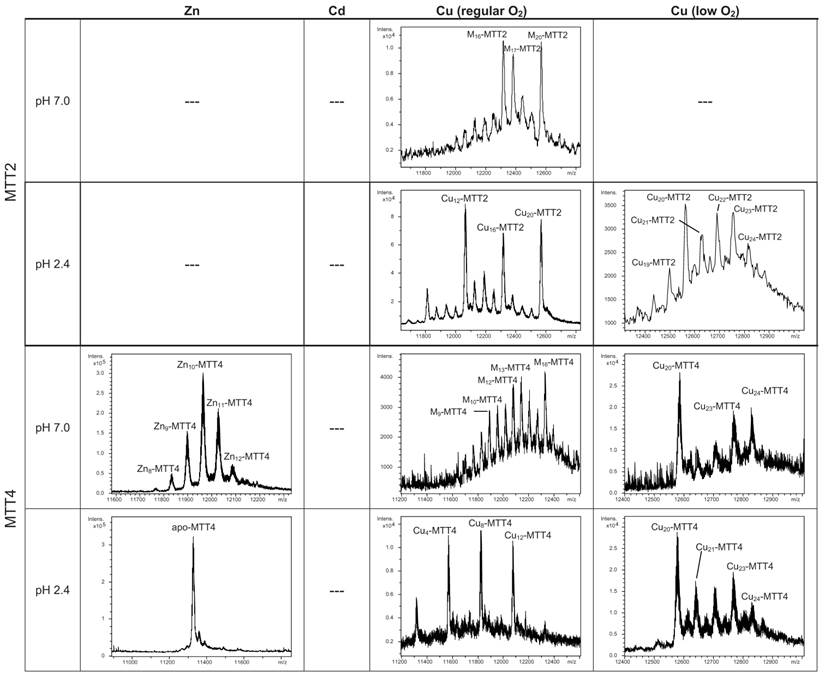
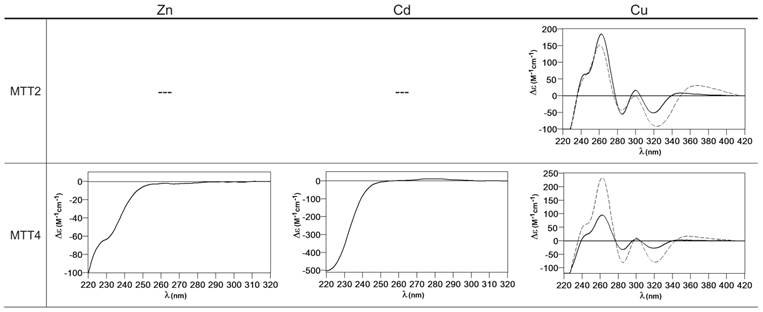
When MTT2 was synthesized by high Cu-enriched cells, the ICP-AES results indicated the total absence of Zn, and therefore all the complexes detected in the acid ESI-MS spectra (Figure 6) were interpreted as homometallic species, major Cu20- and minor Cu21- to Cu23-MTT2. In summary, Cu20-MTT2 was therefore assumed as the principal Cu-containing complex yielded by MTT2, which is also in good agreement with the data estimated in (18). The CD spectra of both Cu-MTT2 preparations (regular and normal aerated cultures) showed very similar profiles, with the typical bands at 260(+) and 285(-) nm of tetrahedrally and/or trigonally coordinated Cu+, as well as absorbances above 300 nm (320-325(-) and 365(+)), which are attributable to digonal Cu+. The latter are consistently more intense in the low aeration sample, which contains homometallic Cu+ complexes (Figure 7).
MTT4, like MTT2, was unable to fold in vivo onto Cd-complexes but, at least it yielded analyzable Zn-MTT4 species, where major Zn10-MTT4 appeared accompanied by several minor species, ranging from Zn8- to Zn12-MTT4. The multiplicity of peaks in the Zn-preparations (Figure 6) and, as a matter of fact, the impossibility of recovering Cd-MTT4 complexes, was highly concordant with the behavior of a typical Cu-thionein. Conversely, MTT4 folded into stable complexes when coordinating Cu+ ions. At regular aeration, the producing cells yielded a mixture of Zn,Cu-containing complexes, as revealed by the ICP-AES analyses (9.0 Cu:4.0 Zn per MTT4) and the divergence of the ESI-MS species detected at neutral (major M16- and M13-MTT4, together with significantly intense M9- to M17-MTT4 peaks) and acid pH (major Cu8-, and minor Cu12- and Cu4-MTT4 peaks) (Figure 6). These results are easily interpreted if assuming the presence of heterometallic ZnxCu4-, ZnyCu8-, and ZnzCu2-MTT4 species (where x, y, and z are a variable number of Zn2+ ions that added to 4, 8 or 12 Cu+ ions end up in the 9-to-17 metal ion content), and maybe some homometallic Cu12-MTT4 species. Contrarily, with a high Cu, MTT4 yields major homometallic Cu20-MTT4, together with higher nucleation species (Figure 6). The CD spectra of the Cu-MTT4 preparations drew the typical Cu-MT fingerprints already observed for the Cu-MTT2 complexes, and as for MTT2, they were more intense for the homometallic Cu-MTT4 than for the heterometallic Zn,Cu-MTT4 samples (Figure 7). Owing to the availability of Zn-MTT4 preparations, it was possible to perform Zn2+/Cu+ replacement studies for this isoform, in order to obtain a deeper insight into its in vitro Cu+ binding abilities (Figures 5G to 5I). It is worth noting that, starting from the uninformative Zn-MTT4 CD spectra (marked as 0 in the titration, Figure 5G), a typical Cu-MT CD profile developed, with absorptions at 260(+), 285(-), and 320(-) nm. Remarkably, when 6 Cu+ equivalents had been added to the initial Zn-MTT4, the CD fingerprint closely resembled that of Cu-MTT4 synthesized in regularly aerated cultures (Figure 5H), also coincident with the composition of the mixture (Figure 5I).
Comparison of the metal binding abilities of MTT2 and MTT4 reveal significant information, because, noteworthy, these two peptides only differ in one amino acid position (#89: Asn in MTT2 and Lys in MTT4, cf. Figure 1). Both MTTs bind up to 20 Cu+, which is consistent with their close similarity and conserved Cys pattern, but several points converge in supporting a more marked Cu-thionein character for MTT2 than for MTT4: i) it was impossible to recover Zn- and Cd-MTT2 complexes, while Zn-MTT4 species are stable; ii) when synthesized under normal Cu, MTT2 is already able to yield Cu20-MTT2 complexes, while this is not the case for MTT4; iii) under these synthesis conditions, MTT2 forms heterometallic species with stable Cu12-cores, while for MTT4, the most stable core is Cu8, with Cu4 and Cu12 as minor ones; iv) in surplus Cu conditions, MTT2 yields homometallic species with a higher Cu stoichiometry than MTT4. These differential Cu-binding features have to be attributed to the unique amino acid substitution, and therefore it is reasonable to conclude that the presence of Asn89 (MTT2) instead of Lys (MTT4) greatly favors the character of Cu-thionein of the polypeptide. This is in total agreement with the situation found in snail MTs, where the comparative analysis of their homologous and metal-specific CuMT and CdMT protein sequences recently revealed the respective major presence of Asn vs. Lys residues in several positions (59).
Circular dichroism spectra of the recombinant preparations of MTT2 and MTT4. The metal-MTT complexes were synthesized in recombinant cultures supplemented with Zn, Cd, or Cu, and in the case of Cu-enriched media, the synthesis was carried out under regular (solid line) and low aeration (dashed line) conditions. (---) denotes that no metal-MTT complexes could be purified from the corresponding cultures.
Conclusions
Overall, the results of the current study show the thorough analysis of the Zn2+, Cd2+ and Cu+ binding abilities of each one of the five metallothionein peptides composing the Tetrahymena thermophila MT system. These allow the polypeptides to be classified as Zn/Cd- or Cu-thioneins, a metal-binding property that is globally concordant with their specificity previously evaluated from gene response criteria. Hence, in this organism divergence evolution of cysteine-rich sequences and of gene expression regulation has led to the generation of two clear Zn/Cd-thioneins (MTT1 and MTT5), an undefined MT (MTT3), and two Cu-thioneins (MTT2 and MTT4). The comprehensive comparison of the recombinant complexes yielded by the encoded peptides towards the three metal ions allows their gradual classification from Zn/Cd-thionein to Cu-thionein as follows: MTT1>MTT5>MTT3>MTT4>MTT2; and vice versa for the Cu- to Zn/Cd-thionein gradation. Strikingly, the MTT3 isoform is an intermediate isoform, which is not particularly suitable for coordination of any of these three metal ions, if considering the poor features of the corresponding metal complexes. Data in the literature on the type of metal ion inducing expression of the T. thermophila MTT genes (17) agree with the classification suggested in this work by the features of the metal-MTT complexes. Although MTT3 was then unambiguously considered as a Cd-thionein, a peculiar behavior was already noted for the MTT3 gene inducibility pattern. Hence, all MTT Cd-thioneins are induced by divalent metal ions, but Cd2+ is the best inducer for MTT1, and Cd2+ is also better than Zn2+ for MTT5. But for the undefined MTT3 isoform, its gene is more responsive to Zn2+ at a short inducibility time, while for long treatments, Cd2+ is the most effective inducer, so that it is tempting to hypothesize that its lack of a definite metal preference responds to a need of plasticity, allowing it to develop diverse physiological tasks. It is captivating to hypothesize on how evolution may have modulated the amino acid sequences of these paralogous sequences in order to achieve such metal binding preferential behavior, since T. thermophila MTs are among the longest MT peptides ever reported. Duplication and subsequent variation of short Cys-rich sequence modules has long been proposed as the basic building mechanism for these long MTs, especially for the three Cd-thioneins, which are far more dissimilar, both in length and in Cys-patterns, than the two Cu-MTTs (17). The MTT Zn/Cd-isoforms are also characterized by the high occurrence of Cys doublets and triplets in their sequences (Figure 1B). Although the former are common in MTs, being, for example a signature for the vertebrate α-domains, the Cys-triplet motif is scarcely found among MTs; but here it appears undoubtedly associated to an increased ability for Cd2+ coordination. No complex modular structure has been defined for the MTT Cu-thioneins (MTT2 or MTT4) beyond the evidence that they encompass repetitions of a (CysLysCysX2-5CysXCys) motif, and thus the total absence of Cys triplets and doublets appears intrinsically related with an optimal Cu-binding performance. Taking into account that MTT2 and MTT4 only differ in one amino acid position, it can be assumed that they have recently differentiated in evolution. It is relevant how this amino acid change increases the Cu-thionein character of MTT2 (Asn) in relation to MTT4 (Lys), consistently with the respective identification of these amino acids as Cu-thionein and Cd-thionein determinants in snail MTs (59). In conclusion, this work confirms how the Tetrahymena MT system constitutes an invaluable model for MT evolutionary studies, a subject that is lately revealing extraordinary convergent strategies, even if analyzed in highly distinct organisms. Hence, the need for high-capacity chelating polypeptides seems to have been tackled by tandem repetition of basic building blocks, as we recently described for the pathogenic fungus Cryptococcus neoformans Cu-thioneins (45,46), and the same amino acids appear to tip the balance in favor of Zn/Cd-thioneins (Lys) or Cu-thioneins (Asn) both in snails (59) and ciliates (this work). Therefore, and despite their complete disparity in protein sequence, MTs from the most diverse organisms seem to have adopted common evolutionary trends in order to achieve their functional differentiation and specialization along the tree of life.
Acknowledgements
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO), grants BIO2012-39682-C02-01 (to SA), -02 (to MC), and grant CGL2008-00317/BOS (to JCG), which are co-financed by the European Union through the FEDER program. Authors from both Barcelona universities are members of the 2014SGR-423 Grup de Recerca de la Generalitat de Catalunya. AE was the recipient of a predoctoral grant from the MINECO (BES-2010-036553). We thank the Centres Científics i Tecnològics (CCiT) de la Universitat de Barcelona (ICP-AES, DNA sequencing) and the Servei d'Anàlisi Química (SAQ) de la Universitat Autònoma de Barcelona (CD, UV-vis, ESI-MS) for allocating instrument time.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708-713
2. Lundin LG. Gene duplications in early metazoan evolution. Semin Cell Dev Biol. 1999;10:523-530
3. Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994;43:125-133
4. Capdevila M, Bofill R, Palacios O, Atrian S. State-of-the-art of metallothioneins at the beginning of the 21stcentury. Coord Chem Rev. 2012;256:46-62
5. Palacios O, Atrian S, Capdevila M. Zn- and Cu-thioneins: a functional classification for metallothioneins? J Biol Inorg Chem. 2011;16:991-1009
6. Capdevila M, Atrian S. Metallothionein protein evolution: a miniassay. J Biol Inorg Chem. 2011;16:977-989
7. Seren N, Glaberman S, Carretero MA, Chiari Y. Molecular evolution and functional divergence of the Metallothioneins gene family in Vertebrates. J Mol Evol. 2014;78:217-233
8. Palmiter R. The elusive function of metallothioneins. Proc Natl Acad Sci USA. 1994;95:8428-8430
9. Blindauer CA, Leszczyszyn OI. Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat Prod Rep. 2010;27:720-741
10. Bofill R, Capdevila M, Atrian S. Independent metal-binding features of recombinant metallothioneins convergently draw a step gradation between Zn- and Cu-thioneins. Metallomics. 2009;1:229-234
11. Waldron JK, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nature Rev Microbiol. 2009;6:25-35
12. Waldron JK, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823-830
13. Palacios O, Pagani A, Perez-Rafael S, Egg M, Höckner M, Brandstätter A, Capdevila M, Atrian S, Dallinger R. Shaping mechanisms of metal specificity in a family of metazoan metallothioneins: evolutionary differentiation of mollusc metallothioneins. BMC Biology. 2011;9:4
14. Palacios O, Perez-Rafael S, Pagani A, Dallinger R, Atrian S, Capdevila M. Cognate and noncognate metal ion coordination in metal-specific metallothioneins: the Helix pomatia system as a model. J Biol Inorg Chem. 2014;19:923-935
15. Nemer M, Wilkinson DG, Travaglini EC, Sternberg EJ, Butt TR. Sea urchin metallothionein sequence: key to an evolutionary diversity. Proc Natl Acad Sci USA. 1985;82:4992-4994
16. Metallothioneins: classification and list of entries. www.uniprot.org/docs/metallo.txt
17. Diaz S, Amaro F, Rico D, Campos V, Benitez L, Martin-Gonzalez A, Hamilton EP, Orias E, Gutierrez JC. Tetrahymena Metallothioneins Fall into Two Discrete subfamilies. PlosOne. 2007;3:e291
18. Gutierrez JC, Amaro F, Diaz S, de Francisco P, Cubas LL, Martin-Gonzalez A. Ciliate metallothioneins: unique microbial eukaryotic heavy-metal-binder molecules. J Biol Inorg Chem. 2011;16:1025-1034
19. Parfrey LW, Lahr DJG, Knoll AH, Katz L. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108:13624-13629
20. Boldrin F, Santovito G, Negrisolo E, Piccinni E. Cloning and Sequencing of Four New Metallothionein Genes from Tetrahymena thermophila and T. pigmentosa. Evolutionary Relationships in Tetrahymena MT Family. Protist. 2003;154:431-442
21. Boldrin F, Santovito G, Gaerteig J, Wlonga D, Cassidy-Hanley D, Clark TG, Piccinni E. Metallothionein Gene from Tetrahymena thermophila with a Copper- Inducible- Repressible Promoter. Eukaryot Cell. 2006;5:422-425
22. Boldrin F, Santovito G, Formigari A, Bisharyan Y, Cassidy-Hanley D, Clark TG, Piccinni E. MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol. 2008;147:232-240
23. Santovito G, Formigari A, Boldrin F, Piccinni E. Molecular and functional evolution of Tetrahymena metallothioneins: New insights into the gene family of Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol. 2007;144:391-397
24. Chang Y, Feng L-F, Xiong J, Miao W. Function comparison and evolution analysis of metallothionein gene MTT2 and MTT4 in Tetrahymena thermophila. Zool Res. 2011;32:476-484
25. Piccinni E, Staudenmann W, Albergoni V, De Gabrieli R, James P. Purification and primary structure of metallothioneins induced by cadmium in the protists Tetrahymena pigmentosa and Tetrahymena pyriformis. Eur J Biochem. 1994;226:853-859
26. Santovito G, Irato P, Palermo S, Boldrin F, Sack R, Hunziker P, Piccinni E. Identification, Cloning and Characterisation of a Novel Copper-Metallothionein in Tetrahymena pigmentosa. Sequencing of cDNA and Expression. Protist. 2001;152:219-229
27. Boldrin F, Santovito G, Irato P, Piccinni E. Metal Interaction and Regulation of Tetrahymena pigmentosa Metallothionein Genes. Protist. 2002;153:283-291
28. Guo L, Fu C, Miao W. Cloning characterization, and gene expression analysis of a novel cadmium metallothionein gene in Tetrahymena pigmentosa. Gene. 2008;423:29-35
29. Piccinni E, Irato P, Coppellotti O, Guidolin L. Biochemical and ultrastructural data on Tetrahymena pyriformis treated with copper and cadmium. J Cell Sci. 1987;88:283-293
30. Piccinni E, Bertaggia D, Santovito G, Miceli C, Kraev A. Cadmium metallothionein gene of Tetrahymena pyriformis. Gene. 1999;234:51-59
31. Fu C, Miao W. Cloning and characterization of a New Multi-Stress Inducible Metallothionein Gene in Tetrahymena pyriformis. Protist. 2006;157:193-203
32. Amaro F, de Lucas M, Martin-Gonzalez A, Gutierrez JC. Two new members of the Tetrahymena multi-stress-inducible metallothionein family: Characterization and expression analysis of T. rostrata Cd/Cu metallothionein genes. Gene. 2008;423:85-91
33. Shuja RN, Shakoori AR. Identification, cloning and sequencing of a novel stress inducible metallothionein gene from locally isolated Tetrahymena tropicalis lahorensis. Gene. 2007;405:19-26
34. Chaundhry R, Shakoori AR. Isolation and characterization of a novel copper-inducible metallothionein gene of a ciliate, Tetrahymena tropicalis lahorensis. J Cell Biochem. 2010;110:630-644
35. Shuja RN, Taimuri SUA, Shakoori FR, Shakoori AR. Efficient expression of truncated recombinant cadmium-metallothionein gene of a ciliate, Tetrahymena tropicalis lahorensis in Escherichia coli. Mol Biol Rep. 2013;40:7061-7068
36. Chang Y, Liu G, Guo L, Liu H, Yuan D, Xiong J, Ning Y, Fu C, Miao W. Cd-Metallothioneins in Three Additional Tetrahymena Species: Intragenic Repeat Patterns and Induction by Metal Ions. J Eukaryot Microbiol. 2014;61:333-342
37. Valls M, Bofill R, Gonzalez-Duarte R, Gonzàlez-Duarte P, Capdevila M, Atrian S. A new insight into Metallothionein (MT) classification and evolution. The in vivo and in vitro metal binding features of Homarus americanus recombinant MT. J Biol Chem. 2001;276:32835-32843
38. Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci USA. 2002;99:3734-3739
39. Dondero F, Cavaletto M, Ghezzi AR, La Terza A, Banni M, Viarengo A. Biochemical Characterization and Quantitative Gene Expression Analysis of the Multi-Stress Inducible Metallothionein from Tetrahymena thermophila. Protist. 2004;155:157-168
40. Gutiérrez JC, Amaro F, Martín-González A. From heavy metal-binders to biosensors: Ciliate metallothioneins discussed. BioEssays. 2009;31:805-816
41. Amaro F, Turkewitz AP, Martin-Gonzalez A, Gutierrez JC. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb Biotechnol. 2011;4:513-522
42. Amaro F, Turkewitz AP, Martin-Gonzalez A, Gutierrez JC. Functional GFP-metallothionein from Tetrahymena thermophila: a potential whole-cell biosensor for monitoring heavy metal pollution and a cell model to study metallothionein overproduction effects. Biometals. 2014;27:195-205
43. Domenech J, Bofill R, Tinti A, Torreggiani A, Atrian S, Capdevila M. Comparative insight into the Zn(II)-, Cd(II)- and Cu(I)-binding features of the protozoan Tetrahymena pyriformis MT1 metallothionein. Biochim Biophys Acta. 2008;1784:693-704
44. Wang Q, Xu J, Chai B, Liang A, Wang W. Functional comparison of metallothioneins MTT1 and MTT2 from Tetrahymena thermophila. Arch Biochem Biophys. 2011;509:170-176
45. Ding C, Festa RA, Chen YL, Espart A, Palacios O, Espin J, Capdevila M, Atrian S, Heitman J, Thiele D. Cryptococcus neoformans copper detoxification machinery is critical for copper virulence. Cell, Host & Microbe. 2013;13:265-276
46. Palacios O, Espart A, Espin J, Ding C, Thiele D, Atrian S, Capdevila M. Full characterization of the Cu-, Zn-, and Cd-binding properties of CnMT1 and CnMT2, two metallothioneins of the pathogenic fungus Cryptococcus neoformans acting as virulence factors. Metallomics. 2014;6:279-291
47. Fogel S, Welch JW, Cathala G, Karin M. Gene amplification in yeast: CUP1 copy number regulates copper resistance. Curr Genet. 1983;7:347-355
48. Horowitz S, Gorovsky MA. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci USA. 1985;82:2452-2455
49. Landt O, Grunert HP, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125-128
50. Pagani A, Villarreal L, Capdevila M, Atrian S. The Saccharomyces cerevisiae Crs5 metallothionein metal-binding abilities and its role in the response to zinc overload. Mol Microbiol. 2007;63:256-269
51. Cols N, Romero-Isart N, Capdevila M, Oliva B, González-Duarte P, González-Duarte R, Atrian S. Binding of excess cadmium(II) to Cd7-metallothionein from recombinant mouse Zn7-metallothionein 1. UV-VIS absorption and circular dichroism studies and theoretical location approach by surface accessibility analysis. J Inorg Biochem. 1997;68:157-166
52. Capdevila M, Cols N, Romero-Isart N, González-Duarte R, Atrian S, González-Duarte P. Recombinant synthesis of mouse Zn3-ß and Zn4-α metallothionein 1 domains and characterization of their cadmium(II) binding capacity. Cell Mol Life Sci. 1997;53:681-688
53. Bofill R, Palacios O, Capdevila M, Cols N, González-Duarte R, Atrian S, González-Duarte P. A new insight into the Ag+ and Cu+ binding sites in the metallothionein β domain. J Inorg Biochem. 1999;73:57-64
54. Bongers J, Walton CD, Richardson DE, Bell JU. Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur. Anal Chem. 1988;60:2683-2686
55. Capdevila M, Domenech J, Pagani A, Tio L, Villarreal L, Atrian S. Zn- and Cd-metallothionein recombinant species from the most diverse phyla may contain sulfide (S2-) ligands. Angew Chem Int Ed Engl. 2005;44:4618-4622
56. Fabris D, Zaia J, Hathout Y, Fenselau C. Retention of Thiol Protons in Two Classes of Protein Zinc Coordination Centers. J Am Chem Soc. 1996;118:12242-12243
57. Perez-Rafael S, Mezger A, Lieb B, Dallinger R, Capdevila M, Palacios O, Atrian S. The metal binding abilities of Megathura crenulata metallothionein (McMT) in the frame of Gastropoda MTs. J Inorg Biochem. 2012;108:84-90
58. Perez-Rafael S, Kurz A, Guirola M, Capdevila M, Palacios O, Atrian S. Is MtnE, the fifth Drosophila metallothionein, functionally distinct from the other members of this polymorphic protein family? Metallomics. 2012;4:342-349
59. Perez-Rafael S, Monteiro F, Dallinger R, Atrian S, Palacios O, Capdevila M. Cantareus aspersus metallothionein metal binding abilities: The unspecific CaCd/CuMT isoform provides hints about the metal preference determinants in Metallothioneins. BBA-Proteins and Proteomics. 2014;1844:1694-1707
Author contact
![]() Corresponding author: Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Av. Diagonal 643, 08028-Barcelona, Spain, Phone: +34 934021501, FAX: +34 934034420, E-mail: satrianedu
Corresponding author: Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Av. Diagonal 643, 08028-Barcelona, Spain, Phone: +34 934021501, FAX: +34 934034420, E-mail: satrianedu

 Global reach, higher impact
Global reach, higher impact