10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(9):1036-1048. doi:10.7150/ijbs.12020 This issue Cite
Research Paper
Identification and Characterization of Candidate Chemosensory Gene Families from Spodoptera exigua Developmental Transcriptomes
College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China /Key Laboratory of Integrated Management of Crop Diseases and Pests (Nanjing Agricultural University), Ministry of Education, China
Received 2015-3-1; Accepted 2015-6-2; Published 2015-7-15
Abstract
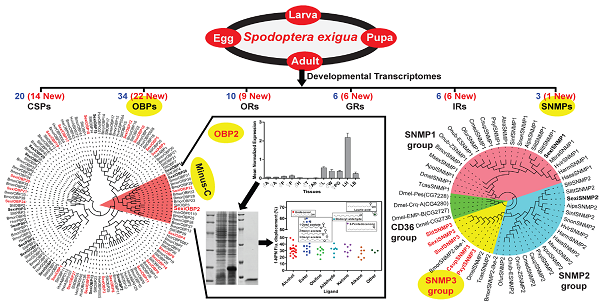
Insect chemosensory genes have been considered as potential molecular targets to develop alternative strategies for pest control. However, in Spodoptera exigua, a seriously polyphagous agricultural pest, only a small part of such genes have been identified and characterized to date. Here, using a bioinformatics screen a total of 79 chemosensory genes were identified from a public transcriptomic data of different developmental stages (eggs, 1st to 5th instar larvae, pupae, female and male adults), including 34 odorant binding proteins (OBPs), 20 chemosensory proteins (CSPs), 22 chemosensory receptors (10 odorant receptors (ORs), six gustatory receptors (GRs) and six ionotropic receptors (IRs)) and three sensory neuron membrane proteins (SNMPs). Notably, a new group of lepidopteran SNMPs (SNMP3 group) was found for the first time in S. exigua, and confirmed in four other moth species. Further, reverse transcription PCR (RT-PCR) and quantitative real time PCR (qPCR) were employed respectively to validate the sequences and determine the expression patterns of 69 identified chemosensory genes regarding to sexes, tissues and stages. Results showed that 67 of these genes could be detected and reconstructed in at least one tissue tested. Further, 60 chemosensory genes were expressed in adult antennae and 52 in larval heads with the antennae, whereas over half of the genes were also detected in non-olfactory tissues like egg and thorax. Particularly, S. exigua OBP2 showed a predominantly larval head-biased expression, and functional studies further indicated its potentially olfactory roles in guiding food searching of larvae. This work suggests functional diversities of S. exigua chemosensory genes and could greatly facilitate the understanding of olfactory system in S. exigua and other lepidopteran species.
Keywords: Spodoptera exigua, developmental transcriptome, chemosensory gene, expression pattern, fluorescence competitive binding assay
Introduction
Insect chemosensory genes, which contain odorant binding proteins (OBPs), chemosensory proteins (CSPs), odorant receptors (ORs), gustatory receptors (GRs), ionotropic receptors (IRs) and sensory neuron membrane proteins (SNMPs), are important molecular target candidates for pest control. In the process of chemosensory reception, each of the protein families is responsible for different physiological steps and thus has unique and distinguishable characteristics and functions [1, 2]. The OBP is a small molecular and soluble protein with a molecular weight of 12-20 kDa. It is secreted into the sensillar lymph by supporting cells and yields about 10 mM concentration [3], and thus a signal peptide sequence of about 20 amino acids generally exists at the N-terminus of protein [4, 5]. Previously six conserved cysteines were considered as the gold signature of the OBP, and these cysteines could form three disulfide bridges to stabilize the tertiary structure. However, less and more than six cysteines were also found in some OBPs [6]. Based on OBP classification of Bombyx mori [6] and Drosophila melanogaster [7], the OBPs are grouped into six sub-families: pheromone binding protein/ general odorant binding protein (PBP/GOBP), antennal binding protein I (ABP I), ABP II, chemical-sense-related lipophilic-ligand-binding protein (CRLBP), Minus-C and Plus-C OBPs. These OBPs are believed to be mainly involved in the uptake, binding and transport as well as receptor activation [8]. Like OBP, the CSP is also a secreted protein but has a smaller molecular weight (10-16 kDa). All CSPs contain four conserved cysteines that are capable of forming two disulfide bridges to stabilize the tertiary structure [9, 10]. This CSP family has been reported to contribute to the recognition of sex pheromone [11] and general odorant [12] as well as other functions such as leg regeneration [13], development [14] and sucking [15].
Apart from these two protein families above, in Lepidoptera there are four other crucial chemosensory-related protein families, i.e. three repertoires of chemosensory receptor super-families of ORs, GRs and IRs as well as SNMPs. These OR, GR and IR were all first identified from D. melanogaster genome by a bioinformatics screen [16-18]. Generally, each of these receptor genes is distributed in different olfactory sensilla, which are tuned to specific odorant molecules [19]. For example, some ORs are specifically distributed in the sex pheromone sensitive trichoid sensilla, and thus are called pheromone receptors (PRs), while others are sensitive to general odorants and called non-pheromone receptors. Compared to other chemosensory families, lepidopteran SNMP is a smaller family that contains only two members of SNMP1 and SNMP2 to date [20, 21]. Functional studies have indicated that SNMP1 is involved in sex pheromone recognition [22] but SNMP2 is still unknown.
With the rapid progress of next-generation sequencing techniques, more and more insect transcriptomes have been sequenced especially pest insects [23-29]. So far, transcriptome sequencing remains to be an efficient approach for gene identification due to the expensiveness of genome sequencing; and moreover even if genome sequencing is completed and extensive manual curation is still required for complex chemosensory genes, like IRs and GRs. The beet armyworm, Spodoptera exigua, is a serious pest of agricultural crops that is mainly distributed in Asia and North America [30]. Its larvae generally undergo five instars and display the maximal feeding activity from the 3rd instars on. In addition, some female moths in this species display calling behaviors (release of sex pheromones) even on the night of emergence, and the percentage of calling females increases from the 1st to 3rd night after emergence; and all calling and mating behaviors occur in the scotophase, mainly during the second half of the scotophase [31]. Similar to other noctuid moths, a highly sophisticated and sensitive olfactory system is crucial for S. exigua larvae and adults various important activities such as host searching, feeding, mating and oviposition. However, so far only 27 chemosensory genes including 12 OBPs, six CSPs, seven ORs and two SNMPs have been identified in this species [32-40]. Among those, five PBP/GOBPs [33], OBP13 [36], ABP2 [38], three CSPs (CSP1, 2 and 3) [40] and five ORs (four PRs and OR3) [35, 37] were functionally characterized by in vitro assays. Although such studies have provided some information of S. exigua chemosensory system, more extensive and systematic studies on its chemosensory mechanisms remain to be required.
Here we integrated and analysed a S. exigua transcriptome from mixed samples of different developmental stages consisting of eggs, 1st to 5th instar larvae, pupae, female and male adults [41]. Using the bioinformatics screen we identified a total of 79 chemosensory genes that contain 54 soluble olfactory proteins with as many as 49 full-length sequences and 25 chemosensory membrane proteins (10 ORs, six GRs, six IRs and three SNMPs). We first used reverse transcription PCR (RT-PCR) to build the large- and extensive-expression patterns for 69 chemosensory genes in different sexes, tissues and stages. Further, we cloned and verified 14 OBP full-length sequences and expression patterns by RACE and quantitative real time PCR (qPCR) strategies. Finally, we functionally characterized one larval head-enriched and biased OBP. This study greatly complements the numbers and diversities of chemosensory genes in S. exigua and provides valuable resources for future studies of chemosensory system in this pest species.
Materials and Methods
Insect rearing and tissue collection
The S. exigua larvae were fed on an artificial diet [42] in the laboratory (Nanjing Agricultural University, Nanjing, China) at 26 ± 1°C and 65 ± 5% relative humidity with a photoperiod of 14: 10 h (light: dark). Pupae were sexed and were separately kept until eclosion. Adults were fed on 10% honey solution.
In the expression pattern analysis by RT-PCR, each tissue was collected from three biological pools as three templates, respectively and each template contains: 1) 30-50 mg 1-d-old eggs; 2) 60 larval heads from 3-d-old larvae equally containing the 1st and 2nd days; 3) 20 pupal heads from 3-d-old pupae, with a sex ratio of 1:1; 4) 100 antennae or 60 tarsi from 2-d-old females and males during 6-8 h of the scotophase, respectively; and 5) 10 thoraxes from 2-d-old female and male moths, respectively. For the expression pattern analysis by qPCR, 1) larval heads were collected from 3-d-old larvae. At least four biological pools were collected and each template consists of six larval heads; and 2) adult tissues were collected from 2-d-old females and males during 6-8 h of the scotophase, respectively, with three biological pools. All collected tissues were immediately frozen in liquid nitrogen and stored at -70°C.
Total RNA extraction and first-strand cDNA synthesis
Total RNA from each tissue was extracted with Trizol® Reagent (Ambion, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA concentration and purify were measured with a NanoDrop™ 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
cDNA synthesis was performed with 1 μg of total RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, Liaoning, China) following the protocols. Briefly, genomic DNA was eliminated with gDNA Eraser at 42°C for 2 min. The reverse-transcription reaction was carried out at 37°C for 15 min and then the reaction was stopped at 85°C for 5 s. RACE templates (5' and 3') were synthesized with a SMART RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) according to the manufacturer's protocols. The synthesized cDNA templates were stored at -20°C.
PCR amplification, cloning and sequencing
In the expression pattern studies of chemosensory genes, S. exigua OBP19, OR1, OR10, GR2 and IR75p were not studied due to their relatively short sequence lengths (< 200 bp). In addition, expression patterns of five SexiPBP/GOBPs were also not performed because they have been extensively conducted in our previous study [33]. RT-PCR reaction of each gene was repeated twice on two independent biological pools, using rTaq DNA polymerase (TaKaRa, Dalian, Liaoning, China) under the following conditions: 94°C for 3 min, 35 cycles of 94°C for 30 s, 58°C or 60°C for 30 s, 72°C for 40 s and final extension for 7 min at 72°C. For 14 S. exigua OBP genes (OBP1-6, OBP8-12 and OBP14-16), RACE PCR was performed with gene-specific primers (GSPs) and Universal Primer A Mix (UPM) or Nested Universal Primer (NUP). The PCR procedure was conducted at 94°C for 3 min, 5 cycles of 94°C for 30 s and 72°C for 1 min, 5 cycles of 94°C for 30 s, 70°C for 30 s and 72°C for 1 min, and followed by 25 cycles of 94°C for 30 s, 68°C for 30 s and 72°C for 1 min and final extension for 10 min at 72°C. Recombinant expression vector was constructed using PrimeSTAR® HS DNA Polymerase (TaKaRa, Dalian, Liaoning, China) with an annealing temperature of 60°C. All the primers are listed in Supplementary Table S1.
PCR products with expected sizes were purified and ligated into cloning vector pEASY-T3 (TransGen, Beijing, China). Positive clones were sequenced. For the construction of SexiOBP2 recombinant expression vector, the verified plasmids were digested with FastDigest® restriction enzymes BamH I and Xho I. The purified products were ligated into the expression vector pET-30a (+), previously digested by the same enzymes. The constructed vector was further sequenced.
Bioinformatics
The S. exigua transcriptome was downloaded from the National Center for Biotechnology Information (NCBI) Transcriptome Shotgun Assembly (TSA) Database (Accession number: GAFU00000000) [41]. Spodoptera litura transcriptome was downloaded from NCBI TSA Database (Accession number: GBBY00000000). Chilo suppressalis (Accession number: SRX497236 and SRX497239) [43] and Sesamia inferens (Accession number: SRX286371) [26] transcriptomes were downloaded from NCBI Sequence Read Archive (SRA) Databases, respectively. B. mori and Plutella xylostella genomic data were downloaded from Silkworm Genome Database (http://silkworm.genomics.org.cn/) [44] and Diamondback Moth Genome Database (http://iae.fafu.edu.cn/DBM/) [45], respectively. Gene identification was carried out with Heliothis virescens [46], Spodoptera littoralis [24, 27, 47] and B. mori [6, 9] chemosensory genes, by TBLASTN program against the local transcriptomes. Next, these identified genes were searched and verified against NCBI non-redundant protein sequences database using TBLASTX program.
Alignments of amino acid sequences were performed by Muscle as implemented in MEGA6.06 program. For the tree of OBPs and CSPs, we selected the OBPs and CSPs from two noctuid species H. virescens [46] and S. littoralis [24, 27, 47] as well as a model moth B. mori [6, 9]. Next, we used SignalP 4.1 Server [48] to remove signal peptides of all OBPs and CSPs. For the tree of SNMPs, we selected the SNMPs from lepidopteran species and two model insects D. melanogaster and Tribolium castaneum. The accession numbers of all the SNMPs are available in Supplementary Table S2. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications in MEGA6.06 [49].
Effect of starvation on S. exigua OBP2 expression
In the starvation experiments, 3-d-old larvae were selected based on SexiOBP2 temporal expression dynamics. First, thirty 3-d-old larvae with the same characteristic and size were picked up, and then were randomly and evenly assigned into five experimental groups. The larvae of every group were individually starved for 24 h. After starvation treatments, larval heads of five groups were dissected, respectively. The same procedure was used to collect larval heads after normally fed on the diet for 24 h.
Quantitative real time PCR
qPCR primers were designed using Beacon Designer 8.13 (PREMIER Biosoft International, CA, USA) (Supplementary Table S1). The amplification efficiencies of target and reference gene primers were evaluated using a 5-fold or 10-fold serial dilution of cDNA templates (adult antennae or larval head). To exactly examine the relative expression levels of target genes, two reference genes of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and elongation factor (EF) [50] were selected to normalize S. exigua OBP gene expression. qPCR was performed on an ABI 7500 (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex Taq™ (TaKaRa, Dalian, Liaoning, China). Each reaction had a total volume of 20 μL, containing 10 μL of SYBR Green PCR Master Mix, 0.2 μM of each primer, 0.4 μL of ROX Reference Dye II, 2 μL of cDNA template and 6.8 μL of nuclease-free water. Each sample was run with three technical replicates on three independent biological pools. The amplification conditions were: 1 cycle of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Data analysis was performed using the Q-GENE statistical analysis package [51, 52].
Fluorescence competitive binding assay
The protein was expressed and purified following our previous protocols [53, 54]. Briefly, the recombinant protein was expressed in a bacterial system with Escherichia coli BL21 (DE3) cells at 37°C until OD600 ≈ 0.60, when expression was induced with a final concentration of 0.20 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After incubating an additional 6 h, cells were harvested by centrifugation. The purification was performed using an affinity chromatography XK 16/20 column with Ni Sepharose High Performance (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The purified target protein was lyophilized and the purity was further analyzed by SDS-PAGE.
In binding assays, all tested chemicals and a fluorescent probe N-Phenyl-1-naphthylamine (1-NPN) were dissolved in HPLC purify grade methanol as 1 mM solution. Binding assays were performed on a SpectraMaxM5 (MD, California, USA) in corresponding wells on the F96 MicroWell™ plate (Nunc, Thermo Fisher Scientific, USA). To measure the binding affinity of SexiOBP2 to 1-NPN, a 2 μM protein solution in Tris buffer (pH 7.4) was titrated by 1 mM 1-NPN to final concentrations of 2-20 μM. The binding of each competitor was tested in the competitive binding assays using 1-NPN as the fluorescent reporter and 2-20 μM concentrations for each competitor. The solution (a total volume of 250 μL) was excited at 337 nm and emission spectra were recorded at 405 nm at 25°C. Three technical replicates for the ligands of less than 70% in 1-NPN fluorescence were conducted.
The data were analyzed according to the previous studies [54, 55]. Briefly, the binding constant (K1-NPN) of SexiOBP2 to 1-NPN and relative Scatchard plot were processed using GraphPad Prism 5 software, assuming that the activity of the protein was 100%. For each competitor, the dissociation constant (Ki) was calculated according to the corresponding IC50 value (the concentrations of ligands halving the initial fluorescence value of 1-NPN), using the following equation: Ki = [IC50]/(1+[1-NPN]/K1-NPN), [1-NPN] being the free concentration of 1-NPN and K1-NPN being the binding constant of the complex protein/1-NPN.
Results
The S. exigua soluble olfactory proteins
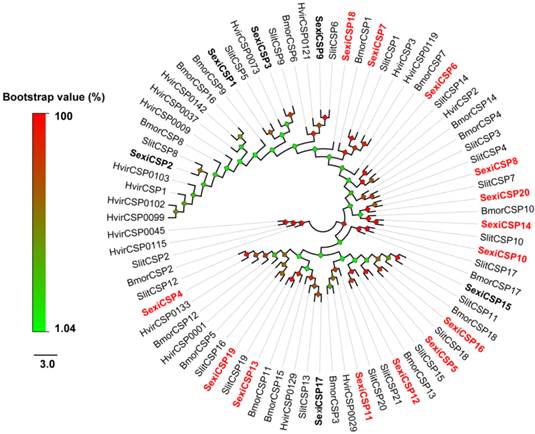
From the S. exigua transcriptomes, a total of 54 soluble olfactory proteins were identified including 34 transcripts encoding SexiOBPs and 20 ones encoding SexiCSPs. Among these genes, 29 SexiOBPs and 20 SexiCSPs were full-length sequences. To name these genes, the reported nomenclatures of SexiPBP/GOBPs [33] and SexiABPs [38] were followed and other SexiOBPs were de novo named using the Arabic numerals 1-29. Similar nomenclature was used for SexiCSPs. Compared to the previous studies in S. exigua, an increase of 22 new candidate SexiOBP and 14 new candidate SexiCSP genes was found (Figure 1 and 2). All newly identified SexiOBP and SexiCSP genes have been deposited in GenBank with accession numbers shown in Supplementary Table S3.
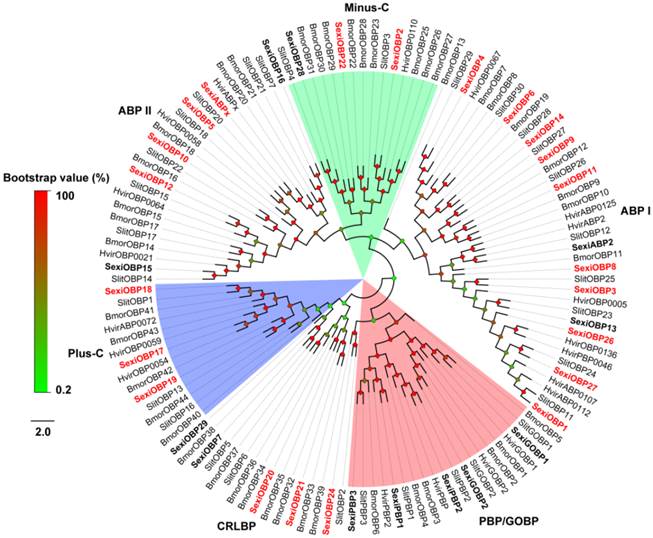
Following the previous studies of D. melanogaster and B. mori, the S. exigua OBPs were classified into six different sub-families, including five PBP/GOBPs, 12 ABP I, six ABP II, five CRLBP, three Plus-C OBPs and three Minus-C OBPs. In the tree, SexiOBP17, 18 and 19 with 10-14 cysteines were members of Plus-C OBPs. SexiOBP2, 22 and 28 with four conserved cysteines were orthologous to the silkworm Minus-C OBPs. Twenty-seven of these SexiOBPs shared over 60% identities to their respective orthologous OBPs from its sibling species S. littoralis, while SexiOBP26 showed a 59% identity to SlitOBP11 and other six SexiOBPs exhibited low identities (< 50%) (Figure 1). In the tree of CSPs, SexiCSPs were clustered into different clades and showed high identities (65-99%) to their respective orthologous S. littoralis CSPs but SexiCSP18 ortholog was not found in S. littoralis. In particular, eight of SexiCSPs shared over 90% identities to their respective orthologous SlitCSPs (Figure 2).
The S. exigua chemosensory membrane proteins
From the S. exigua transcriptomes, a total of 22 chemosensory receptor genes containing 10 ORs, six GRs and six IRs were identified. All these genes except for SexiIR76b were partial sequences. For the 10 SexiORs, only one candidate pheromone receptor SexiOR11 was identified (Supplementary Table S3 and Supplementary Table S4). For the six SexiGRs, SexiGR2 was orthologous to B. mori “CO2” receptors, SexiGR6 was one member of “GR43-like” receptors and the rest four ones were clustered into “Bitter” receptors (data not shown). None of “Sugar” receptors were identified. For the SexiIR family, two potential co-receptors SexiIR25a and 76b were found, and the others were grouped into antennal-conserved IR clade (data not shown).
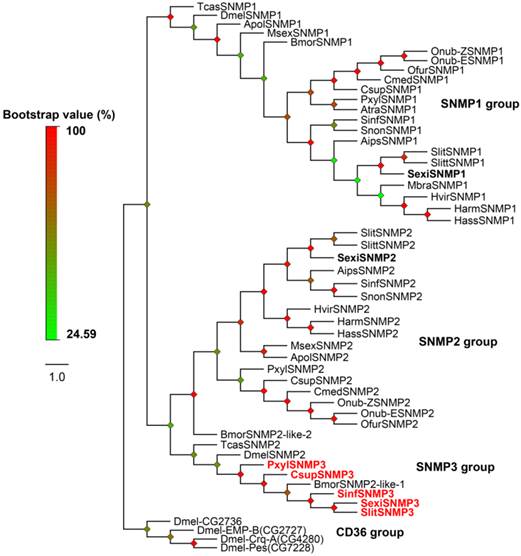
In Lepidoptera, two members of SNMPs have been reported as SNMP1 and SNMP2. Interestingly, except for SexiSNMP1 and 2, one novel candidate SNMP was identified from the S. exigua transcriptomes. Based on the phylogenetic analysis, NCBI Blast result and sequence characteristic, we named it as SexiSNMP3. The SexiSNMP3 shared 25% and 27% identities to SexiSNMP1 and 2, respectively. In order to verify the fact that the novel SNMP3 orthologs commonly exist in other lepidopteran species, we downloaded and analysed other public transcriptomes from several moths and genomes from B. mori and P. xylostella. As a result, SNMP3 orthologs were successfully found from C. suppressalis, S. litura, S. inferens and P. xylostella (Supplementary Table S4). S. litura SNMP3 (Accession number: KP729048) was a full-length sequence and shared a high identity (~87%) with SexiSNMP3, while the SNMP3s from other three species were only partial sequences. The phylogenetic tree of insect SNMPs clearly showed that insect SNMPs were classified into three distinguishable conserved groups: SNMP1, SNMP2 and SNMP3 groups. Specially, newly identified five lepidopteran SNMP3s, together with reported BmorSNMP2-like-1, TcasSNMP2 and DmelSNMP2 formed the SNMP3 group (Figure 3).
Neighbor-joining tree of candidate S. exigua OBPs with other lepidopteran OBPs. Signal peptides of all OBPs were removed based on SignalP 4.1 Server. According to B. mori and D. melanogaster OBP classification, candidate S. exigua OBPs are divided into six different clades: PBP/GOBP, ABP I, ABP II, CRLBP, Minus-C and Plus-C OBPs. S. exigua OBPs are in bold and newly identified OBP genes are highlighted with red letters. Bootstrap values are shown by the legend with a colour gradient from green to red. Bmor, B. mori; Hvir, H. virescens; Sexi, S. exigua and Slit, S. littoralis.
Neighbor-joining tree of candidate S. exigua CSPs with other lepidopteran CSPs. Signal peptides of all CSPs were removed based on SignalP 4.1 Server. S. exigua CSPs are in bold and newly identified CSP genes are highlighted with red letters. Bootstrap values are shown by the legend with a colour gradient from green to red. Bmor, B. mori; Hvir, H. virescens; Sexi, S. exigua and Slit, S. littoralis.
Neighbor-joining tree of candidate S. exigua SNMPs with other insect SNMPs. Based on the tree and sequence characteristics, SNMPs are divided into three different groups: SNMP1, SNMP2 and SNMP3 groups. Candidate S. exigua SNMPs are in bold and newly identified SNMP genes are highlighted with red letters. Bootstrap values are shown by the legend with a colour gradient from green to red. In the CD36 group, EMP-B, Epithelial Membrane Protein, isoform B; Crq-A, Croquemort isoform A and Pes, Peste. Aips, Agrotis ipsilon; Apol, Antheraea polyphemus; Atra, Amyelois transitella; Bmor, B. mori; Csup, C. suppressalis; Cmed, Cnaphalocrocis medinalis; Harm, H. armigera; Hass, H. assulta; Hvir, H. virescens; Mbra, Mamestra brassicae; Msex, Manduca sexta; Ofur, Ostrinia furnacalis; Onub-Z/E, Ostrinia nubilalis Z/E race; Pxyl, P. xylostella; Sinf, S. inferens; Sexi, S. exigua; Slit, S. littoralis; Slitu, S. litura; Snon, Sesamia nonagrioides; Dmel, D. melanogaster and Tcas, T. castaneum.
Tissue-, sex- and stage-expression pattern of S. exigua chemosensory genes
Expression pattern studies with RT-PCR were first built on crucial chemosensory organs, sexes and developmental stages. Results showed that all tested SexiOBP and SexiCSP genes could be detected in at least one tissue tested. For 28 SexiOBPs studied in RT-PCR experiments, 24 OBP genes were expressed in larval heads with the antennae and 23 ones in adult antennae. In particular, SexiOBP3, 4 and 13 appeared to be specifically or highly expressed in larval heads while SexiOBP7, 12, 15, 16 and ABPx were abundant in adult antennae. For 20 SexiCSPs expression patterns, half of them were detected in all tested tissues and none of tissue specific-expression SexiCSPs were found. Eighteen SexiCSP genes appeared to be expressed in larval heads and 17 ones in adult antennae. For eight SexiORs expression patterns, SexiOR5, 8 and 11 expression appeared to be specific in female and male antennae. For three SexiGRs expression pattern studies, SexiGR3 and 4 expression was not detected in any tested tissues, possibly due to the low expression or incorrect assemblies. SexiGR5 exhibited a weak band in female and male antennae. For five SexiIRs expression pattern studies, a lepidopteran specific SexiIR87a showed a specific expression in adult antennae and two co-receptors SexiIR25a and 76b were detected in all tested tissues. Finally, expression pattern results of three SexiSNMPs showed that SexiSNMP1 was specially expressed in female and male antennae while SexiSNMP2 and 3 displayed a broad expression (Figure 4).
To further validate the results of expression patterns above, qPCR was employed to study 14 SexiOBPs. We first used the RACE strategies to identify the open reading frames of these OBP genes. The cDNA sequences of all these genes are available in GenBank (Supplementary Table S3). Further, qPCR results of 14 SexiOBP genes were well consistent with their RT-PCR results. Specially, SexiOBP2 displayed a higher expression in larval head relative to other tissues. Four SexiOBPs expression (OBP5, 12, 15 and 16) appeared to be specific or high in adult antennae. SexiOBP6 and 8 expression was abundant in proboscis. In addition, some SexiOBPs were predominantly expressed in leg and wing such as SexiOBP1 and 9 (Figure 5).
Functional study of S. exigua OBP2
SexiOBP2 was selected for further functional studies, since it displayed a much higher expression in larval heads with the antennae than other tissues in qPCR analysis, suggesting a potential role in larval olfaction. qPCR was conducted to first determine the relative expression levels of SexiOBP2 in different days of larvae. Results showed that SexiOBP2 showed relatively low expression at the 2nd and 3rd days, and high expression after the 3rd day. Interestingly, at the 7th day SexiOBP2 expression remarkably dropped to very low level (Figure 6A).
To address the issue whether SexiOBP2 is involved in larval olfactory activities such as food searching, the effect of starvation on SexiOBP2 expression was also determined. The larvae were subjected to starvation treatment at the 3rd day, and the expression levels were measured at the 4th day. As a result, after 24 h starvation SexiOBP2 expression was significantly increased in the larval heads compared to the normal fed larvae for 24 h (Figure 6B).
Next, to study SexiOBP2 potential functions in larval olfaction, the protein was expressed in a prokaryotic expression system and showed a high yield with about 15 mg/L culture. The protein was present mainly in soluble forms. Purification was carried out by an affinity chromatography column and an apparent band of about 18 kDa was observed. To rule out the possible effect of His-tag on subsequent binding assays, the His-tag was removed by digestion with recombinant enterokinase. Finally, an expected target protein of about 12 kDa was obtained (Supplementary Figure S1).
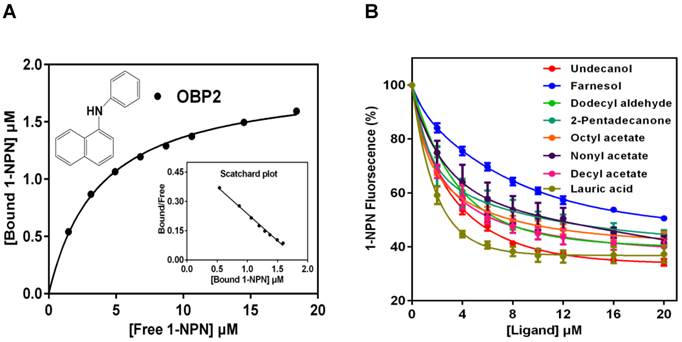
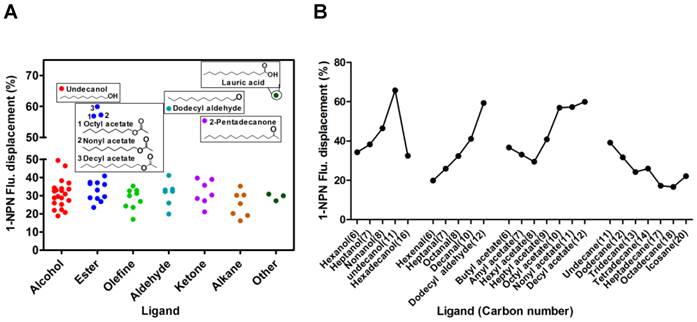
With the purified SexiOBP2, a fluorescence competitive binding assay was further employed to study the ligand binding properties of SexiOBP2 to different host volatiles. The binding of SexiOBP2 and 1-NPN were first measured. Results showed that SexiOBP2 could strongly bind 1-NPN with a binding constant (K1-NPN) of 3.80 ± 0.18 μM. A saturation and a linear Scatchard plot were observed, suggesting a single binding site and no allosteric effect (Figure 7A). Next, using 1-NPN as the fluorescent probe the binding of SexiOBP2 to 72 odorants was examined. In general, SexiOBP2 showed weak binding to most ligands tested. Lauric acid was the best ligand to SexiOBP2 (Ki = 2.16 μM). In addition, three linear acetates (octyl, nonyl and decyl acetate), dodecyl aldehyde, undecanol and 2-pentadecanone could be also strongly bound by SexiOBP2 (Ki < 9.0 μM) (Figure 7B and Supplementary Table S5). Further analyses of ligand-structural characteristics indicated that compared to other odorants, the linear odorants with 10-12 carbon atoms and a functional group were better ligands for SexiOBP2 including undecanol, dodecyl aldehyde, lauric acid, octyl acetate, nonyl acetate and decyl acetate (Figure 8A and 8B).
Expression pattern of candidate S. exigua chemosensory genes in different developmental stages. E, egg; LH, larval head; PH, mixed pupal heads; ♂A, male antenna; ♀A, female antenna; ♂Ta, male tarsus; ♀Ta, female tarsus; T, mixed thoraxes and NC, negative control using sterilised water as the template. SexiGAPDH was used as the quality and quantity control for all cDNA templates.
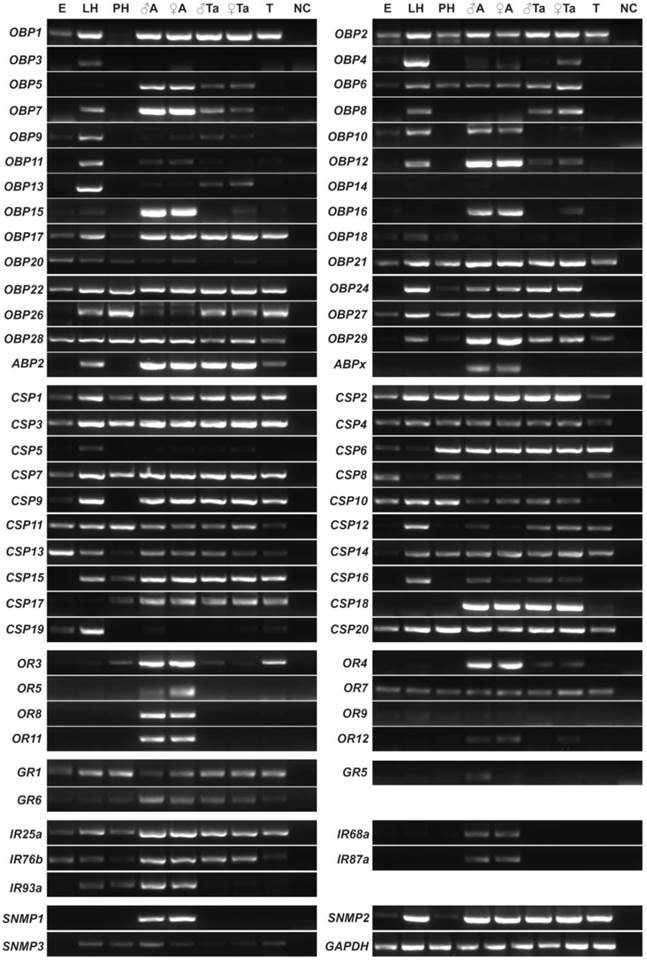
qPCR analysis of 14 candidate S. exigua OBP genes in different tissues of both sexes and stages. Expression level of each gene was calculated relative to two reference genes of GAPDH and EF. Error bars represent standard error of three independent biological pools, with three replicates of each template. ♂A, male antenna; ♂P, male proboscis; ♂H, male head without antenna and proboscis; ♂T, male thorax; ♂Ab, male abdomen; ♂L, male leg; ♂W, male wing; ♀A, female antenna; ♀P, female proboscis; ♀PG, female pheromone gland; LH, larval head and LB, larval body.
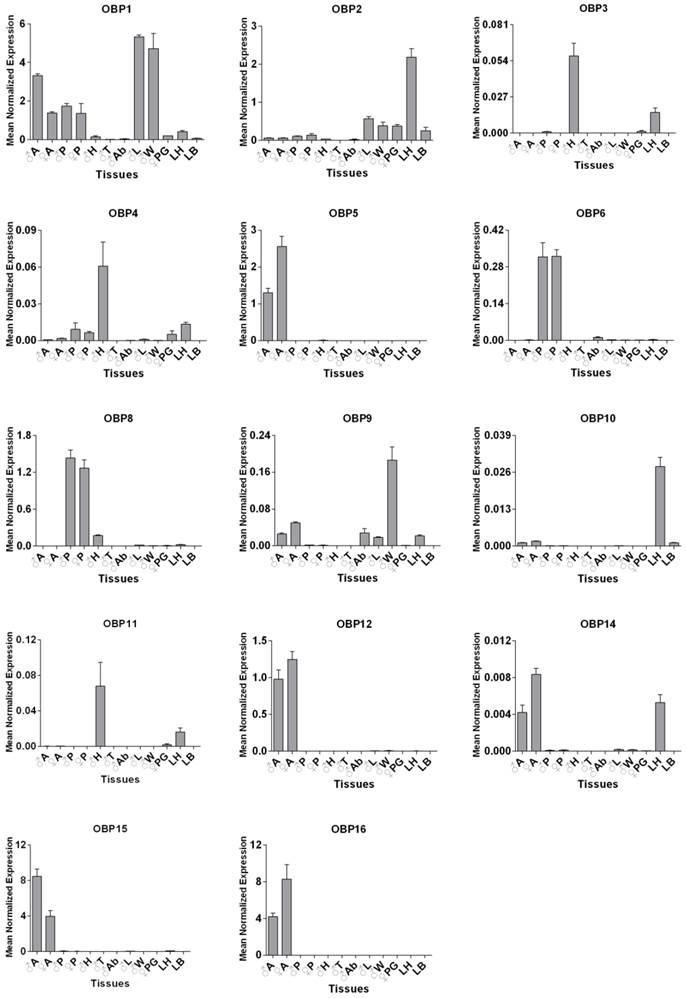
qPCR analysis of S. exigua OBP2 in larval heads of different days (A) and effect of starvation on S. exigua OBP2 expression (B). In Figure 6B, the larvae from the 3rd day (0 h) were treated for 24 h starvation (24 h-S). The normal feeding larvae were selected as the control (24 h-F). Expression level of the gene was calculated relative to the reference genes of GAPDH and EF. Error bars represent standard error of at least four independent biological pools, with three replicates of each template. Significant differences among different days or treatments were present using different letters on the bars (one-way ANOVA followed by Duncan multiple range tests, P < 0.05).
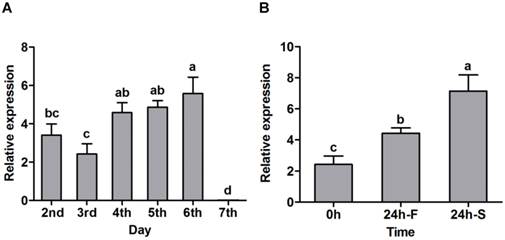
Binding of S. exigua OBP2 to 1-NPN and selected ligands. (A) Binding curves of 1-NPN to SexiOBP2 and relative Scatchard plot. Binding constant (N = 3) is 3.80 μM (SEM 0.18). (B) Competitive binding curves of selected ligands to SexiOBP2. The protein and 1-NPN concentrations are both 2.0 μM. Every data point represents mean ± SEM (N = 3).
The relationship of S. exigua OBP2 and Ligand structures. (A) Binding affinities of all tested ligands to SexiOBP2. These ligands whose 1-NPN fluorescent displacement values of SexiOBP2 are over 50% were present with structures. Based on the functional groups of chemicals, the ligands were classified into seven groups: alcohol, ester, olefine, aldehyde, ketone, alkane and other. (B) Binding comparison of SexiOBP2 to the linear ligands with different carbon-chain numbers. The 1-NPN fluorescent displacements of SexiOBP2 to ligands were calculated at the maximal concentration of 20 μM as percent of the values in the absence of ligands. All these related information is listed in Supplementary Table S5.
Discussion
In this study, 22 new OBP and 14 new CSP genes from the S. exigua transcriptomes were identified, leading to a total of 34 OBPs and 20 CSPs in this species. The number of S. exigua OBPs is very close to that of its sibling species S. littoralis (36 OBPs), and CSP number is only one less than the 21 in S. littoralis [27]. However, OBPs and CSPs in the two species were all identified from the transcriptomic data while B. mori genome contains 46 OBPs and 24 CSPs [6, 9, 56]. Thus, we suggest that at least some OBPs or CSPs of lower transcription abundances have been not identified from S. exigua. As for chemosensory receptor genes, fewer members were found from the transcriptomes. For example, we only identified 10 putative ORs, far less than the normal number of 50-60 ORs in other moths reported [28, 29, 57]. The main reason might contribute to the samples used for the transcriptome sequencing. As we know, ORs as well as other chemosensory receptor genes are specifically or predominantly expressed in chemosensory organs at very low levels, but the public transcriptomic data of S. exigua was obtained from a mixed sample consisting of equal RNA amount of egg, larva, pupa, female and male adults [41]. In this case, the relative abundances of chemosensory receptor transcripts were definitely very low, and thus most those genes could not be identified. To found more receptor genes, it would be better to sequence only the chemosensory tissues (particularly adult antennae).
Lepidopteran SNMPs contain two conserved groups of SNMP1 and SNMP2 [20, 21]. Previously, S. exigua SNMP1 and 2 were identified by their respective orthologs and PCR strategies [34]. Here we identified a new group of candidate SNMP, which has never been reported in noctuid species and thus named it as SNMP3. This SNMP3 in S. exigua shared only less than 30% identities with SNMP1 and 2, suggesting that it is not an isoform of SNMP1 or 2 but a new SNMP candidate. From four other lepidopteran species, orthologs of SexiSNMP3 were also found, which provides evidence for the existence of the third SNMP group in Lepidoptera. Based on our phylogenetic tree, the gene that was previously submitted to the GenBank as BmorSNMP2-like-1 (Accession number: XP_004933211) is actually B. mori SNMP3. It appeared that lepidopteran SNMP3 was the orthologs of DmelSNMP2 in D. melanogaster and TcasSNMP2 in T. castaneum. To confirm this, we conducted a search against the D. melanogaster and T. castaneum genome databases, and did not found one more SNMP from these two species. Thus, these two species may lose one SNMP gene that is orthologous to lepidopteran SNMP2 due to the evolutionary pressure.
In the analyses of expression patterns by RT-PCR, most of S. exigua OBPs and CSPs were expressed in olfactory organs like adult antennae and larval heads, suggesting their functional roles in adult and/or larval olfaction. However, nearly half of SexiOBPs or CSPs were also detected in non-olfactory tissues such as egg and thorax, indicating other functions of these genes in addition to the olfaction. As for chemosensory receptors, several new receptor genes are attractive to us. For example, adult antennal-specific SexiOR5 and 8 as well as SexiIR87a (a lepidopteran-specific IR) are possibly involved in adult chemosensory reception. Their orthologous genes were also reported from H. armigera [29], H. assulta [28] and S. littoralis [27], implying their conserved functions that deserve to be explored. Finally, newly identified SexiSNMP3 were detected in several tissues, suggesting functional diversities and further studies on its functions are required in future.
For moth species, larva is the most important developmental stage that imposes the damage on host plants, and thus is the major target stage in pest management. Particularly, from the 3rd instars on, the larva enters into the period of maximal food searching and feeding activity. Larval antenna is the principle olfactory organ that senses host volatiles, locates host plants and finally feeds the plants [27, 57]. The functional studies of larvae antennal-specific or high expression olfactory genes will be of significance for larva-based pest control. In our present study, SexiOBP2 displayed a highly head-biased expression in larvae but very low levels in adult tissues, suggesting its involvements in larval olfaction. Moreover, the temporal expression profile showed that the SexiOBP2 expression of 4- to 6-d-old larvae were higher than the 2- to 3-d-old larvae, which is consistent with the food searching activity of the larvae (4-d-old larvae are approximately equivalent to 3rd instar larvae under the laboratory conditions). At the 7th day, SexiOBP2 showed a sharp decrease in the expression, which might be explained by the fact that most larvae are sluggish in feeding and ready for pupation. This also indicates SexiOBP2 role in larval reception of food related odorants. The SexiOBP2 role in larval olfaction is further supported by the starvation experiments, in which SexiOBP2 showed a significant up-regulated expression after the starvation treatments, as seen the case for AlinOBP13 gene from Adelphocoris lineolatus [58]. Finally, the result of binding assays showed that lauric acid, which has been isolated and identified from S. exigua host plants corn [59] and Momordica charantia [60], was the best one of tested ligands to SexiOBP2. This provides further evidence for SexiOBP2 role in the host searching and feeding of S. exigua larvae.
Conclusion
From the S. exigua transcriptomes of different developmental stages, a total of 79 chemosensory genes were identified with an increase of 22 OBPs, 14 CSPs, nine ORs, six GRs, six IRs and one SNMP. In particular, we identified, for the first time, a novel SNMP group, SNMP3 in S. exigua and confirmed its existence in other lepidopteran species. This finding provides new insights into lepidopteran SNMP evolution and functional diversities. Further, the large- and extensive-expression profile studies not only give a clear map of S. exigua chemosensory genes, but also facilitate functional studies on these genes in future. Finally, functional studies suggest that the larval head-enriched and biased SexiOBP2 plays an olfactory role possibly associated with food searching in S. exigua larvae.
Supplementary Material
Figure S1, Tables S1-S5.
Acknowledgements
We are grateful to Master students Yan Liu, Dong-Juan Niu and Shuang-Mei Li (Nanjing Agricultural University) for their help in collecting tissues. This work is supported by a Special Fund for Agro-scientific Research in the Public Interest (201203036), grants from the National Natural Science Foundation (31071978, 31372264), and the Academic Innovative Program of Jiangsu Higher Education Institutions (CXZZ13_0304), China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698-711
2. Kaissling KE. Kinetics of olfactory responses might largely depend on the odorant-receptor interaction and the odorant deactivation postulated for flux detectors. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2013;199:879-96
3. Klein U. Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae). Insect Biochem. 1987;17:1193-204
4. Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol Part B Biochem Mol Biol. 1995;111:503-14
5. Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161-3
6. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10:332
7. Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357-69
8. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
9. Gong DP, Zhang HJ, Zhao P, Lin Y, Xia QY, Xiang ZH. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007;37:266-77
10. Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658-76
11. Jacquin-Joly E, Vogt RG, Francois MC, Nagnan-Le Meillour P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem Senses. 2001;26:833-44
12. Liu R, He X, Lehane S, Lehane M, Hertz-Fowler C, Berriman M. et al. Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour. Insect Mol Biol. 2012;21:41-8
13. Kitabayashi AN, Arai T, Kubo T, Natori S. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem Mol Biol. 1998;28:785-90
14. Maleszka J, Foret S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol. 2007;217:189-96
15. Liu YL, Guo H, Huang LQ, Pelosi P, Wang CZ. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J Exp Biol. 2014;217:1821-6
16. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327-38
17. Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830-4
18. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149-62
19. Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 2010;11:188-200
20. Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J. et al. The insect SNMP gene family. Insect Biochem Mol Biol. 2009;39:448-56
21. Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. 2001;49:47-61
22. Pregitzer P, Greschista M, Breer H, Krieger J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol Biol. 2014;23:733-42
23. Glaser N, Gallot A, Legeai F, Montagne N, Poivet E, Harry M. et al. Candidate chemosensory genes in the stemborer Sesamia nonagrioides. Int J Biol Sci. 2013;9:481-95
24. Jacquin-Joly E, Legeai F, Montagne N, Monsempes C, Francois MC, Poulain J. et al. Candidate chemosensory genes in female antennae of the noctuid moth Spodoptera littoralis. Int J Biol Sci. 2012;8:1036-50
25. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A. 2011;108:7449-54
26. Zhang YN, Jin JY, Jin R, Xia YH, Zhou JJ, Deng JY. et al. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One. 2013;8:e69715
27. Poivet E, Gallot A, Montagne N, Glaser N, Legeai F, Jacquin-Joly E. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PloS one. 2013;8:e60263
28. Xu W, Papanicolaou A, Liu NY, Dong SL, Anderson A. Chemosensory receptor genes in the oriental tobacco budworm Helicoverpa assulta. Insect Mol Biol. 2014;24:253-63
29. Liu NY, Xu W, Papanicolaou A, Dong SL, Anderson A. Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics. 2014;15:597
30. C.A.B. (Commonwealth Agricultural Bureaux). Distribution maps of pests. Pest: Spodoptera exigua (Hübner), London. Series A, Map no. 302. 1972
31. Dong SL, Du JW. Diel rhythms of calling behavior and sex pheromone production of beet armyworm, Spodoptera exigua (Lepidoptera: Noctuidae). Insect Sci. 2001;8:89-96
32. Zhu JY, Ze SZ, Yang B. Identification and expression profiling of six chemosensory protein genes in the beet armyworm, Spodoptera exigua. J Asia Pac Entomol. 2015;18:61-6
33. Liu NY, Yang F, Yang K, He P, Niu XH, Xu W. et al. Two subclasses of odorant-binding proteins in Spodoptera exigua display structural conservation and functional divergence. Insect Mol Biol. 2015;24:167-82
34. Liu C, Zhang J, Liu Y, Wang G, Dong S. Expression of SNMP1 and SNMP2 genes in antennal sensilla of Spodoptera exigua (Hubner). Arch Insect Biochem Physiol. 2014;85:114-26
35. Liu C, Liu Y, Guo M, Cao D, Dong S, Wang G. Narrow tuning of an odorant receptor to plant volatiles in Spodoptera exigua (Hubner). Insect Mol Biol. 2014;23:487-96
36. Jin R, Liu NY, Dong SL. A larval specific OBP able to bind the major female sex pheromone component in Spodoptera exigua (Hübner). J Integr Agric. 2014
37. Liu C, Liu Y, Walker WB, Dong S, Wang G. Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem Mol Biol. 2013;43:747-54
38. Zhang T, Liu NY, Dong SL. cDNA cloning, tissue distribution and ligand binding characteristics of antennal binding protein 2 from the beet armyworm, Spodoptera exigua (Lepidoptera: Noctuidae). Acta Entomol Sin. 2012;55:499-509
39. Xiu WM, Dong SL. Molecular characterization of two pheromone binding proteins and quantitative analysis of their expression in the beet armyworm, Spodoptera exigua Hubner. J Chem Ecol. 2007;33:947-61
40. Gong L, Luo Q, Rizwan-ul-Haq M, Hu MY. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. Bull Entomol Res. 2012;102:600-9
41. Li H, Jiang W, Zhang Z, Xing Y, Li F. Transcriptome analysis and screening for potential target genes for RNAi-mediated pest control of the beet armyworm, Spodoptera exigua. PLoS One. 2013;8:e65931
42. Huang CX, Zhu LM, Ni JP, Chao XY. A method of rearing the beet armyworm Spodoptera exigua. Entomol Knowl. 2002;39:229-31
43. Cao D, Liu Y, Wei J, Liao X, Walker WB, Li J. et al. Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int J Biol Sci. 2014;10:846-60
44. Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B. et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306:1937-40
45. You M, Yue Z, He W, Yang X, Yang G, Xie M. et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 2013;45:220-5
46. Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29
47. Legeai F, Malpel S, Montagne N, Monsempes C, Cousserans F, Merlin C. et al. An expressed sequence tag collection from the male antennae of the noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics. 2011;12:86
48. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth. 2011;8:785-6
49. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725-9
50. Zhu X, Yuan M, Shakeel M, Zhang Y, Wang S, Wang X. et al. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PLoS One. 2014;9:e84730
51. Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439-40
52. Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques. 2002;32:1372-4 6, 8-9
53. Liu NY, Liu CC, Dong SL. Functional differentiation of pheromone-binding proteins in the common cutworm Spodoptera litura. Comp Biochem Physiol Part A Mol Integr Physiol. 2013;165:254-62
54. Liu NY, He P, Dong SL. Binding properties of pheromone-binding protein 1 from the common cutworm Spodoptera litura. Comp Biochem Physiol B Biochem Mol Biol. 2012;161:295-302
55. Calvello M, Guerra N, Brandazza A, D'Ambrosio C, Scaloni A, Dani FR. et al. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell Mol Life Sci. 2003;60:1933-43
56. Heliconius Genome C. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94-8
57. Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R. et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881-90
58. Sun L, Xiao HJ, Gu SH, Zhou JJ, Guo YY, Liu ZW. et al. The antenna-specific odorant-binding protein AlinOBP13 of the alfalfa plant bug Adelphocoris lineolatus is expressed specifically in basiconic sensilla and has high binding affinity to terpenoids. Insect Mol Biol. 2014;23:417-34
59. Yang G, Wiseman BR, Isenhour D, Espelie K. Chemical and ultrastructural analysis of corn cuticular lipids and their effect on feeding by fall armyworm larvae. J Chem Ecol. 1993;19:2055-74
60. Sarkar N, Mukherjee A, Barik A. Olfactory responses of Epilachna dodecastigma (Coleoptera: Coccinellidae) to long-chain fatty acids from Momordica charantia leaves. Arthropod-Plant Interactions. 2013;7:339-48
Author contact
![]() Corresponding authors: Shuang-Lin Dong; Tel/Fax: +86-025-84399062; E-mail: sldongedu.cn or Fei Li; Tel/Fax: +86-025-84399025; E-mail: lifeiedu.cn
Corresponding authors: Shuang-Lin Dong; Tel/Fax: +86-025-84399062; E-mail: sldongedu.cn or Fei Li; Tel/Fax: +86-025-84399025; E-mail: lifeiedu.cn

 Global reach, higher impact
Global reach, higher impact