10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(9):1073-1087. doi:10.7150/ijbs.12065 This issue Cite
Research Paper
Cathepsin B Cysteine Proteinase is Essential for the Development and Pathogenesis of the Plant Parasitic Nematode Radopholus similis
1. Laboratory of Plant Nematology and Research Center of Nematodes of Plant Quarantine, Department of Plant Pathology, South China Agricultural University, Guangzhou 510642, China;
2. College of Forestry, Northeast Forestry University, Haerbin 150040, China
† These authors contributed equally to this work
Received 2015-3-7; Accepted 2015-5-21; Published 2015-7-15
Abstract
Radopholus similis is an important plant parasitic nematode which severely harms many crops. Cathepsin B is present in a wide variety of organisms, and plays an important role in many parasites. Understanding cathepsin B of R. similis would allow us to find new targets and approaches for its control. In this study, we found that Rs-cb-1 mRNA was expressed in esophageal glands, intestines and gonads of females, testes of males, juveniles and eggs in R. similis. Rs-cb-1 expression was the highest in females, followed by juveniles and eggs, and was the lowest in males. The maximal enzyme activity of Rs-CB-1 was detected at pH 6.0 and 40 °C. Silencing of Rs-cb-1 using in vitro RNAi (Soaking with dsRNA in vitro) not only significantly inhibited the development and hatching of R. similis, but also greatly reduced its pathogenicity. Using in planta RNAi, we confirmed that Rs-cb-1 expression in nematodes were significantly suppressed and the resistance to R. similis was significantly improved in T2 generation transgenic tobacco plants expressing Rs-cb-1 dsRNA. The genetic effects of in planta RNAi-induced gene silencing could be maintained in the absence of dsRNA for at least two generations before being lost, which was not the case for the effects induced by in vitro RNAi. Overall, our results first indicate that Rs-cb-1 plays key roles in the development, hatching and pathogenesis of R. similis, and that in planta RNAi is an effective tool in studying gene function and genetic engineering of plant resistance to migratory plant parasitic nematodes.
Keywords: Radopholus similis, Cathepsin B, development, pathogenesis, RNAi, transgenic tobacco
Introduction
The banana burrowing nematode, Radopholus similis, is an important plant parasitism nematode with a wide host range [1]. R. similis severely harms various fruit trees, ornamental plants and other agronomic and horticultural crops [2-4], and leads to significant growth reduction and severe economic losses [4-7]. Therefore, it is listed as a quarantine pest in many countries and regions [8, 9]. Although R. similis greatly damages agriculture and has drawn much attention and research efforts, effective measures to control the nematode are still lacking. Currently, chemical nematicides are the major approaches to control R. similis, but are being more and more restricted due to their high cost and harm to ecological environment [10]. Therefore, it is particularly important to explore new approaches for controlling the nematode through studying the parasitism and pathogenesis related genes of R. similis.
Urwin et al. [11] first investigated gene functions of plant parasitic nematodes, Globodera pallida and Heterodera glycines using in vitro RNAi (Soaking with dsRNA in vitro). Subsequently, the same method has been successfully applied to study gene functions of various plant parasitic nematodes [12-19]. However, the effect of gene silencing induced by in vitro RNAi is temporary and non-inheritable in sedentary plant nematodes [20]. Instead, in planta RNAi is used to silence genes involved in the development, parasitism and pathogenesis of nematodes under field conditions. The use of in planta RNAi against pests or pathogens was first described for root-knot nematodes [21]. It has also been used to study the control methods of many pests including the Rice stripe virus [22], Rice tungro bacilliform virus [23], Diabrotica virgifera virgifera [24], Myzus persicae [25, 26], Nilaparvata lugens [27], Helicoverpa armigera and Spodoptera exigua [28, 29] and some sedentary plant parasitic nematodes [30-34]. There are no reports so far on silencing genes related to the development and pathogenesis of R. similis and other migratory plant parasitic nematodes using in planta RNAi for controlling the nematodes.
Cathepsin is a cysteine protease that is present in a variety of organisms. In parasites, the most common and important cathepsins are cathepsin L(EC 3.4.22.15) and cathepsin B (EC 3.4.22.1) [35]. At present, cathepsin L gene (cl) has drawn much attention and research efforts in plant parasitic nematodes, such as H. glycines [36], H. avenae (GenBank No: ACJ13100), H. schachtii (ACJ13098), Meloidogyne incognita (CAD89795), G. pallid (AAY45896), G. virginiae (ACJ13094), G. Mexicana (ACJ13096), Rotylenchulus reniformis (AAY45870) and Bursaphelenchus xylophilus (ACH56225). Cathepsin B (CB) plays key roles in reproduction, development, invasion, pathogenesis and immune evasion [35, 37, 38]. It is a promising target for the control of animal parasites. However, the cathepsin B gene (cb) has rarely been studied, and only cb of B. xylophilus (GU130153) and R. similis [39] are cloned. The functions of cb of plant parasitic nematodes are largely unknown, even though Li et al. [16] have reported the relationship between Rs-cb-1 and the reproductive capacity of R. similis.
In this study, we investigated the expression and localization of Rs-cb-1 in R. similis and the relative enzyme activity of the Rs-CB-1. We also studied the roles of Rs-cb-1 in the development and pathogenesis of R. similis using in vitro RNAi. To investigate the effect of plant-mediated RNAi on the inhibition of Rs-cb-1 expression and control of R. similis, the transgenic tobacco expressing Rs-cb-1 dsRNA was generated. The persistence and inheritance of gene silencing induced by in vitro RNAi and in planta RNAi were compared and investigated.
Materials and Methods
Nematode cultivation and extraction
The banana burrowing nematode was collected from roots of ornamental plants Philodendron sp. and cultured in vitro on carrot disks at 25oC in a dark incubator. The carrot disks were prepared as described by Reise et al. [40], and the nematodes inoculation and cultivation were performed as described previously [17, 41]. According to the method of Zhang et al. [17], cultured nematodes were extracted in a beaker.
Plant materials
Seedlings of Anthurium andraeanum used in this study were prepared as described previously [17]. Tobacco (Nicotiana benthamiana) seeds were surface sterilized and sowed in MS medium (pH 5.8) solidified with 0.3% phytagel [42, 43]. The aseptic tobacco seeds germinated and the seedlings were cultured in a 25°C chamber (16 h-light /8h-dark photoperiod) [34].
RNA extraction , cDNA synthesis and PCR amplification of Rs-cb-1
Total RNA was extracted and the quality was verified as described previously [17]. The cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). The full-length Rs-cb-1 was amplified using primers Rscb-S1 and Rscb-A1 (Supplementary Table S1). The purified PCR product was ligated to the pMD18-T vector (Takara) and transformed into Escherichia coli JM109. Positive clones were sequence confirmed and the recombinant plasmid pMD18-Rscb was extracted for later use.
In situ hybridization
Sense (ISHS-T7S1, ISHS-A1) and antisense (ISHA-S2, ISHA-T7A2) primers (Supplementary Table S1) were designed to amplify a 317-bp fragment of Rs-cb-1. The purified PCR product was served as the template to synthesize digoxigenin (DIG)-labeled sense and antisense RNA probes using DIG RNA labeling mix (Roche). In situ hybridization was performed as described previously [44, 45]. After hybridization, the stained nematodes were examined and photographed with microscopy (Nikon 90i).
qPCR analysis of Rs-cb-1 transcript levels in different development stages of R. similis
About 100 eggs, juveniles, females and males of R. similis respectively were used for RNA extraction using RNeasy Micro kit (Qiagen). RNA was treated and quantified as described previously [45]. The RNA from each sample was used as the template for cDNA synthesis using the ReverTra Ace qPCR RT kit (TOYOBO). Primers qPCR-F1 and qPCR-R1 (Supplementary Table S1) were designed to detect the expression level of Rs-cb-1. β-actin was amplified as a reference gene in all experiments using primers Actin-F and Actin-R (Supplementary Table S1) [46]. qPCR was performed on qPCR machine and the initial data analyses were carried out using Bio-Rad CFX-96 manager software as described previously [17, 45]. All experiments were performed in triplicate with three biological replicates.
Western blot and enzymatic assay
The Rs-cb-1 gene coding region was amplified (Not including the signal peptide) using primers Pe-F and Pe-R (Supplementary Table S1) with restriction enzyme sites. The PCR product was digested with BamHI and XhoI and cloned into the expression vector pET-32a (Novagen). Recombinant plasmid pET-Rs-cb-1 was transformed into E. coli JM109 for sequence confirmation and propagation, and then extracted and introduced into E. coli BL21(DE3) for expression. Expression of recombinant protein was examined on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie brilliant blue staining after the cells were induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG). The recombinant fusion protein His-Rs-CB-1 with His-tag at the N-terminus was purified using Ni Sepharose 6 Fast Flow (GE Healthcare). The purity of purified recombinant protein was confirmed by SDS-PAGE.
For western blot, the separated proteins were transferred onto a nitrocellulose (NC) membrane. The NC membrane was blocked by incubation with 5% bovine serum albumin (BSA) for 2 h at room temperature, followed by washing with Tris-buffered saline (TBS)-Tween 20 (TBST) containing 0.05% Tween 20 [47]. Then the membrane was incubated overnight with the diluted (1:1000) primary antibody RGS-His (Qiagen) at 4°C. After being washed with TBST, the membrane was incubated with the diluted (1:3000) horseradish peroxidase conjugated goat anti-mouse IgG (Sigma) secondary antibody for 1 h at room temperature. After a final wash with TBST, signals on the membrane were detected and photographed.
The recombinant protein His-Rs-CB-1 was treated with enterokinase to remove the His-tag, and then purified as described above. The enzyme activity of Rs-CB-1 at different pH values (ranging from 3 to 10) and temperature (35°C to 60°C) were tested using fluorescent substrate Z-Arg-Arg-7-amido-4-methylcoumarin hydrochloride (Z-Arg-Arg-AMC) (Sigma) as described previously [48]. Approximately 1.6 µg purified Rs-CB-1 protein was added into 1 mL assay buffer (100 mM sodium phosphate, pH 6.2, 1 mM DTT and 1 mM EDTA) containing Z-Arg-Arg-AMC(10 µM). The mixture was incubated at 37°C for 1 h. The reaction was terminated with the addition iodoacetic acid to a final concentration of 2 mM. The release of AMC was measured with excitation and emission wavelengths of 360 nm and 460 nm, respectively. The experiment was performed in triplicate.
Synthesis of Rs-cb-1 dsRNA
A 537-bp fragment from Rs-cb-1 ORF was amplified using primers Rscb-T7S/Rscb-A and Rscb-S/Rscb-T7A (Supplementary Table S1) containing a T7 promoter. The sense and antisense single-stranded RNA (ssRNA) were transcribed using ScriptMAXTM Thermo T7 Transcription Kit (TOYOBO). The dsRNA was synthesized and purified as described previously [45, 49]. Non-endogenous control egfp dsRNA (enhanced green fluorescent protein gene) was generated with the primers eGFP-T7S/eGFP-A and eGFP-S/eGFP-T7A (Supplementary Table S1) as described above.
Rs-cb-1 dsRNA treatment and silence detection
For each silence assay, about 500 mixed stage nematodes were washed with DEPC water, soaked in Rs-cb-1 dsRNA solution (2.0 mg/mL), and shaken slightly in a dark rotary incubator at 25°C for 12 h, 24 h, 36 h, 48 h and 72 h, respectively. Non-endogenous egfp dsRNA solution (2.0 mg/mL) was used as a control. Meanwhile, untreated nematodes were used as a blank control [17]. The treated nematodes were washed with DEPC water and the RNA was then extracted. qPCR was used to analyze the transcript suppression of Rs-cb-1 in R. similis as described above.
Embryonic development, hatching and post-embryonic development of R. similis
The eggs of Radopholus similis were washed from the surface of carrot callus, the single cell (1-cell) stage eggs were observed and collected by a dropper under the inverted microscope (Nikon Ti-U), and then washed with sterile water in centrifuge tube. These 1-cell stage eggs were respectively soaked with Rs-cb-1 dsRNA and egfp dsRNA for different durations, treated with streptomycin sulfate (3 g/L) for 30 min, and then washed with sterile water. After that, the tests were performed as the following: (i) Eggs in different treatment groups were placed in concave slides and cultivated in a dark incubator at 25°C with moisture. Development of the eggs was observed and recorded every 12 h under the inverted microscope till hatching; (ii) Eggs with different treatment were placed in Petri dishes (3 cm in diameter, 10-15/dish) with water and cultivated at 25°C. After a week, eggs were observed every 12 h till hatching and the hatching rate were calculated (hatching rate = number of hatched eggs/total number of eggs); (iii) Eggs with different treatment were inoculated onto seven carrot callus (20 eggs each) and cultivated at 25°C, respectively. After the second stage juveniles (J2) appeared, nematodes and eggs on the carrot callus were isolated and observed every other day under the microscope, and the duration of post-embryonic development of nematodes were recorded. Five biological replicates were performed.
Pathogenicity test of nematodes
The mixed stage nematodes were treated with Rs-cb-1 dsRNA, egfp dsRNA, M9 buffer and sterile water for 12, 24, 36, 48, and 72h, respectively. The selected seedlings of A. andraeanum were same in height (about 10 cm) and growth condition, and each seedling was inoculated with 1,000 treated nematodes and cultivated in greenhouse (26 ± 1°C, 60-80% relative humidity and 16 h-light/8 h-dark photoperiod) [29]. After 60 days, three growth parameters (Plant4 height, fresh shoot weight and fresh root weight) of these plants were measured and recorded. The rhizosphere nematodes were isolated and counted as described previously [17, 50]. The symptoms of infected roots were photographed. Five biological replicates were performed.
Construction of plant RNAi vectors and production of transgenic plants
A 382-bp fragment from the Rs-cb-1 ORF was amplified using primers RNAi-F and RNAi-R (Supplementary Table S1) with restriction enzyme sites. The digested PCR fragment was inserted at inverted repeats into the XhoI/NcoI and XbaI/BamHI sites of the binary vector pFGC5941 to form a plant RNAi vector. The constructed vector pFGC5941-Rscb2 contains a CaMV35S promoter, a sense and antisense fragment of Rs-cb-1 cDNA, a CHSA intron and an OCS terminator, and the inverted repeats of the target gene is separated by CHSA intron. A similar vector pFGC5941-egfp was constructed as a control using primers eGFP-F and eGFP-R (Supplementary Table S1). The plasmids pFGC5941-Rscb2, pFGC5941-egfp and an empty pFGC5941 vector were transformed into Agrobacterium tumefaciens strain EHA105 by the frozen-thaw method [51], respectively, and then introduced into tobacco plants by co-cultivation of Agrobacterium with tobacco leaf discs [52]. The plant transformation was performed as described previously [53]. The well-rooted plantlets were transplanted into soil and grown in greenhouse to obtain seeds.
Molecular confirmation of transgenic plants
Genomic DNA was isolated from kanamycin-tolerant T0 generation Rs-cb-1 transgenic tobacco leaves using the CTAB method [54], and checked by PCR and sequencing using primers RNAi-F/RNAi-R and CHSA-F/OCS-R, respectively (Supplementary Table S1). The DNA from T0 generation egfp transgenic plants, empty transformation vector plants and wild-type tobacco plants were used as the controls, and the egfp transgenic tobacco plants were checked using primers egfp-F/egfp-R and CHSA-F/OCS-R (Supplementary Table S1).
For Southern blot analysis, primers RNAi-F1 and RNAi-R1 (Supplementary Table S1) were designed to amplify the DIG-labeled probe. About 15 µg genomic DNA of Rs-cb-1 transgenic plants were digested with NdeI. The digested DNA was separated on a 0.8% agarose gel, and then transferred to a Hybond-N+ membrane (Amershan) [27]. Hybridization and detection processes were performed using the Dig High Primer DNA Labeling and Detection Starter Kit I (Roche). The equal amounts of DNA from egfp transgenic plants and empty transformation vector tobacco plants were used as the controls.
Total RNAs of T0 generation PCR- and Southern-positive Rs-cb-1 transgenic plants were extracted and checked by RT-PCR using primers RNAi-F and RNAi-R (Supplementary Table S1). Positive transgenic tobacco plants were grown in greenhouse. After 2 months, twenty tobacco seeds (T1 generation) collected from the single plant were germinated in pots. Genomic DNA of these Rs-cb-1 transgenic plants were extracted and examined by PCR using primers RNAi-F and RNAi-R (Supplementary Table S1) for genetic stability. The positive T1 generation plants were grown in greenhouse to obtain T2 generation homozygous transgenic plants for further experiments.
Resistance analysis of T2 generation Rs-cb-1 transgenic tobacco plants to R. similis
The selected seedlings of Rs-cb-1 transgenic plants were same in height (about 20 cm) and growth condition, and each seedling was inoculated with 2,000 mixed stage nematodes and cultivated in greenhouse. The egfp transgenic plants, empty transformation vector plants and wild-type tobacco plants were used as the controls. 75 d after inoculation, the resistance of transgenic plants were examined according to the method as described in "Pathogenicity test of nematodes" in this study. Nematodes within the roots were stained with acid fuchsin as described previously [55], and observed under a microscope. Five biological replicates were performed.
Rs-cb-1 expression of R. similis in T2 generation transgenic plants
One hundred mixed stage nematodes isolated from the roots of T2 generation Rs-cb-1 transgenic tobacco plants were washed with DEPC water and used for RNA extraction. qPCR was used to assess the expression levels of Rs-cb-1 in R. similis as described above. The nematodes isolated from the roots of T2 generation egfp transgenic plants, empty transformation vector plants and wild-type tobacco plants were used as the controls.
Persistence and inheritance of the RNAi effect
The mixed stage nematodes soaked in Rs-cb-1 dsRNA for 48 h or isolated from the roots of T2 generation Rs-cb-1 transgenic tobacco plants were washed with sterile water and transferred into 15-mL centrifuge tubes. These nematodes were maintained in water for 1, 3, 5, 7, 9, 11, 13, and 15 d, respectively. Then 100 females were removed for qPCR analysis as described above. The remaining nematodes were maintained in water at 25ºC and the water was changed daily [20]. Meanwhile, untreated nematodes were used as a control.
About 100 female nematodes (P0 generation) soaked in Rs-cb-1 dsRNA for 48 h or isolated from T2 generation Rs-cb-1 transgenic tobacco roots were sterilized and used to inoculate carrot callus. After cultured for 20 d at 25°C, the F1 generation juveniles and immature females were isolated from carrot callus and used for the following experiments: (I) Total RNAs were extracted from 100 females treated differently (F1), and qPCR was used to assess the expression levels of Rs-cb-1 in R. similis as described above. (II) A total of 30 females (F1) were inoculated onto carrot callus, and the reproduction was evaluated after 30 d; (III) Each seedling of wild-type tobacco plants was inoculated with 400 mixed stage nematodes (F1) and cultivated in greenhouse. After 45 d, three growth parameters were measured and recorded. The rhizosphere nematodes were isolated and counted. Plants inoculated with untreated nematodes were used as the control. Five biological replicates were performed; (IV) About 100 females (F1) were cultured on carrot callus to obtain F2 generation nematodes. Total RNAs were extracted from these nematodes(F2), and qPCR was used to assess Rs-cb-1 expression as described above.
Data analysis
Statistical analysis was performed using SAS 9.2 (SAS Institute, Cary, NC, USA). All data in this study were subjected to analysis of one-way variance (ANOVA) and tested for differences among treatments at 5% level using Duncan's Multiple Range Test (DMRT).
Results
Cloning, tissue localization and expression analysis of Rs-cb-1
The full-length cDNA sequence of Rs-cb-1 was amplified and confirmed by sequencing (Supplementary Fig. S1). The sequencing result was consistent with that in GenBank (GU360972), and the recombinant plasmid pMD18-Rscb including an 1071-bp intact ORF of Rs-cb-1 was extracted for later use.
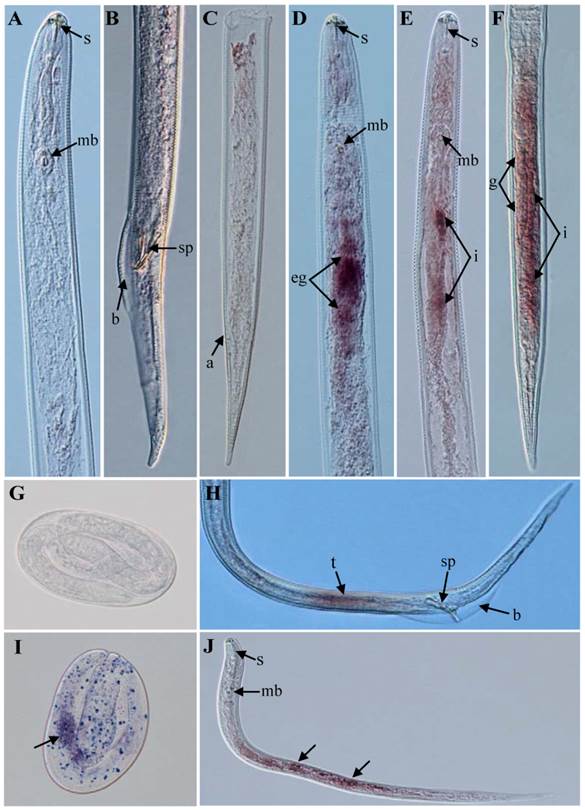
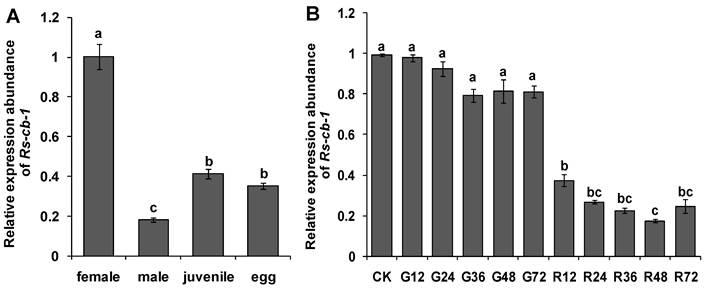
The results of in situ hybridization indicated that the Rs-cb-1 mRNA was present in the oesophageal glands, intestines and gonads of females (Fig. 1D, E, F), the testes of males (Fig. 1H), and in the juveniles and eggs of R. similis (Fig. 1I, J). No hybridization signal was detected in nematodes and eggs after they were incubated with the control DIG-labeled sense probe (Fig. 1A, B, C, G). The qPCR results showed that the Rs-cb-1 mRNA transcript was present in all developmental stages, and the highest expression levels were detected in females. The expression in juveniles, eggs and males accounted for 41.2%, 35.1% and 18.1% of the expression level in females, respectively. Rs-cb-1 expression in juveniles and eggs were significantly higher than that in males (p < 0.05), and no singnificant difference (p > 0.05) between juveniles and eggs (Fig. 2A).
Enzyme activity assay of recombinant protein Rs-CB-1
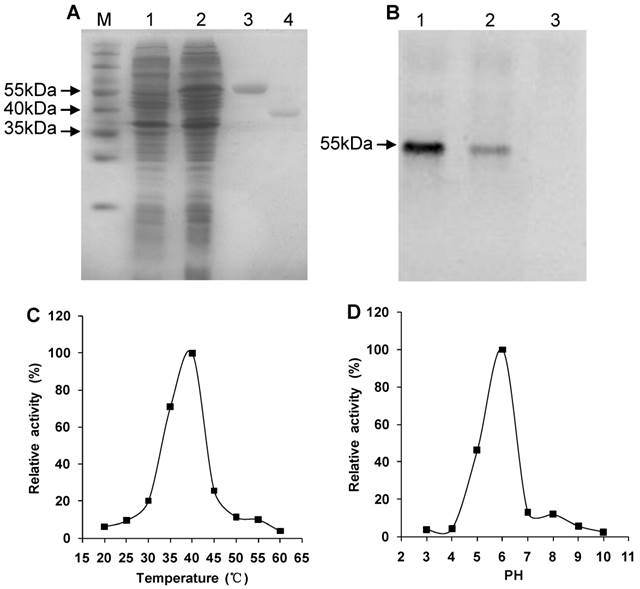
SDS-PAGE showed that the recombinant His-Rs-CB-1 was expressed in E. coli BL21(DE3) after IPTG induction. The recombinant fusion protein was purified and showed a single band of approximately 55 kDa. After treated with enterokinase, the purified protein showed a single band of approximately 40 kDa, which was consistent with the theoretical molecular mass of Rs-CB-1 (Fig. 3A).
Western blot analysis showed that a specific 55-kDa band was recognized by antibodies, which was consistent with the theoretical molecular mass of His-Rs-CB-1, but the non-induced recombinant protein failed to react with the antibodies, indicating that the recombinant protein His-Rs-CB-1 was expressed correctly in E. coli (Fig. 3B).
At pH 6.0, the enzyme activity of Rs-CB-1 was increased with the increase in temperature, and the optimal temperature was 40°C (100%). When temperature was above 40°C, the enzyme activity was reduced with the increase in temperature, and was the lowest at 60°C (4.01%) (Fig. 3C). At 40°C, the optimal pH of the enzyme activity was detected at pH 6.0 (100%), and the enzyme retained approximately 3.97%, 4.41%, 46.50%, 12.90%, 12.03%, 5.77% and 2.63% of its activity at pH 3.0, 4.0, 5.0, 7.0, 8.0, 9.0 and 10.0, respectively (Fig. 3D).
The effect of RNAi silence on Rs-cb-1 expression
The Rs-cb-1 dsRNA and non-endogenous control egfp dsRNA were synthesized (Supplementary Fig. S1). After treated with Rs-cb-1 dsRNA for different times, the expression level of Rs-cb-1 in R. similis was detected by qPCR. Compared with relative expression levels of Rs-cb-1 in the corresponding control nematodes treated with egfp dsRNA, the relative expression of Rs-cb-1 in nematodes treated with Rs-cb-1 dsRNA was decreased by 61.8%, 71.0%, 71.7%, 78.8% and 69.7% when treated for 12 h, 24 h, 36 h, 48 h and 72 h, respectively (Fig. 2B). Rs-cb-1 expression in nematodes treated with Rs-cb-1 dsRNA were significantly lower than those in untreated and egfp dsRNA treated nematodes (p < 0.05), and the silencing efficiency was enhanced with the increasing treatment time within a certain range of time. The silencing efficiency at 48 h was the highest. Rs-cb-1 expression showed no significant difference (p > 0.05) among untreated and egfp dsRNA treated nematodes.
Tissue localization of Rs-cb-1 mRNA in Radopholus similis using in situ hybridization. (A-C, G) No signal in nematodes and eggs were hybridized with digoxigenin-labeled sense Rs-cb-1 RNA probe. Rs-cb-1 was located in the esophageal glands (D) and intestines (E) of females. (F) Rs-cb-1 was located in the gonads and intestines of females. (H) Rs-cb-1 was located in the testes of males. (I, J) Rs-cb-1 was located in the eggs and juveniles, respectively. a, anus; b, bursa; g, gonads; i, intestine; mb, medium bulb; eg, esophageal glands; s, stylet; sp, spicules; t, testis.
Expression of the Rs-cb-1 in Radopholus similis. (A) Expression of the Rs-cb-1 in about 100 females, males, juveniles and eggs, respectively. (B) Expression of the Rs-cb-1 in R. similis treated with Rs-cb-1 double-stranded (ds) RNA. CK, expression of Rs-cb-1 in untreated nematodes; G12-G72, expression of Rs-cb-1 in control nematodes treated with non-endogenous egfp dsRNA for 12 h, 24 h, 36 h, 48 h and 72 h, respectively; R12-R72, expression of Rs-cb-1 in nematodes treated with Rs-cb-1 dsRNA for 12 h, 24 h, 36 h, 48 h and 72 h, respectively. Bars indicate standard errors of mean data (n=3) and different letters indicate significant differences (p<0.05) among different treatments.
Western blotting and enzyme activity assay of the recombinant protein. (A) Expression and purification of the recombinant protein. Protein was analyzed by SDS-PAGE and stained with Coomassie blue. M, protein marker (Thermo); Lane 1, protein extract from induced Escherichia coli cells harboring the empty pET-32a vector by 1.0 mM IPTG; Lane 2, protein extract from induced E.coli cells containing the recombinant pET-Rs-CB-1 by 1.0 mM IPTG; Lane 3, purified His-Rs-CB-1 recombinant Protein; Lane 4, purified Rs-CB-1 protein without the His-tag. (B) Western boltting analysis of the recombinant His-Rs-CB-1 protein. Lane 1-2, protein from induced E.coil with pET-Rs-CB-1 by IPTG; Lane 3, protein from non-induced E.coil with pET-Rs-CB-1 by IPTG. (C) Effect of temperature on Rs-CB-1 activity in base buffer ranging from 20 to 60 °C at a pH of 6.0. Relative activity is expressed as a percentage of maximum activity at 40 °C. (D) Effect of pH on Rs-CB-1 activity in base buffer ranging from 3 to 10 at a temperature of 40 °C. Relative activity is expressed as a percentage of maximum activity at pH 6.0. Enzyme activity was measured after incubation with Z-Arg-Arg-AMC for 1 h.
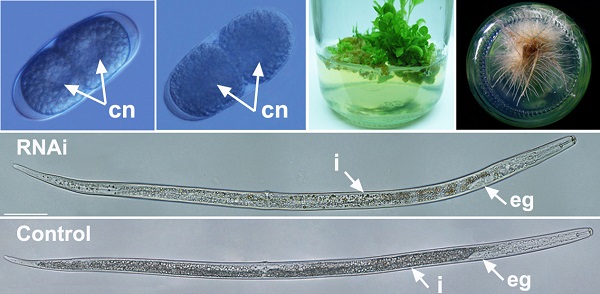
Silencing of Rs-cb-1 inhibited embryonic development and hatching of R. similis
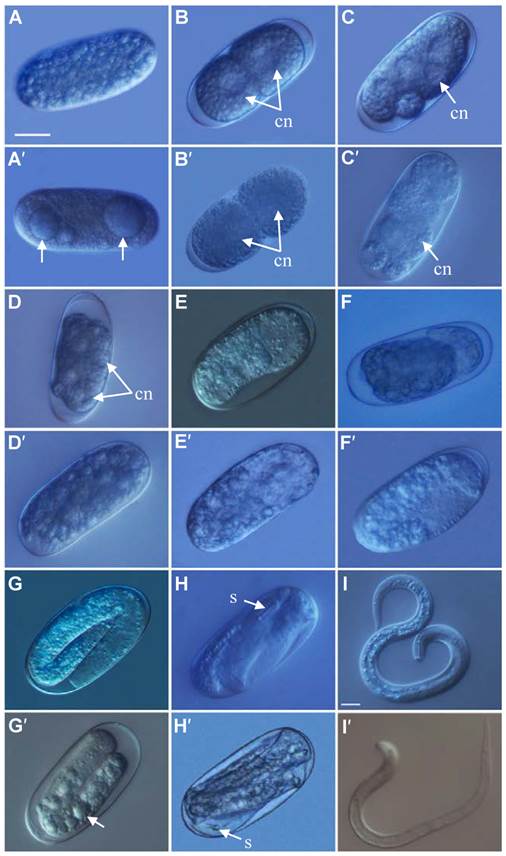
After treated with Rs-cb-1 dsRNA and egfp dsRNA (as a control) for 48 h, Rs-cb-1 dsRNA treated eggs showed abnormal embryogenesis compared with control eggs. These eggs contained diffused and foam-like substances in irregular sizes that were separated from egg shells. No obvious cell nuclei were observed from 2-cell to multi-cell stage (Fig. 4A′-D′). Although rearrangement also occurred in the eggs and there were inside and outside layers of the cells, cells were diffused at the blastula stage (Fig. 4E′). Some eggs were able to develop slowly from gastrula stage to the first stage juveniles(J1), and there were obvious foam-like substances in the eggs (Fig. 4F′, G′). The tissues in the body cavity of J2 without hatching were diffused as well. Although some eggs hatched, the development of J2 was inhibited, and even death was observed (Fig. 4H′, I′). In the control, these eggs were filled with small round particles in regular sizes from 1-cell to multi-cell stage. The cell nuclei were clearly observed from 2-cell to multi-cell stage (Fig. 4A-D). At blastula stage, the egg cells rearranged into the inside and outside layers, which formed the large and dark endoderm and the small and bright ectoderm, respectively (Fig. 4E). Eggs continued to develop during the gastrula stage and the linear larva could be observed clearly at J1, but the stylet was not obvious (Fig. 4F, G). During the development of J1 to J2, stylet developed normally and could be observed clearly (Fig. 4H). Then the juveniles continuously pressed the egg shell using their heads and stylets till hatching of J2 (Fig. 4I). The vast majority of eggs could normally develop and hatch.
The phenotypic observation of the embryonic development of Radopholus similis. The nematode eggs soakeded with non-endogenous egfp (A-I) and Rs-cb-1 dsRNA (A′-I′) for 48 h. (A, A′) 1-cell stage. (B, B′) 2-cell stage. (C, C′) 5-cell stage. (D, D′) Multi-cell stage. (E, E′) Blastula stage. (F, F′) Gastrula stage. (G, G′) J1 stage (the first-stage juvenile). (H, H′)J2 stage. (I, I′) Hatching. cn, cell nucleus; s, stylet. Scale bar= 20 μm.
After soaked in Rs-cb-1 dsRNA for 12 h, 24 h, 36 h, 48 h and 72 h, the average time of embryonic development of eggs were 17, 18, 19, 19.5 and 20 d, respectively, which were significantly (p < 0.05) delayed for 6-9 d compared with the control treatments (untreated or egfp dsRNA treated eggs). There was no significant difference between untreated and egfp dsRNA treated eggs (p > 0.05) (Supplementary Fig. S2A). Soaking treatment with Rs-cb-1 dsRNA also caused inhibition of embryonic development of most eggs. After treated with Rs-cb-1 dsRNA for 12 h, 24 h, 36 h, 48 h and 72 h, 90.9%, 95.8%, 95.1%, 100%, and 100% of the eggs were in 1-cell to multi-cell stage after 7 d of incubation, respectively. Only a small proportion of eggs were able to develop into blastula stage. After egfp dsRNA treatments for 12 h, 24 h, 36 h, 48 h and 72 h or without any treatment, only 10.4%, 10.8%, 9.5%, 10.6%, 12.6% and 9.8% of eggs were in 1-cell to multi-cell stage, respectively. Most of eggs developed into gastrula stage (Supplementary Fig. S2C). After 11 d of incubation, the percentage of eggs in 1-cell to multi-cell stage were 47.4%, 49.6%, 65.7%, 71.8%, and 74.6% upon Rs-cb-1 dsRNA treatment for 12 h, 24 h, 36 h, 48 h and 72 h, respectively. The vast majority of eggs was still in blastula stage and could not complete the embryonic development, and even death was observed. In contrast, most of the eggs with egfp dsRNA treatment for 12 h, 24 h, 36 h, 48 h and 72 h or without any treatment developed into J2 stage, and the percentages were 77.2%, 75.2%, 73.7%, 73.1%, 72.2% and 76.5%, respectively (Fig. S2D ).
The hatching rate of eggs treated with Rs-cb-1 dsRNA were significantly lower than the control treatments (untreated and egfp dsRNA treated eggs) (p < 0.05). The hatching rate of eggs treated with Rs-cb-1 dsRNA for 48 h and 72 h were 32% and 32.4% respectively, and which were significantly lower than that of eggs treated for 12 h-36 h (p < 0.05). Except for egfp dsRNA treatment for 72 h, there was no significant difference in hatching rates among the control treatments (p > 0.05) (Supplementary Fig. S2B). Therefore, Rs-cb-1 dsRNA soaking treatment significantly inhibited the embryonic development of R. similis. Most of the eggs could not complete the embryonic development and some even died, which led to low hatching rate.
Silencing of Rs-cb-1 inhibited post-embryonic development of R. similis
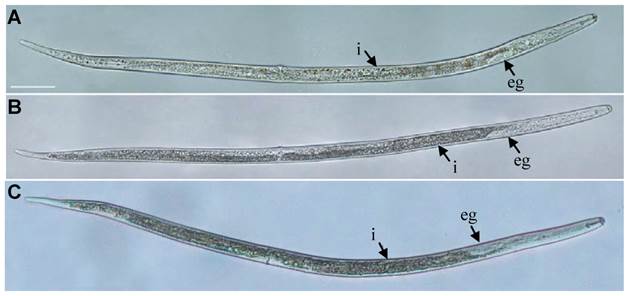
The post-embryonic development (From hatching to adults) of R. similis treated with Rs-cb-1 dsRNA for 48 h was not normal. The tissues in the body cavity were diffused and had fuzzy boundaries (Fig. 5A). In contrast, the post-embryonic development of egfp dsRNA treated for 48 h and untreated nematodes were normal. The tissues were distinguishable with clear boundaries, and almost filled the entire body cavity (Fig. 5B, C). Rs-cb-1 dsRNA soaking treatment inhibited the post-embryonic development of R. similis. After treated with Rs-cb-1 dsRNA for 12 h, 24 h, 36 h, 48 h and 72 h, the average times of post-embryonic development of R. similis were 14.5, 15.6, 16.3, 17.0 and 17.3 d, respectively, which were significantly (p < 0.05) delayed for 5-8 d compared with the control treatments (untreated and egfp dsRNA treated nematodes). The duration of post-embryonic development increased with the increase of Rs-cb-1 dsRNA soaking time, but no significant difference between the 48 h and 72 h (p >0.05). There was no significant difference among the control treatments (p > 0.05) (Supplementary Fig. S3).
Rs-cb-1 silencing significantly reduced the pathogenicity of R. similis
60 d after inoculation, the anthurium plants inoculated with R. similis treated with Rs-cb-1 dsRNA showed much better above-ground and root growth compared with control treatments (egfp dsRNA treated), 48 h and 72 h treatments grew normally without obvious disease symptoms (Supplementary Fig. S4A). In the control treatment groups, the growth of plants were weaker than that of healthy plants, and root rot was observed and there was no significant difference among them (Supplementary Fig. S4B). We also inoculated plants with nematodes that had been treated with M9 buffer and sterile water, or untreated nematodes as controls. The results of these treatments were similar to those of the egfp dsRNA treatments (Supplementary Fig. S4C, D).
Influence of Rs-cb-1 silence on the post-embryonic development of Radopholus similis. (A-C) The phenotypic observation of the post-embryonic development of R. similis. (A) Treated with Rs-cb-1 dsRNA for 48 h. (B) Untreated nematode. (C) Treated with non-endogenous egfp dsRNA for 48 h. i, intestine; eg, esophageal glands. Scale bar= 50 μm.
Anthurium plants inoculated with nematodes treated with Rs-cb-1 dsRNA had significantly greater height, fresh shoot weight, and fresh root weight than those inoculated with untreated or egfp dsRNA treated nematodes (control treatments), but was still significantly lower than those of healthy plants (p < 0.05). There was a significant difference between the soaking time of 48 h-72 h and 12 h (p < 0.05), but no significant difference among the control treatments (p > 0.05) (Supplementary Fig. S5A, B, C). The nematode number in the rhizosphere of anthurium inoculated with Rs-cb-1 dsRNA treated nematodes was significantly lower than that of the control treatments (p < 0.05). A large number of nematodes were isolated from the rhizosphere of control treatments, but no significant difference among them (p > 0.05) (Supplementary Fig. S5D). Taken together, the pathogenicity of R. similis was reduced significantly after treatment with target-specific Rs-cb-1 dsRNA, and that of Rs-cb-1 dsRNA for 48 h was the lowest among them, whereas the pathogenicity of R. similis was not impacted upon treatment with non-specific target egfp dsRNA.
Generation and molecular analysis of transgenic tobacco plants
The plant RNAi vector pFGC5941-Rscb2 expressing hairpin Rs-cb-1 dsRNA was constructed (Fig. 6A). The transgenic tobacco plants were generated from transformed callus tissues (Fig. 6C-F). A total of 69 independent kanamycin-resistant transgenic tobacco plants were obtained (39 Rs-cb-1 transgenic plants, 19 egfp transgenic plants and 11 empty vector transgenic plants). The growth morphology of these transgenic plants was indistinguishable from that of the wild-type tobacco plants (Supplementary Fig. S6).
Independently derived T0 generation transgenic lines were analyzed by PCR, and two 382-bp and 752-bp fragments were amplified from Rs-cb-1 transgenic tobacco plants, two 315-bp and 685-bp fragments were amplified from egfp transgenic plants, and a 407-bp fragment was amplified from empty transformation vector plants, while wild-type plants did not show specific amplification (Fig. 6G, H). These results indicated that the hairpin dsRNAs were inserted into tobacco genomic DNA successfully. Southern blot analysis showed that PCR-positive Rs-cb-1 transgenic plants had one to multiple copies of the target coding sequences (Plant No. 2 had a single copy insertion, plant No. 3 had two copies of the insertion, and plant No. 6 had multiple copies of the insertion). Conversely, genomic DNA from egfp transgenic plants and empty transformation vector plants failed to show any hybridization signals with the probe (Fig. 6I).
Generation and molecular analysis of transgenic tobacco plants. (A) Construction of the plant RNAi vector expressing hairpin Rs-cb-1 dsRNA in transgenic plants. (B) Independent transgenic lines were analyzed by RT-PCR using primers of RNAi-F/RNAi-R (lanes 2, 3 and 6, RNA from the 2, 3 and 6 Rs-cb-1 transgenic lines). (C-F) Development of transgenic tobacco plants expressing Rs-cb-1 dsRNA. (C) Putative transformed calli growing on selection medium. (D) Transgenic plantlets germinated from transformed tobacco calli. (E) Transgenic plants growing on rooting medium. (F) Hardened transgenic plant growing in an illumination incubator. (G, H) Independent derived transgenic lines were analyzed by PCR amplifications of the genomic DNA using primers of RNAi-F/RNAi-R, CHSA-F/OCS-R or eGFP-F/eGFP-R (lanes 1-10, Independent Rs-cb-1 transgenic lines; lanes e1-e5, Independent egfp transgenic lines). (I) Southern blot analysis of NdeI-digested genomic DNA from leaves of the T0 generation transgenic plants (lanes 1-2, DNA from empty transformation vector plant and e5 egfp transgenic plant as a control; lanes 3-5, DNA from the 2, 3 and 6 Rs-cb-1 transgenic lines). M, DNA marker (DL2000); B, Blank control without template; V, Empty transformation vector plant; W, Wild type plant (negative control); P, Positive plasmid control.
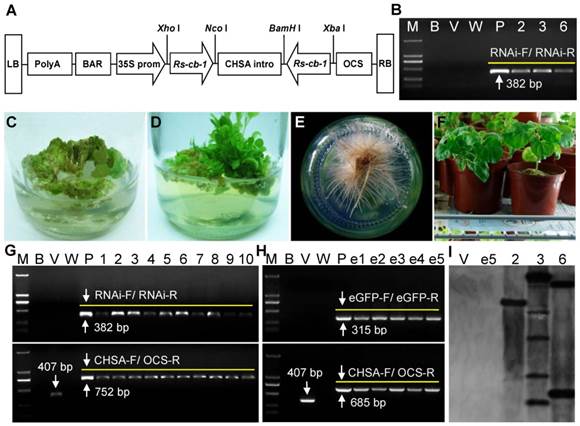
RT-PCR analysis with RNAs isolated from Rs-cb-1 transgenic plants No. 2, 3 and 6 showed a 382-bp fragment corresponding to the sequence of Rs-cb-1; while wild-type and empty transformation vector plants did not show specific amplification (Fig. 6B). These results indicated that the integrated Rs-cb-1 dsRNA was successfully expressed. Genetic stability analysis of T1 generation Rs-cb-1 transgenic plants (No. 2, 3 and 6) showed that the ratio of positive and negative Rs-cb-1 transgenic plants was 3:1. A 382-bp fragment corresponding to the sequence of Rs-cb-1 was amplified from positive plants and no specific bands was amplified from negative plants. These results indicated that the integrated Rs-cb-1 could be inherited steadily in tobacco genomic DNA (Supplementary Fig. S7). Kerschen et al. [56] and Peng et al. [57] have shown that the effectiveness of transgene-induced RNAi in single-copy lines is higher than in other lines. Therefore, the single copy Rs-cb-1 transgenic plant (No. 2) was chosen for further analyses.
Resistance to R. similis is improved in T2 generation transgenic tobacco plants expressing Rs-cb-1 dsRNA
Homozygous T2 generation plants expressing Rs-cb-1 dsRNA (No. 2), together with control plants (egfp transgenic plants, empty transformation vector plants and wild-type tobacco plants), were inoculated with nematodes to evaluate whether the resistance to R. similis was improved. 75 d after inoculations, there was no significant difference (p > 0.05) in height and fresh shoot weight between Rs-cb-1 transgenic plants and uninoculated wild-type tobacco plants (CK) (Supplementary Fig. S8A, B); however, the fresh root weight of Rs-cb-1 transgenic plants (6.96 g) was significantly lower (p < 0.05) than that of CK (8.18 g) (Supplementary Fig. S8C). The three growth parameters of Rs-cb-1 transgenic plants were significantly greater than those of control plants inoculated with R. similis (p < 0.05). In addition, the degree of root damage was much less and there was no obvious root rot in Rs-cb-1 transgenic plants compared with the control plants (Supplementary Fig. S8F). The number of nematodes in the rhizosphere and per gram root of Rs-cb-1 transgenic plants were 960 and 43, respectively, which were significantly lower than that of control plants (p < 0.05) (Supplementary Fig. S8D, E). There was no significant difference among the inoculated control plants in these pathogenicity measures (p > 0.05) (Supplementary Fig. S8), and the roots were severely damaged (Supplementary Fig. S8F). The results clearly confirmed that resistance to R. similis was significantly improved in transgenic tobacco plants expressing Rs-cb-1 dsRNA.
Rs-cb-1 mRNA levels in R. similis are significantly suppressed by feeding with T2 generation transgenic tobacco plants expressing Rs-cb-1 dsRNA
The Rs-cb-1 mRNA levels in R. similis fed with Rs-cb-1 transgenic plants were significantly reduced by 77.7%, 78.9% and 79.9% compared with that in egfp transgenic plants, empty transformation vector plants and wild-type tobacco plants, respectively. There was no significant difference among three control groups (p > 0.05) (Supplementary Fig. S8G). Therefore, we concluded that suppression of Rs-cb-1 expression in R. similis by feeding with T2 generation transgenic plants expressing Rs-cb-1 dsRNA caused reduction in pathogenicity.
Persistence and inheritance of Rs-cb-1 silencing induced by in vitro RNAi and in planta RNAi
After recovered in water for 1-7 d, the expression levels of Rs-cb-1 in R. similis treated with Rs-cb-1 dsRNA for 48 h were reduced by 58.3-72.3% compared with the untreated control (CK) group, and there was a significant difference (p < 0.05) between treated and untreated control groups, but no significant difference among the different recovery times (p > 0.05). However, recovery time of 9 d, 11 d and 13 d induced the increase of Rs-cb-1 expression levels to 297%, 409% and 158% of the CK group, respectively, and there was a significant difference (p < 0.05). Recovery time of 15 d induced Rs-cb-1 expression levels to the normal level (Supplementary Fig. S9A). The expression levels of Rs-cb-1 in R. similis which were isolated from Rs-cb-1 transgenic plants and recovered in water for 1-15 d were reduced by 70.7-78.8%, and all the treatments were significantly lower than the CK group (p < 0.05), but no significant difference among the different recovery times (p > 0.05) (Supplementary Fig. S9B).
The qPCR results revealed that compared with the CK group nematodes, the expression levels of Rs-cb-1 in F1 generation nematodes derived from Rs-cb-1 dsRNA soaking treatment showed no significant difference (p > 0.05), but that in F1 nematodes derived from Rs-cb-1 transgenic plants was reduced by 65.4%, and there was a significant difference (p < 0.05) (Supplementary Fig. S9C). After being cultured on carrot callus for 30 d, the F1 nematodes derived from Rs-cb-1 transgenic plants showed significantly lower (p < 0.05) reproduction than that from Rs-cb-1 dsRNA soaking treatment and the CK group, but no significant difference (p > 0.05) between the latter two groups (Supplementary Fig. S9D). After being inoculated with F1 nematodes derived from Rs-cb-1 transgenic plants for 45 d, the plant height, fresh shoot weight and fresh root weight of tobacco plants were significantly greater (p < 0.05) than those of the Rs-cb-1 dsRNA soaking treatment and the CK group, and the number of rhizosphere nematodes was significantly lower (p < 0.05) than that of Rs-cb-1 dsRNA soaking treatment and the CK group, but no significant difference (p > 0.05) between the latter two groups (Supplementary Fig. S9E-H). The qPCR results revealed that the expression level of Rs-cb-1 in F2 nematodes derived from Rs-cb-1 transgenic plants was reduced by 36.4% compared with the CK group, which was significantly lower than that of Rs-cb-1 dsRNA soaking treatment and the CK group (p < 0.05), but no significant difference between the latter two groups (p > 0.05) (Supplementary Fig. S9I).
These results indicated that in vitro RNAi-induced Rs-cb-1 silencing only lasted for a limited time and could be recovered, F1 nematodes had normal expression levels of Rs-cb-1, and had normal reproduction and pathogenicity. The expression level of Rs-cb-1 in R. similis isolated from transgenic plants was significantly inhibited after recovery in water, and the expressions of Rs-cb-1 in F1 and F2 nematodes were still inhibited, and reproduction capability and pathogenicity of F1 nematodes were reduced significantly. Overall, in planta RNAi-induced Rs-cb-1 silencing could be maintained in the absence of dsRNA for at least two generations before being lost, which was not the case for the effects induced by in vitro RNAi.
Discussion
Cathepsin B plays vital roles in various parasites [35, 37, 38]. However, systematic characterization of cb has not yet been reported in plant parasitism nematodes. To determine the roles of cb in R. similis, we investigated the expression and localization of Rs-cb-1, and confirmed the gene function by In vitro RNAi. We also confirmed that transgenic tobacco plants expressing Rs-cb-1 dsRNA not only significantly suppressed target gene expression, but also greatly reduced the pathogenicity of R. similis. This study demonstrates the feasibility of in planta RNAi in controlling this pathogen by targeting the cathepsin B gene.
Previous studies have shown that cb is mainly expressed in the intestines, reproductive system and eggs of some animal parasites and free living nematodes [37, 58-60]. Furthermore, cb is also expressed at different development stages of A. cantonensis and Trichinella spiralis. The expression level of cb is significantly higher in the fourth-stage larvae and the fifth-stage larvae than that in the third-stage larvae and adult worms in A. cantonensis [61], and is the highest during the adult stage in T. spiralis [62]. In this study, we confirmed that the expression and location of Rs-cb-1 were conformed to the characteristics of cathepsin B and the biological functions of R. similis. The secretions of esophageal glands produced by plant parasitic nematodes are thought to play key roles throughout parasitism [63, 64]. The expression of Rs-cb-1 in esophageal glands and intestines may help R. similis to neutralize plant defense, complete the infection, and digest the host cells quickly to provide nutrients necessary for metabolism and other physiological functions. The Rs-cb-1 mRNA is also present in the reproductive system and eggs, which indicates that CB may play important roles in reproduction, cell differentiation, embryo formation and development in R. similis. The CB protein is necessary for the females of R. similis not only to destroy the host defense response and complete the infection, but also to obtain the necessary nutrients for metabolism and complete the reproduction. Therefore, the expression level of Rs-cb-1 is the highest in females. The expression levels of Rs-cb-1 in juveniles are higher than that in eggs, but no significant difference between them, likely because the CB protein plays an equally important role in destroying host defense response, establishing parasitic relationship, cell differentiation and embryo formation in R. similis. Males of R. similis with a degraded stylet and esophagus are non-parasitical, and the number of males is much less than females. Their activities may just include mating with females, but the nematodes can reproduce through parthenogenesis, the male is not necessary for the reproduction of R. similis. Therefore, the expression level of Rs-cb-1 is the lowest in males.
Some investigations have demonstrated that transcripts having <100% complementarity with an siRNA can be targeted for knockdown by the RNAi pathway, this phenomenon is referred to as off-target effects (OTEs) [65-68]. In our preliminary experiment, the expression level of Cathepsin S (GenBank No: EU659125, Rs-cs), Cathepsin L (EU659124, Rs-cl), fatty acid and retinol binding protein (JN968974, Rs-far-1), Calreticulin (JX067552, Rs-crt) and beta-1,4-endoglucanase-1 (EU414839, Rs-eng-1) in R. similis were also detected by qPCR after the nematodes were treated with Rs-cb-1 dsRNA for 48 h. The results showed that Rs-cb-1 expression in nematodes treated with Rs-cb-1 dsRNA was significantly lower (p < 0.05) than that in the control nematodes, but there were no significant differences (p > 0.05) in the expression levels of other five genes between Rs-cb-1 dsRNA treated nematodes and the control nematodes (result not shown). These results indicated that OTEs did not appear in the process of RNAi research in this study.
In this study, we demonstrated that Rs-cb-1 dsRNA soaking treatment significantly inhibited the embryonic development of R. similis. Most of the treated eggs could not complete embryonic development and the duration of nematodes from J2 to mature female was significantly prolonged. These results indicated that Rs-cb-1 was essential in embryonic and post-embryonic development of R. similis. The roles of CB in the development of plant parasitic nematodes have not yet been reported, although CB proteinases are known to be involved in the development, invasion, immunoregulation and pathogenesis in some insects and animal parasites[61, 69-73]. In this study, we confirmed that Rs-cb-1 silencing significantly reduced the pathogenicity of R. similis to anthurium, indicating that Rs-cb-1 was involved in pathogenesis. This is the first report of the key roles of cb gene in the development, hatching and pathogenesis in plant parasitic nematodes.
To further confirm the function of Rs-cb-1 and to explore the possibility of using this target gene to control R. similis, we obtained transgenic tobacco plants expressing Rs-cb-1 dsRNA. Using in planta RNAi, we confirmed that the expression levels of Rs-cb-1 in nematodes were significantly suppressed, and that the reproduction and pathogenicity of nematodes were significantly reduced in transgenic plants. These results further underpinned the roles of Rs-cb-1 in reproduction and pathogenesis of R. similis. Previous studies have shown that the resistance to M. incognita and M. javanica is significantly improved in transgenic tobacco plants expressing specific dsRNA [21,30,34]. Parasitism gene 16D10 of M. incognita has been analyzed using in planta RNAi, and a wide resistance against four major root-knot nematode species is obtained by expressing 16D10 dsRNA in transgenic Arabidopsis [32]. Steeves et al. [31] have demonstrated that MSP (major sperm protein) transgenic soybean plants significantly reduces the reproductive potential of H. glycines. Klink et al. [33] have also shown that the number of H. glycines females in transgenic soybean roots reduces by 81-93%. All these works were using in planta RNAi to study gene functions of sedentary plant parasitic nematodes. Here we first report the use of in planta RNAi in studying the functions of development and pathogenesis genes of migratory plant parasitic nematodes and its application in controlling plant parasitic nematodes.
In planta RNAi is an effective technology in studying gene functions and genetic engineering of plant resistance to pathogens. The first demonstration of the inheritability of in planta RNAi-induced gene silencing in sedentary plant parasitic nematodes was made by Steeves et al. [31]. In their study, cyst nematodes infecting transgenic soybean plants expressing specific dsRNA targeting a major sperm protein gene from H. glycines showed a 68% reduction in eggs g-1 root tissue. Interestingly, the progeny nematodes hatched from the eggs showed a 75% reduction in eggs g-1 root tissue when allowed to infect wild-type susceptible soybean plants. Our study showed that the expression levels of Rs-cb-1 in F1 R. similis isolated from Rs-cb-1 transgenic plants was still inhibited, its reproduction and pathogenicity were reduced significantly, and the RNAi effect was maintained in F2 nematode. This is the first demonstration of the inheritability of in planta RNAi-induced gene silencing in migratory plant parasitic nematodes. In summary, we demonstrate that in planta RNAi is suitable for studying different types of plant parasitic nematodes, which overcomes the limitations of in vitro RNAi. In planta RNAi is a promising tool for studying gene function and genetic engineering of plant resistance to plant parasitic nematodes, and has wide application prospects.
Supplementary Material
Table S1, Figures S1-S9.
Acknowledgements
We thank Dr. Guozhang Kang (The National Engineering Research Centre for Wheat, Henan Agricultural University, China) for contributing the binary vector pFGC5941, Prof. Huaping Li (South China Agricultural University) for contributing the seeds of Nicotiana benthamiana and Agrobacterium tumefaciens strain EHA105. This work was funded by National Natural Science Foundation of China (No.31071665 and No. 31371920).
Competing Interests
The authors have declared that no competing interest exists.
References
1. O'Bannon JH. Worldwide dissemination of Radopholus similis and its importance in crop production. J Nematol. 1977;9:16-25
2. Luc M, Sikora RA, Bridge J. Plant parasitic nematodes in subtropical and tropical agriculture. Wallingford, UK: CABI Publisher. 2005
3. Richardson PN, Grewal PS. Nematode pests of glasshouse crops and mushrooms. Evans K, Trudgill DL, Webster JM, (Eds). Plant parasitic nematodes in temperate agriculture. Wallingford: CABI International. 1993:515-6
4. Ssango F, Speijer PR, Coyne DL. et al. Path analysis: a novel approach to determine the contribution of nematode damage to East African Highland banana (Musa spp, AAA) yield loss under two crop management practices in Uganda. Field Crop Res. 2004;90:177-87
5. Sarah JL. Banana nematodes and their control in African. Nematropica. 1989;19:199-216
6. Musabyimana T, Saxena RC. Efficacy of neem seed derivatives against nematodes affecting banana. Phytoparasitica. 1999;27:43-9
7. Tripathi L. Genetic engineering for improvement of Musa production in Africa. Afr J Biotechnol. 2003;2:503-8
8. Cotton J, Van Riel H. Quarantine, problems and solutions. Evans K, Trudgill DL, Webster JM, (Eds). Plant parasitic nematodes in temperate agriculture. Wallingford: CAB International. 1993:593-607
9. Smith IM, Charles LMF. Distribution Maps of Quarantine Pests for Europe. Wallingford, UK: CABI Publisher. 1998
10. Jeger MJ, Waller JM, Johanson A. et al. Monitoring in banana pest management. Crop Prot. 1996;15:391-7
11. Urwin PE, Lilley CJ, Atkinson HJ. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microbe In. 2002;15:747-52
12. Rosso MN, Dubrana MP, Cimbolini N. et al. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol Plant Microbe In. 2005;18:615-20
13. Bakhetia M, Charlton W, Atkinson HJ. et al. RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol Plant Microbe In. 2005;18:1099-106
14. Shingles J, Lilley CJ, Atkinson HJ. et al. Meloidogyne incognita: molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp Parasitol. 2007;115:114-20
15. Chen Q, Rehman S, Smant G. et al. Functional analysis of pathogenicity proteins of the patato cyst nematode Globodera rostochiensis using RNAi. Mol Plant Microbe In. 2005;18:621-5
16. Li Y, Xie H, Xu CL. et al. RNAi effect of Cathepsin B gene on reproduction of Radopholus similis. Scientia Agricultura Sinica. 2010;43:1608-16
17. Zhang C, Xie H, Xu CL. et al. Differential expression of Rs-eng-1b in two populations of Radopholus similis (Tylenchida: Pratylenchidae) and its relationship to pathogenicity. Eur J Plant Pathol. 2012;133:899-910
18. Li XD, Zhuo K, Luo M. et al. Molecular cloning and characterization of a calreticulin cDNA from the pinewood nematode Bursaphelenchus xylophilus. Exp Parasitol. 2011;128:121-6
19. Tan JACH, Jones MGK, Fosu-Nyarko J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp Parasitol. 2013;133:166-78
20. Bakhetia M, Urwin PE, Atkinson HJ. qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol Plant Microbe In. 2007;20:306-12
21. Michaeli S, Kenigsbuch D, Livneh O. et al. Plants resistant to cytoplasm-feeding parasites. WIPO Patent No. 2005019408. 2005
22. Ma J, Song Y, Wu B. et al. Production of transgenic rice new germplasm with strong resistance against two isolations of Rice stripe virus by RNA interference. Transgenic Res. 2011;20:1367-77
23. Tyagi H, Rajasubramaniam S, Rajam MV. et al. RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants. Transgenic Res. 2008;17:897-904
24. Baum JA, Bogaert T, Clinton W. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322-6
25. Pitino M, Coleman AD, Maffei ME. et al. Silencing of aphid genes by dsRNA feeding from plants. PLoS One. 2011;6:e25709
26. Mao JJ, Zeng FR. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res. 2014;23:145-52
27. Zha W, Peng X, Chen R. et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One. 2011;6:e20504
28. Mao YB, Cai WJ, Wang JW. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307-13
29. Zhu JQ, Liu SM, Ma Y. et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PloS One. 2012;7:e38572
30. Yadav BC, Veluthambi K, Subramaniam K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasit. 2006;148:219-22
31. Steeves RM, Todd TC, Essig JS. et al. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct Plant Biol. 2006;33:991-9
32. Huang GZ, Allen R, Davis EL. et al. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A. 2006;103:14302-6
33. Klink VP, Kim KH, Martins V. et al. A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta. 2009;230:53-71
34. Hu L, Cui R, Sun L. et al. Molecular and biochemical characterization of the β-1,4-endoglucanase gene Mj-eng-3 in the root-knot nematode Meloidogyne javanica. Exp Parasitol. 2013;135:15-23
35. Malagón D, Díaz-López M, Benítez R. et al. Cathepsin B- and L-like cysteine protease activities during the in vitro development of Hysterothylacium aduncum (Nematoda: Anisakidae), a worldwide fish parasite. Parasitol Int. 2010;59:89-92
36. Urwin PE, Lilley CJ, McPherson MJ. et al. Characterization of two cDNAs encoding cysteine proteinases from the soybean cyst nematode Heterodera glycines. Parasitology. 1997;114:605-13
37. Meemon K, Grams R, Vichasri-Grams S. et al. Molecular cloning and analysis of stage and tissue-specific expression of cathepsin B encoding genes from Fasciola gigantica. Mol Biochem Parasitol. 2004;136:1-10
38. Sripa J, Pinlaor P, Brindley PJ. et al. RNA interference targeting cathepsin B of the carcinogenic liver fluke, Opisthorchis viverrini. Parasitol Int. 2011;60:283-8
39. Li DL, Li Y, Xie H. et al. Cloning and analysis of cathepsin B gene of Radopholus similis. J ZheJiang Univ. 2013;39:26-33
40. Reise RW, Huettel RN, Sayre RM. Carrot callus tissue for culture of endoparasitic nematodes. J Nematol. 1987;19:387-9
41. Fallas GA, Sarah JL. Effect of storage temperature on the in vitro reproduction of Rahodpholus similis. Nematropica. 1994;24:175-7
42. Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum. 1962;15:473-97
43. Chaudhury A, Qu R. Somatic embryogenesis and plant regeneration of turf-type bermudagrass: Effect of 6-benzyladenine in callus induction medium. Plant Cell Tiss Org. 2000;60:113-20
44. De Boer JM, Yan Y, Smant G. et al. In-situ Hybridization to Messenger RNA in Heterodera glycines. J Nematol. 1998;30:309-12
45. Cheng X, Xiang Y, Xie H. et al. Molecular Characterization and Functions of Fatty Acid and Retinoid Binding Protein Gene (Ab-far-1) in Aphelenchoides besseyi. PLoS One. 2013;8:e66011
46. Jacob J, Vanholme B, Haegeman A. et al. Four transthyretin-like genes of the migratory plant-parasitic nematode Radopholus similis: Members of an extensive nematode-specific family. Gene. 2007;402:9-19
47. Biswas S, Sharma S, Saroha A. et al. Identification of novel autoantigen in the synovial fluid of rheumatoid arthritis patients using an immunoproteomics approach. PLoS One. 2013;8:e56246
48. Mendieta L, Pico A, Tarrago T. et al. Novel peptidyl aryl vinyl sulfones as highly potent and selective inhibitors of cathepsins L and B. ChemMedChem. 2010;5:1556-67
49. Hannon GJ. RNAi: A guide to gene silencing. New York, USA: Cold spring harbor laboratory publisher. 2003
50. Kaplan DT, Vanderspool MC, Opperman CH. Sequence tag site and host range assays demonstrate that Radopholus similis and R. citrophilus are not reproductively isolated. J Nematol. 1997;29:421-9
51. Chen H, Nelson RS, Sherwood JL. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques. 1994;16:664-70
52. Sunilkumar G, Vijayachandra K, Veluthambi K. Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci. 1999;141:51-8
53. Horsch RB, Fry JE, Hoffmann NL. et al. A simple and general method for transferring genes into plants. Science. 1985;227:1229-31
54. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic acids Res. 1980;8:4321-6
55. Bybd DW, Kirkpatrick T, Barker KR. An improved technique for clearing and staining plant tissues for detection of nematodes. J Nematol. 1983;15:142-3
56. Kerschen A, Napoli CA, Jorgensen RA. et al. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004;566:223-8
57. Peng H, Zhai Y, Zhang Q. et al. Establishment and functional analysis of high efficiency RNA interference system in rice. Scientia Agricultura Sinica. 2006;39:1729-35
58. Pratt D, Armes LG, Hageman R. et al. Cloning and sequence comparisons of four distinct cysteine proteases expressed by Haemonchus contortus adult worms. Mol Biochem Parasitol. 1992;51:209-18
59. Ray C, McKerrow JH. Gut-specific and developmental expression of a Caenorhabditis elegans cysteine protease gene. Mol Biochem Parasitol. 1992;51:239-49
60. Cheng M, Yang X, Li Z. et al. Cloning and characterization of a novel cathepsin B-like cysteine proteinase from Angiostrongylus cantonensis. Parasitol Res. 2012;110:2413-22
61. Han YP, Li ZY, Li BC. et al. Molecular cloning and characterization of a cathepsin B from Angiostrongylus cantonensis. Parasitol Res. 2011;109:369-78
62. Zhan JH, Yao JP, Liu W. et al. Analysis of a novel cathepsin B circulating antigen and its response to drug treatment in Trichinella-infected mice. Parasitol Res. 2013;112:3213-22
63. Hussey RS, Davis EL, Baum TJ. Secrets in secretions: genes that control nematode parasitism of plants. Braz J Plant Physiol. 2002;14:183-94
64. Vanholme B, De Meutter J, Tytgat T. et al. Secretions of plant-parasitic nematodes: a molecular update. Gene. 2004;332:13-27
65. Jackson AL, Bartz SR, Schelter J. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635-7
66. Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599-606
67. Birmingham A, Anderson EM, Reynolds A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199-204
68. Fedorov Y, Anderson EM, Birmingham A. et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188-96
69. Yamamoto Y, Takahashi SY. Cysteine proteinase from Bombyx eggs: role in programmed degradation of yolk proteins during embryogenesis. Comp Biochem Phys B. 1993;106:35-45
70. Cho WL, Tsao SM, Hays AR. et al. Mosquito cathepsin B-like protease involved in embryonic degradation of vitellin is produced as a latent extraovarian precursor. J Biol Chem. 1999;274:13311-21
71. Rabossi A, Stoka V, Puizdar V. et al. Novel aspartyl proteinase associated to fat body histolysis during Ceratitis capitata early metamorphosis. Arch Insect Biochem. 2004;57:51-67
72. Lustigman S, McKerrow JH, Shah K. et al. Cloning of a cysteine protease required for the molting of Onchocerca volvulus third stage larvae. J Biol Chem. 1996;271:30181-9
73. Law RHP, Smooker PM, Irving JA. et al. Cloning and expression of the major secreted cathepsin B-like protein from juvenile Fasciola hepatica and analysis of immunogenicity following liver fluke infection. Infect Immun. 2003;71:6921-32
Author contact
![]() Corresponding author: Prof. Xie Hui, Tele: 86-20-38297286, Fax: 86-20-38297286, E-mail: xiehuiedu.cn
Corresponding author: Prof. Xie Hui, Tele: 86-20-38297286, Fax: 86-20-38297286, E-mail: xiehuiedu.cn

 Global reach, higher impact
Global reach, higher impact