10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(11):1281-1295. doi:10.7150/ijbs.12528 This issue Cite
Research Paper
Structure-Based Analysis of the Ligand-Binding Mechanism for DhelOBP21, a C-minus Odorant Binding Protein, from Dastarcus helophoroides (Fairmaire; Coleoptera: Bothrideridae)
1. Hubei Insect Resources Utilization and Sustainable Pest Management Key Laboratory, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, P. R. China
2. College of Informatics, Huazhong Agricultural University, Wuhan 430070, P. R. China
3. Department of Horticulture, Beijing Vocational College of Agriculture, Beijing 102442, PR China
Received 2015-4-28; Accepted 2015-8-17; Published 2015-9-15
Abstract
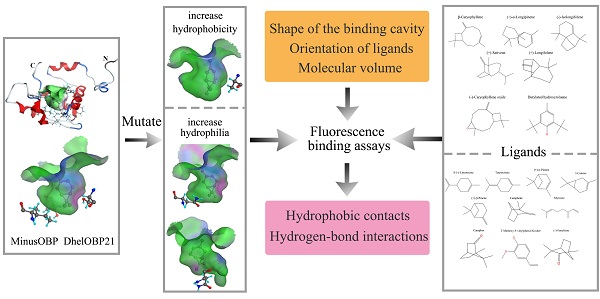
Odorant binding proteins (OBPs) transport hydrophobic odor molecules across the sensillar lymph to trigger a neuronal response. Herein, the Minus-C OBP (DhelOBP21) was characterized from Dastarcus helophoroides, the most important natural parasitic enemy insect that targets Monochamus alternatus. Homology modeling and molecular docking were conducted on the interaction between DhelOBP21 and 17 volatile molecules (including volatiles from pine bark, the larva of M. alternatus, and the faeces of the larva). The predicted three-dimensional structure showed only two disulfide bridges and a hydrophobic binding cavity with a short C-terminus. Ligand-binding experiments using N-phenylnaphthylamine (1-NPN) as a fluorescent probe showed that DhelOBP21 exhibited better binding affinities against those ligands with a molecular volume between 100 and 125 ų compared with ligands with a molecular volume between 160 and 185 ų. Molecules that are too big or too small are not conducive for binding. We mutated the amino acid residues of the binding cavity to increase either hydrophobicity or hydrophilia. Ligand-binding experiments and cyber molecular docking assays indicated that hydrophobic interactions are more significant than hydrogen-bonding interactions. Although hydrogen-bond interactions could be predicted for some binding complexes, the hydrophobic interactions had more influence on binding following hydrophobic changes that affected the cavity. The orientation of ligands affects binding by influencing hydrophobic interactions. The binding process is controlled by multiple factors. This study provides a basis to explore the ligand-binding mechanisms of Minus-C OBP.
Keywords: Odorant-binding proteins, fluorescence competitive binding assays, molecular docking, site-directed mutagenesis, molecular volume, hydrophobic interactions, Dastarcus helophoroides.
Introduction
A sensitive chemosensory system is essential for insects to accomplish many important physiological behaviors, such as the detection of food, predators, hosts, oviposition sites, and mates [1, 2]. Many proteins have been found to be involved in odorant reception in the antennae, the major chemosensory organ. In these proteins, odorant-binding proteins (OBPs) can bind, transport and deliver exogenous odorant molecules across the lymph to odorant receptors (ORs) on the dendrite membrane of sensory neurons. OBPs are a family of small (13-16 kDa) water-soluble proteins that are abundantly expressed in the sensillum lumen [3-6]. OBPs that bind and convey pheromones are known as pheromone-binding proteins (PBPs) [7]. Additionally, OBPs can convey signals from general odorants, so they are termed general odorant binding proteins (GOBPs) [8].
Many PBPs or GOBPs have been previously structurally characterized. The typical characteristics shared among many of these proteins, such as a six α-helix core, an internal cavity, and three disulfide bridges, have been used to define a “classical OBP” group [9]. Some non-classical OBPs have also been identified, including Minus-C (which contains only four conserved cysteines) [10] and Plus-C OBPs (which contain more than six conserved cysteines) [11]. Several 3D structures of classical OBPs have been determined to date, such as the classic Anopheles gambiae AgamOBP1 [12, 13], AgamOBP4 [14], Aedes aegypti AaegOBP1 [15], and Culex quinquefasciatus CquiOBP1 [16]. Their observed structures are diverse, with a wide variety of cavities shapes and positions, solvent accessibility, and amino acids sequences [17]. These factors could influence ligand binding. Bombyx mori BmorGOBP2, Drosophila melanogaster Lush, and Antheraea polyphemus ApolPBP1 are monomeric when they bind ligands and have a single entrance that opens up into a larger binding pocket [18-20]. AgamOBP1 contains one continuous hydrophobic channel that runs through the dimer interface; notably, DEET (N,N-diethyl-m-toluamide) binds more favorably to the dimeric form of AgamOBP1 [13]. Similar ligand-binding pockets have been reported for CquiOBP1 bound to MOP ((5R,6S)-6-acetoxy-5-hexadecanolide) and AaegOBP1 bound to PEG (polyethylene glycol) [15, 16]. Interactions between OBPs and ligands mainly involved hydrophobic (CquiOBP1 with MOP) and hydrogen (BmorGOBP2 with bombykol and LUSH with alcohol) bonding [16, 18, 19, 21].
Ligand release also varied in a structure-dependent manner. BmorPBP has a C-terminal tail that is long enough to form a helix that could fit into the binding pocket and occupy the bombykol-binding site at a pH lower than 5.5, whereas at a pH higher than 5.5 the same C-terminus region can form an elongated stretch outside of the binding site, making it accessible to pheromone molecules [22]. A similar ligand-release mechanism has been observed for a PBP from the giant silk moth Antheraea polyphemus [20, 23]. However, the AgamOBP1 C-terminus lacks a C-terminal extension that is long enough to form a helix that could displace a ligand. However, it could form a wall in the binding pocket. The reduction in pH causes the C-terminal loop to open, exposing the binding tunnel to solvent [13]. Additionally, ligand release could involve sensory neuron membrane proteins (SNMPs) with a local pH, as the Drosophila SNMP can directly capture pheromone molecules on the surface of OSN (olfactory sensory neuron) cilia. It is possible that these ligands could be retrieved from odorant-binding proteins in the extracellular milieu, which could facilitate the transfer of such ligands to odor receptor complexes [24].
However, crystal structures of Minus-C OBPs have been rarely reported. Apis mellifera AmelOBP14 was the first 3D structure of a member of the C-minus class OBPs, which is characterized by only two disulfide bonds [25]. The cavity walls were principally formed of hydrophobic residues, and three hydrophilic residues were also part of the cavity wall. Its C-terminus segment formed an external seventh helix at the interface between the protein exterior and internal cavity, which completely enclosed the cavity. Unlike the proteins mentioned above, no biological dimers were observed in the crystal. Hydrophobic contacts and hydrogen bonds have played an important role in binding to 1-NPN, eugenol, and citralva ligands [25]. In addition to the structural study, two Minus-C OBPs—HarmOBP17 and HarmOBP18—with a medium sized C-terminus display higher binding at pH 5.0 than at pH 7.4, and the mutant OBPs (with the C-terminus eliminated) exhibited much lower binding affinities compared with the intact OBPs [26]. These data suggest that many diverse mechanisms are likely to exist for ligand binding and release for different OBPs.
Using molecular docking methods on homology models of OBPs, we could predict the presence of cavities in ligand binding proteins and indicate the amino acid residues that formed the cavities based on the reported 3D protein structures [6]. By mutating residues in the binding site, the physicochemical properties can be changed, and the ligand binding affinity can be altered. Many key residues in the binding site of the protein have shown to function in different OBPs [27-31]
Dastarcus helophoroides (Fairmaire) (Coleoptera: Bothrideridae) is a prominent biological control agent against Monochamus alternatus, which is a quarantine pest that transmits the pine wood nematode, Bursaphelenchus xylophulus (Steiner et Buhrer) Nickle (Nematoda:Aphelenchoididae) [32, 33]. Dastarcus helophoroides parasitizes mature larvae, pupae and teneral adults of M. alternatus. Adult females of D. helophoroides lay eggs in clusters on gallery walls that can be bored into tree stems by longhorned beetle host larvae, and then the hatchlings seek out and paralyze the host [34]. Chemical cues are involved in host location by D. helophoroides [32], and 23 OBPs have been identified from the antenna transcriptome of D. helophoroides [35]. Herein, we cloned Minus-C DhelOBP21 (GenBank accession no. KF984184) and expressed and purified the protein in vitro. We selected 17 volatiles (including volatiles from pine bark, the larva of M. alternatus, and the faeces of the larva) for tests of binding affinities with DhelOBP21. With support of dimensional structure modeling and molecular docking, the characteristics of the binding cavity and ligands were analyzed, including the molecular volume, hydrophobic interactions, hydrogen bonding patterns, and orientation of ligands. With reference to the previous crystal structure studies, especially that of AmelOBP14, we designed mutant proteins with an aim to change the cavity hydrophobicity. A comparison of the binding affinities with the volatiles isolated from the forest showed that the molecular volume and hydrophobic interactions play major roles in the binding mechanisms, while hydrogen bond interactions were less important. The binding process can be controlled by multiple factors, so the previous site-specific mutagenesis studies may be not comprehensive [28, 31]. These results provide helpful data about our structure-based understanding of the ligand-binding mechanism of Minus-C OBPs.
Materials and Methods
Insects
D. helophoroides adults were provided by the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry. Laboratory colonies of D. helophoroides originated from parasitized larvae and pupae of M. alternatus.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from 50 antennae and heads from male and female D. helophoroides (1:1) using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol, and the RNA concentration was determined using an ultraviolet spectrophotometer (Eppendorf BioPhotometer Plus, Hamburg, Germany) before reverse transcription. Then, cDNA was prepared from total RNA by reverse transcription using an RT-PCR system (Promega, Beijing, China) according to the manufacturer's protocol.
Polymerase Chain Reaction (PCR)
Aliquots of 1 mL crude cDNA were amplified in a Bio-Rad Gene CyclerTM thermocycler with gene-specific primers as follows: DhelOBP21F: 5-CGGGATCCAATGAAATCTTTCGCTT-3 and DhelOBP21R: 5-CCCTCGAGTTAAGCTAAGCTAATGTG-3. The restriction enzymes sites for BamHI and XhoI in the forward and reverse primers are underlined, respectively. The PCR conditions consisted of an initial 3 min step at 94°C followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 1 min and a final 10 min step at 72°C.
Cloning and Sequencing
PCR products were ligated into a pMD-18T vector using a 1:5 (plasmid:insert) molar ratio and were incubated 0.5 h at 4°C. Ligation products were transformed into DH5α Escherichia coli competent cells and grown on LB (lysogeny broth) solid medium with 10 mg/mL ampicillin. Positive colonies were selected and grown in LB liquid medium with ampicillin and then sequenced.
Cloning of DhelOBP21 in Expression Vectors
The pMD-18T plasmid containing positive clones and pET-17b plasmid were digested with BamHI and XhoI restriction enzymes for 3 h at 37°C. Digested products were separated on an agarose gel. Target fragments were purified and ligated into a digested pET-17b plasmid; recombinant plasmids were transformed into DH5α E. coli competent cells and grown on LB solid medium with 10 mL ampicillin (50 mg/mL). Selected colonies were grown in LB liquid medium with ampicillin and then were sequenced. Then, BL21 (DE3) pLysS E. coli competent cells were transformed with correct recombinant plasmids. A single clone was identified and cultivated overnight, in LB liquid medium that included ampicillin, in a shaker at 200 rpm and 37°C. The resulting plasmids were sequenced and were found to encode the mature proteins.
Recombinant Protein Expression and Purification
A single positive clone was used to inoculate 5 mL Luria-Bertani broth containing 50 µg/mL ampicillin overnight at 37°C. The culture was diluted to 1 L in fresh medium, and bacteria were cultured for 2-3 h at 37°C until the culture reached an optical density value of 0.6-0.8 at 600 nm. Protein expression was induced by the addition of IPTG (isopropyl-beta D-thiogalactopyranoside) to a final concentration of 0.5 mM. Cells were grown for another 4 h at 37°C, then were harvested by centrifugation (10,000 rpm, 10 min) and sonicated. After centrifugation (10,000 rpm, 20 min), the expressed OBPs, present as inclusion bodies, were solubilized in 10 mL 8 M urea and 1 mM DTT in 50 mM Tris buffer, pH 7.4, then were treated with 250 mL 100 mM cystine in 0.5 M NaOH and 5 mL of 5 mM cysteine in 100 mM Tris buffer pH 8.0. The solution was dialyzed five times against Tris buffer, pH 7.4. Protein purification was accomplished in 30 mM Tris buffer, pH 7.4, by combinations of chromatographic steps on anion-exchange resins, such as DE-52, QFF or Mono-Q (GE-Healthcare, Beijing, China). All purification steps were monitored by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis). Finally, proteins were dialyzed three times against Tris buffer, pH 7.4 for fluorescence binding assays. The effects of pH on binding were evaluated at pH 5.0 and in Tris buffer, pH 5.0.
Molecular Modeling and Ligand Docking
Delta-BLAST was performed with the DhelOBP21 sequence against the Protein Data Bank (PDB; http://www.rcsb.org) on the NCBI server (NCBI; http://blast.ncbi.nlm.nih.gov/Blast.cgi). As an improved algorithm of Blast, Delta-BLAST [36] searches a database of pre-constructed PSSMs (position-specific scoring matrix) before searching against a protein-sequence database, to yield a better homologous protein profile. The profile returned by BLAST was subjected to ClustalW2 analysis to obtain a Multiple Sequence Alignment and phylogram.
Then, homology modeling was performed with Molecular Operating Environment (MOE, version 2012.10) [37] as follows. The homologous protein profile was first realigned according sequence similarity and secondary structure. Next, DhelOBP21 was aligned to the alignment obtained above. The best protein was selected, judged by homology, evolution, sequence similarity, the number of Cys (cysteine), the phylogram and several techniques for structure determination, as the template to build a 3D model of DhelOBP21. In the modeling procedure, we set the 'maximum number of mainchain models' to 50 and the 'sidechain samples at temperature 300 K' to 5. In the model refinement section, 'intermediates' and the 'final model' were set to “fine”, and AMBER99 was selected as the force field, while other parameters were set to default.
After building the model, it was subjected to sufficient stereochemical refinement and energy minimization according the electrostatic solvation energy, which was calculated using the Generalized Born/Volume Integral methodology. Second, a further refinement was performed based on Protonate 3D in MOE. Notably, in molecular docking, reproduction of the complex crystal structure is both a necessary prerequisite and a challenging issue. Protonate 3D is a powerful tool that can assign ionization states and position hydrogens in a macromolecular structure based on their 3D coordinates (typically from a crystal structure). In our experience in drug design, it is a state-of-the-art way to optimize a protein, especially a protein pocket. Generally, if a protein is processed with Protonate 3D before docking, such a problem can be solved smoothly.
Subsequently, the stereochemical structure of the model was checked in MOE, involving dihedral angles ψ against φ, bond lengths, bond angles, dihedrals, rotamers, and atom clashes. The best DehlOBP21 model with the lowest electrostatic solvation energy and optimal geometric properties was selected for follow-up molecular docking analysis.
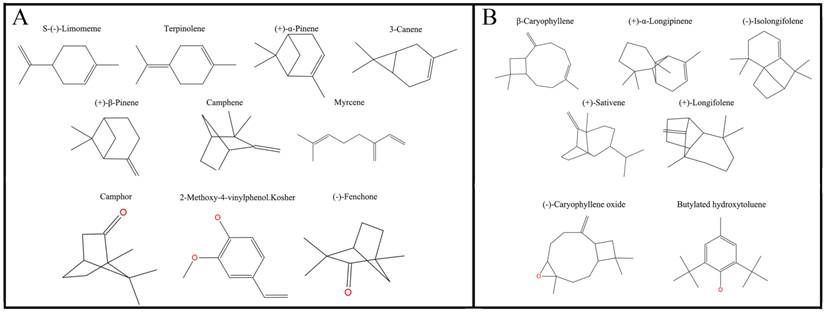
After the tertiary structure was obtained, 17 [S-(-)-Limomeme, Terpinolene, (+)-α-pinene, 3-canene, (+)-β-pinene, myrcene, camphene, β-caryophyllene, (+)-α-longipinene, (-)-isolongifolene, (+)-sativene, (+)-longifolene, (-)-caryophyllene oxide, butylated hydroxytoluene, camphor, 2-methoxy-4-vinylphenol Kosher, (+)-fenchone] volatiles and 1-NPN were docked into the pocket of DhelOBP21, which was established as above employing the Surflex-Dock suite embedded in Sybyl-X (version 2.0) [38].
In the Docking Suite, Surflex-Dock was selected as the docking mode and a Multi-Channel Surface was set as the protomol generation mode. Subsequently, 'bloat' was set to 2Å, the 'additional starting conformations per molecule' were increased to 10, and 'density of search' was set to 6. Additionally, 'consider ring flexibility' was also checked. All of these parameters are set to improve docking accuracy. Finally, 'minimum RMSD (root-mean-square deviation) between final poses' was set to 0.5 Å to explore additional docking poses and to achieve higher accuracy. Other parameters were set to default.
The binding patterns of ligands to the OBP receptor were explored, and key amino acid residues were identified. The mechanism of interaction between signal molecules and DhelOBP21 was analyzed. On the basis of the docking results, the key residues in the cavity of the DhelOBP21 were mutated to yield several mutants of DhelOBP21 and re-docked with the volatiles mentioned above to study the differences of binding mode between the wild-type and mutant types.
Calculation of the Properties of Ligands and OBP
More than 30 molecular descriptors, such as molecular weight, volume, solvent accessible surface area (SASA), polarSASA, number of hydrogen-bond acceptors and donors (HBA, HBD), and logD, were calculated using Pipeline Pilot (version 8.5) after ligands were standardized with hydrogen and a charge was added. The volume of the protein binding site was also calculated with MolCAD in Sybyl-X.
Preparation of Site-Directed Mutants
The three mutants of DhelOBP21, S67A (mutation of amino acid, serine to alanine at position 67), I84N (mutation of amino acid, isoleucine to asparaginate at position 84) and T119N (mutation of amino acid, threonine to asparaginate at position 119), were developed using the Fast Mutagenesis System (TransGen, Beijing, China). The DhelOBP21-pET17b recombinant plasmids were used as template. The primers were designed manually and mutation sites are underlined as listed below.
Ser67 to Ala67: TCG to GCG for DhelOBP21-S67A mutant.
Forward primer: 5-AACACCTCTTCTGCTTCGCGAAGAAGGCTGG-3.
Reverse primer: 5-CGAAGCAGAAGAGGTGTTCCATCAACTTGGG-3.
Ile84 to Asn84: ATC to AAC for DhelOBP21-I84N mutant.
Forward primer: 5-ATATCCAAACTGACGTGAACAAGGCCAAGCT-3.
Reverse primer: 5-TTCACGTCAGTTTGGATATCACCGGCTTCGT-3.
Thr119 to Asn119: ACC to AAC for DhelOBP21-T119N mutant.
Forward primer: 5-CAGAAGACCGCATTCGATAACATCAAATGTTA-3.
Reverse primer: 5-TTATCGAATGCGGTCTTCTGTGGGGTTGCCT-3.
The PCR conditions were 95°C for 3 min for initial denaturation, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s and final extension at 72°C for 2 min. Selected mutants were sequenced. The same expression vector and competent cells were used as for DhelOBP21-WT (the wild-type protein). The expression and purification mutant genes and proteins were performed as described for wild-type protein.
Fluorescence binding assays
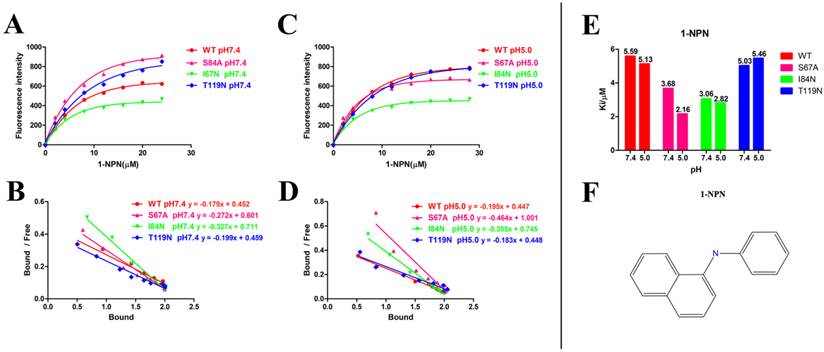
Fluorescence binding assays were performed to determine the binding affinity of DhelOBP21 for various volatiles using 1-NPN as a fluorescent probe. The 1-NPN and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). All ligand stock solutions were prepared in spectrophotometric-grade methanol. To measure the binding constants for 1-NPN, a 2 mM solution of protein in 30 mM Tris-HCl, pH 7.4 was added with aliquots of 1 mM 1-NPN, pH 7.4 at room temperature. The 1-NPN/protein mixture was excited using an excitation wavelength of 337 nm, and the fluorescence intensity was recorded between 360 and 600 nm using a RF-5301PC fluorescence spectrophotometer (Shimadzu, Kyoto, Japan) with a 1 cm light path and a quartz cuvette. The saturation curves of the binding of 1-NPN by DhelOBP21 were constructed, and the dissociation constant, Kd, of the binding reaction was calculated by performing a Scatchard analysis of data using Prism 5 software (GraphPad, La Jolla, CA, USA). The binding analyses were performed based on the assumption that the protein had 100% activity and that the stoichiometry of binding was 1:1 at saturation. The affinity of various volatile ligands was measured in competitive binding assays. Aliquots of competitor ligand were added to a sample containing 2 μM DhelOBP21 and a standard concentration of 1-NPN. A reduction in the relative fluorescence intensity indicated that the competitor displaced 1-NPN from the binding site of DhelOBP21. Binding data were collected during three independent measurements. The Ki, which represents Kd of the competitor, was determined based on the IC50 value (the concentration of competitor that halved the initial fluorescence level). The Ki was calculated according to the following equation: Ki = [IC50]/1 + [1-NPN]/K1-NPN, where [1-NPN] is the free concentration of 1-NPN and K1-NPN is the Kd of the 1-NPN-DhelOBP binding reaction determined in the Scatchard analysis [39]. To make the analysis more easily visualized, we calculated 1/Ki*1000, for which a bigger value indicates a stronger binding capacity.
Results
Cloning and Sequence Analysis of DhelOBP21
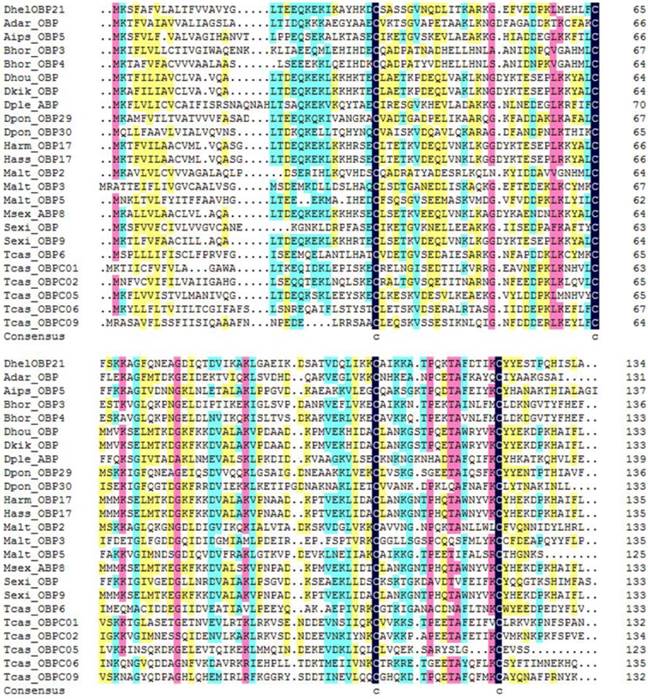
DhelOBP21 was obtained from the antennal transcriptome. Using gene-specific primers, a full-length cDNA encoding DhelOBP21 was cloned. Sequence analysis showed that the full-length Open Reading Frame (ORF) consists of 405 nucleotides that encode 135 amino acid residues, with a predicted MW of 14.87 kD. For DhelOBP21, SignalP predicted a peptide with 18 amino acid residues, and ExPASy (http://web.expasy.org/compute_pi/) predicted an isoelectric point of 8.60. We conducted an alignment of the amino acid sequence of DhelOBP21 with the corresponding OBPs from other species (Fig. 1). DhelOBP21 has less than six Cys residues and belongs to the Minus-C insect OBP subfamily with a common pattern X33-C1-X30-C2-X39-C3-X16-C4-X12 (X denotes any amino acid).
Molecular Modeling and Molecular Docking
Using Delta-blast, nine homologous proteins were obtained with a sequence similarity cutoff of 20%, including Tenebrio molitor THP12 (1C3Y), Anopheles gambiae AgamOBP1 (2ERB), Culex quinquefasciatus odorant-binding protein (2L2C), Aedes aegypti AaegOBP1 (3K1E), Culex quinquefasciatus CquiOBP1 (3OGN), Anopheles gambiae AgamOBP07 (3R1O), Apis mellifera AmelOBP5 (3R72), Apis mellifera AmelOBP14 (3RZS), and Apis mellifera AmelOBP14 (3S0G).
Alignment of DhelOBP21 (KF984184) from Dastarcus helophoroides as well as other insects. Anopheles darlingi (AdarOBP, ETN64377.1) \ Agrotis ipsilon (AipsOBP5, AGR39568.1) \ Batocera horsfieldi (BhorOBP3, ADD82416.1) \ Batocera horsfieldi (BhorOBP4, ADD82417.1) \ Dendrolimus houi (DhouOBP,AII00983.1) \ Dendrolimus kikuchii (DkikOBP, AII01006.1) \ Danaus plexippus (DpleOBP, EHJ66992.1) \ Dendroctonus ponderosae (DponOBP29, AGI05182.1) \ Dendroctonus ponderosae (DponOBP30, AGI05176.1) \ Helicoverpa armigera (HarmOBP17, AFI57166.1) \ Helicoverpa assulta (HassOBP17, AGC92792.1) \ Monochamus alternatus (MaltOBP2, AHA39267.1) \ Monochamus alternatus (MaltOBP3, AHA39268.1) \ Monochamus alternatus (MaltOBP5, AHA39270.1) \ Manduca sexta (MsexABP8, AAL60426.1) \ Spodoptera exigua (SexiOBP, ADY17884.1) \ Spodoptera exigua (SexiOBP9, AGH70105.1) \ Tribolium castaneum (TcasOBP6, EFA04594.1) \ Tribolium castaneum (TcasOBP01, EFA07544.1)\ Tribolium castaneum (TcasOBP02, EFA07545.1) \ Tribolium castaneum (TcasOBP05, EFA07543.1) \ Tribolium castaneum (TcasOBP06, EFA07548.1) \ Tribolium castaneum (TcasOBP09, EFA07429.1).
1C3Y (Tenebrio molitor, THP12) was selected as the template for homology modeling for the following reasons. (1) Classifying according to the 'Structural Classification of Proteins', 1C3Y belongs to the family of insect PBP/OBPs. (2) We noted that Spinelli et al. proposed that 1C3Y is not a proper template for AmelOBP14 [25] and argued that the best characterized OBPs are those of Lepidoptera, such as B. mori, and of Dipterans. By contrast, in our study, the evolution and taxonomy analyzed both D. helophoroides and T. molitor, which are members of Coleoptera, and A. mellifera of Hymenoptera. Hence THP12 (1C3Y) might represent a more reasonable template for DhelOBP21 than other proteins from Hymenoptera, Lepidoptera, or Dipterans, such as AmelOBP14, BmorGOBP2, or AgamOBP1, respectively (3) The sequence similarity between 1C3Y and DhelOBP21 is 42% (E-value: 2e-21), which is the significantly higher than the others (27% or lower) and this difference may be significant. (4) According to the traditional classification of the OBP and considering the highly conserved Cys residues, both the DhelOBP21 and 1C3Y have four Cys residues while the other eight proteins have five (3RZS) or more Cys (2ERB, 2L2C, 3K1E, 3OGN, 3R72 with six, 3S0G with seven and 3R1O with eight) residues. (5) DhelOBP21 was perfectly aligned to 1C3Y without gaps, including as many as 46 conserved residues. There were a few gaps when DhelOBP21 was aligned to other proteins (Fig. S1), such as AmelOBP14 (3RZS).
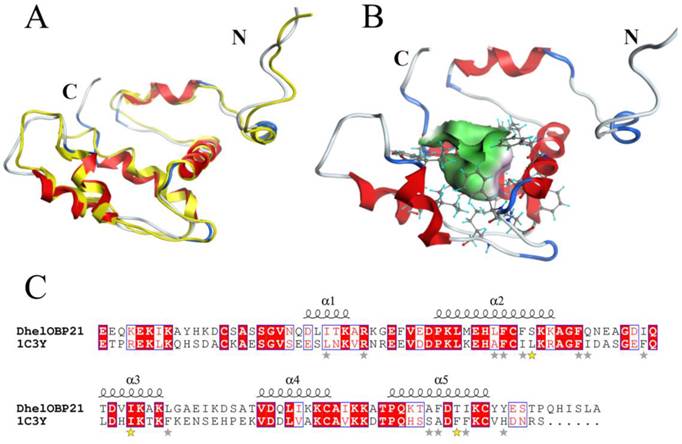
Based on the stereo-chemical optimization and energy minimization performed with MOE, a first-rank model with the minimum energy among the 250 intermediate models was inspected using the stereo-chemical quality evaluation tool in MOE—Protein Geometry. A pairwise RMSD of alpha C between the template 1C3Y and DhelOBP21 was 1.856 Å (Fig. 2A). As shown in Fig. S2, 97.2% of residues (107 residues) were located in the allowed region and 2.8% (three residues, Glu, Gln, and Ala) were located near the marginal region in a Ramachandran map, along with other stereochemical indices (including bond lengths, bond angles, and dihedrals), indicating that its overall stereochemical quality was generally reliable and acceptable.
To gain insights into the recognition mechanism between volatiles and DhelOBP21, 17 reported volatiles that could be recognized by D. helophoroides and 1-NPN were docked into the pocket of DhelOBP21 and mutant protein models. The docking was performed with Surflex-Dock in Sybyl-X. Based on the predicted docked structures, most ligands bound to the protein overlap at the center of the pocket. For 2-methoxy-4-vinylphenol Kosher and (+)-Fenchone, a hydrogen bond was formed between Ser67 and oxygen atoms in the two ligands. The oxygen atom acted as a hydrogen bond donor in the former ligand and as an acceptor in the later ligand. The other volatiles were not endowed with a polar atom-like oxygen or nitrogen to participate in hydrogen bond formation.
To alter the hydrophobicity of the binding cavity, Ser67, Ile84 and Thr119 were mutated to Ala, Asn and Asn, respectively. The first mutation aimed to increase hydrophobicity, while the other two mutations were designed increase the possible formation of polar interactions, such as hydrogen bonds.
Predicted structure of DhelOBP21
DhelOBP21 has four Cys residues and belongs to the Minus-C insect OBP class (Fig. 2B-C). A total of two disulfide bridges could be observed between Cys residues 34-65 and 105-122. After removal of the signal peptide (18 amino acid residues), the positions of those Cys residues were found to be similar to Cys residues 17-49 and 88-106 of AmelOBP14. However, a comparison with the core of six α-helices of AmelOBP14 and based on data from ESPript3.0 (http://espript.ibcp.fr/ESPript/ESPript/) and MOE2012, the binding cavity of DhelOBP21 is formed by five α-helices, with residues 24-30 in α1, 38-52 in α2, 62-70 in α3, 79-86 in α4, and 93-103 in α5. DhelOBP21 has a short C-terminus instead of an exposed C-terminal helix. The N-terminus is long, but did not form a helix. Additionally, DhelOBP21 have five helices unlike other insect OBPs, and consists of six α-helices. In the crystal structure of THP12, it can be observed to contain six helices, and the first α-helix in the N-terminus consists of only four amino acid residues (Fig. 2A). Because the conformation of THP12 in the crystal structure may be a conformation adopted after ligand release from the pocket, the first α-helix in the N-terminus could be longer than the current one during conformational transformation. The corresponding first α-helix in the N-terminus in the resulting model, DhelOBP21, is therefore too short to be recognized by artificial software, such as ESPript3.0 and MOE2012, which are not yet sufficiently sophisticated. It has been speculated that before the first α-helix in DhelOBP21, there should be an α-helix that was so short that it was unexpectedly interpreted as a random coil by the prediction software.
At the center of the protein core, the cavity walls are principally formed of hydrophobic residues: Ile45, Leu63, Phe64, Phe66, Phe72, Ile79, Ile84, Leu88, Ala116, Phe117, and Ile120. One basic residue is Arg49. Additionally, four hydrophilic residues are also part of the cavity wall: Ser67, Gly73, Thr119, and Tyr124. Only the oxygen atom of Ser67 points towards the cavity center, which forms the only polar surface of the cavity.
Ligand Characteristics and Fluorescence Binding Assays for DhelOBP21-WT
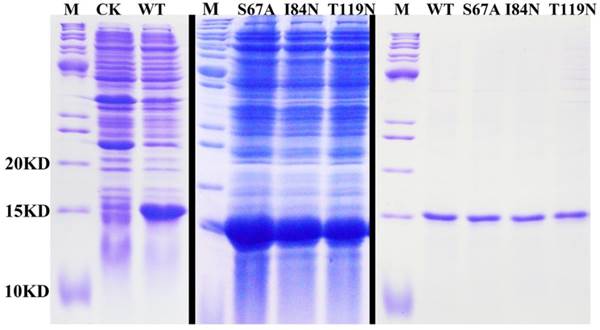
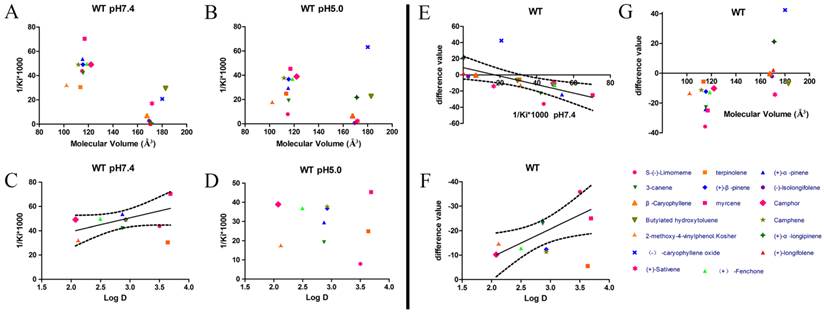
After protein purification, the identity and integrity of the recombinant proteins were confirmed using SDS-PAGE (Fig. 3). Those proteins were used in the following fluorescence binding assays. The ligand-binding assays of 1-NPN of DhelOBP21 and the mutant are shown in Fig. 4. The binding affinities (indicated by 1/Ki*1000) of DhelOBP21 and the mutant are shown in Table 1. Some binding and structural studies have shown remarkable plasticity in the ligand-binding site of OBP [40]. We must first consider the molecular volumes of the ligands. Based on the molecular volume and hydrophobicity of these ligands, they were divided into two major groups (Fig. 5): (1) ligands with a molecular volume between 100 and 125 ų; and (2) ligands with a molecular volume between 160 and 185 ų. By plotting these values in different pH and molecular volumes (Fig. 6A-B), one could find that the ligands with a molecular volume between 100 and 125 ų had a stronger binding ability than the ligands with a molecular volume between 160 and 185 ų at pH 7.4. Hydrophobic contacts with the cavity wall residues have been reported for AmelOBP14 bound to citralva. Considering the high hydrophobicity of the cavity, ligands with a molecular volume between 100 and 125 ų were selected to plot against the LogD of these ligands, which denotes hydrophobicity of ligands at different pH values. We found that ligands that are more hydrophobic have a stronger binding affinity at pH 7.4 (Fig. 6C). However, when the pH reduced to 5.0, this trend was not obvious. Furthermore, this difference was obtained by subtracting the binding ability at pH 7.4 from the corresponding value at pH 5.0 to study the influence of pH. Similarly, we plotted these values (Fig. 6D). Compared with the binding ability at pH 7.4, most ligands have a lower binding ability at pH5.0, except for (+)-α-longipinene and (-)-caryophyllene oxide. We also found that pH had a stronger influence on those ligands that had a stronger binding ability at pH 7.4 (Fig. 6E-F).
Binding data (indicated by 1/Ki(uM)*1000) of the DhelOBP21 and its mutant with different ligands.
| Ligand | WT | S67A | I84N | T119N | ||||
|---|---|---|---|---|---|---|---|---|
| pH7.4 | pH5.0 | pH7.4 | pH5.0 | pH7.4 | pH5.0 | pH7.4 | pH5.0 | |
| S-(-)-Limomeme | 43.80 | 7.95 | 21.26 | 60.57 | 33.99 | 43.73 | 42.54 | 45.88 |
| Terpinolene | 30.54 | 24.93 | 19.58 | 52.63 | 46.78 | 17.96 | 23.95 | 45.92 |
| (+)-α-Pinene | 53.73 | 29.52 | 34.44 | 60.36 | 59.36 | 47.90 | 28.03 | 28.73 |
| 3-Canene | 42.10 | 19.26 | 16.04 | 43.37 | 14.99 | 21.33 | 5.99 | 40.60 |
| (+)-β-Pinene | 49.12 | 36.80 | 29.17 | 64.70 | 26.40 | 26.79 | 38.73 | 14.94 |
| Myrcene | 70.37 | 45.37 | 25.69 | 82.00 | 22.26 | 24.72 | 34.05 | 25.22 |
| Camphene | 49.02 | 37.72 | 4.65 | 82.51 | 4.45 | 32.92 | 9.10 | 35.95 |
| β-Caryophyllene | 6.91 | 6.59 | 7.16 | 0.00 | 0.00 | 0.00 | 0.93 | 0.00 |
| (+)-α-Longipinene | 0.42 | 21.27 | 6.65 | 5.37 | 1.04 | 0.00 | 0.08 | 20.29 |
| (-)-Isolongifolene | 2.72 | 0.68 | 0.51 | 0.89 | 0.00 | 0.00 | 0.00 | 0.00 |
| (+)-Sativene | 16.60 | 1.93 | 1.64 | 14.18 | 2.03 | 0.05 | 6.11 | 4.66 |
| (+)-Longifolene | 0.14 | 2.31 | 2.09 | 0.00 | 0.35 | 0.00 | 0.24 | 3.27 |
| (-)-Caryophyllene oxide | 20.80 | 63.25 | 37.77 | 88.38 | 19.63 | 45.89 | 26.45 | 27.52 |
| Butylated hydroxytoluene | 29.55 | 22.69 | 14.80 | 88.15 | 22.74 | 50.42 | 57.06 | 42.67 |
| Camphor | 49.11 | 38.94 | 20.11 | 79.19 | 29.95 | 38.28 | 24.82 | 37.04 |
| 2-Methoxy-4-vinylphenol.Kosher | 31.18 | 16.99 | 13.97 | 64.47 | 10.53 | 28.32 | 29.01 | 22.05 |
| (-)-Fenchone | 49.72 | 36.93 | 39.04 | 128.47 | 9.00 | 35.96 | 28.86 | 46.58 |
3-D structure model and docking of DhelOBP21. (A) Superimposed secondary structures of DhelOBP21 and the template 1C3Y. The model of DhelOBP21 and crystal structure of 1C3Y are shown in red and yellow, respectively. (B) Predicted 3D model of DhelOBP21. The centre is the binding cavity within the ligands. The green area expresses hydrophobicity and red area hydrophilia. (C) Sequence alignment of DhelOBP21 and template. The secondary elements for DhelOBP21 are shown above the sequences. α-helices are displayed as squiggles. Identical residues are highlighted in white letters with a red background. Residues with similar physicochemical properties are shown in red letters and a blue frame. The stars are amino acid of forming the cavity, and the stars in yellow are mutation sites.
SDS-PAGE analyses showing the expression and purification of the recombinant OBPs. M, molecular marker; CK, bacterial cells before induction by IPTG. The first two pictures show the bacterial cells after induction by IPTG. The last picture shows the purified protein of DhelOBP21-WT, S67A, I84N, T119N.
Ligand-binding assays of the 1-NPN of DhelOBP21 and mutants. (A, B) Binding curve for 1-NPN to DhelOBP21 and mutants at pH 7.4 and pH 5.0. (C, D) Scatchard plot of these OBPs at pH 7.4 and pH 5.0. (E) Comparison of Ki-NPN of these protein pH 7.4 and pH 5.0.
Plane structure and classification of ligands. (A) The ligands with the molecular volume between 100 and 125 ų. (B) The ligands with the molecular volume between 160 and 185 ų.
Binding affinities (indicated by 1/Ki*1000) assays of DhelOBP21-WT. (A, B) Relationship between molecular volume and Ki at pH 7.4 and pH 5.0. (C, D) Relationship between LogD and Ki at pH 7.4 and pH 5.0. (E) Relationship between difference value and Ki at pH 7.4. (F) Relationship between difference value and LogD. (G) Relationship between difference value and molecular volume. The dashed lines express confidence interval.
Ligand Characteristics and Fluorescence Binding Assays for Mutant Proteins
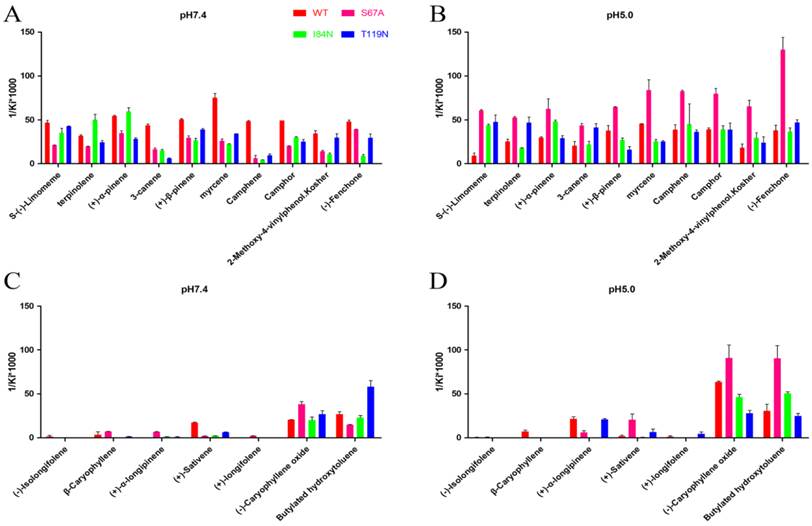
To gain deeper insights into the relationship between binding affinity and the nature of cavity, the binding profiles of mutant proteins S67A, I84N, and T119N were compared with DhelOBP-WT at pH 7.4 and 5.0 (Fig. 7). At pH 7.4, only (-)-caryophyllene oxide exhibited a stronger binding ability when the mutant protein S67A was compared with DhelOBP-WT, whereas the others are weaker. The binding affinity of terpinolene with mutant protein I84N was better than with DhelOBP-WT, and a slight enhancement was achieved between (+)-α-pinene and protein I84N compared with the wild-type protein. Mutant protein T119N had a greater binding ability towards Butylated hydroxytoluene and equivalent binding affinity for S-(-)-limomeme and 2-methoxy-4-vinylphenol Kosher compared with DhelOBP-WT. The binding abilities of the other ligands were reduced to a variable extent, especially for three ligands—3-canene, myrcene and camphene—for which their binding abilities declined significantly. By contrast, at pH 5.0, mutant protein S67A showed a stronger binding ability against most ligands. The binding performance of ligand S-(-)-limomeme was clearly weaker for DhelOBP-WT compared with the three mutant proteins. Binding of Camphene, Camphor, and (-)-Fenchone with DhelOBP-WT or mutant proteins I84N or T119N did not show any apparent difference. Compared with the binding abilities of DhelOBP-WT, butylated hydroxytoluene and (+)-α-pinene showed relatively higher affinities towards the mutant protein I84N, whereas terpinolene and 3-canene showed a relatively higher affinity towards the mutant protein T119N. Additionally, the effects of pH for the four proteins were each distinct. In addition to (-)-Caryophyllene oxide, DhelOBP-WT displayed weaker binding affinity at pH 5.0 than at pH 7.4, especially the S-(-)-limomeme. However, mutant protein S64A displayed a higher binding affinity at pH 5.0 than at pH 7.4. The mutant protein I84N showed a different change for each ligand, as did the mutant protein T119N.
Binding site structure
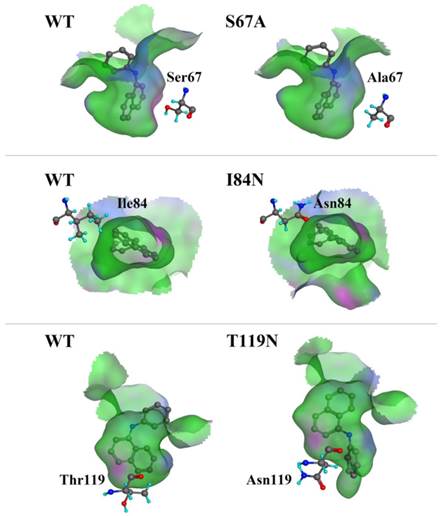
We compared the binding cavity shapes using 1-NPN as a reference (Fig. 8). We found that the cavity shape was different, as was the pose of 1-NPN. It did not establish any hydrogen bonds, but instead maintained binding through hydrophobic contact with the cavity. By contrast, we found that the mutant protein S67A was more spacious and had a more hydrophobic cavity, so the binding ability of 1-NPN was the strongest. Because it induced spatial changes in conformation, the mutant I84N showed enhanced binding capacity. Although the cavity of mutant T119N became larger, a new area also appeared in the corner. Additionally, the polar atom “N” could enter the binding cavity, but it interacted with the hydrophobic area. Therefore, its binding ability did not change compared with the cavity of the wild-type protein.
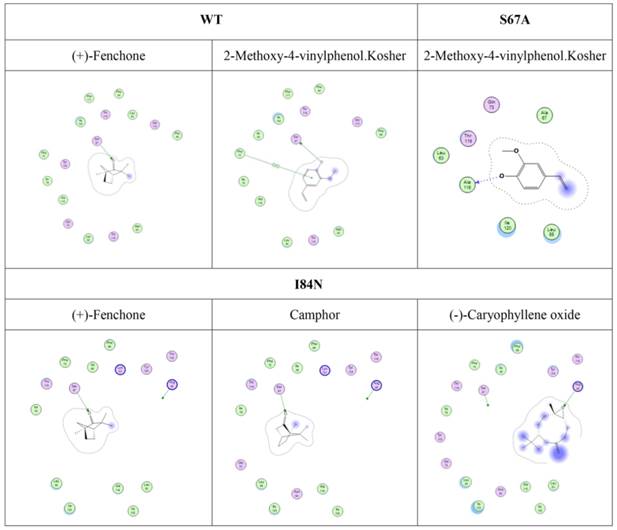
Furthermore, although hydrogen bonding can be predicted by molecular docking (Fig. 9), these ligands did not have a stronger binding ability than some other ligands. Compared with mutant proteins I84N and T119N (Table 2), which enhanced the hydrophilia of the binding cavity, the binding capacity did not increase. Despite the increased polar surface area of the T119N cavity, molecular docking did not show evidence of hydrogen bonding with any ligand. One reason is that the formation of hydrogen bonds is related to both the ligand orientation and the steric hindrance in the cavity. The optimal orientations of specific ligands whose binding affinities changed with the four proteins were calculated using molecular docking. In this analysis, we visually observed that the ligands showed different orientations in the four cavities (Fig. S3).
The properties of ligands
| Ligand | Molecular Mass | Molecular Volume | Log D at pH7.4 | Log D at pH5.0 | Molecular Surface Area | Molecular Polar Surface Area | Molecular SASA | Molecular Polar SASA |
|---|---|---|---|---|---|---|---|---|
| S-(-)-Limomeme | 136.1252005 | 114.9 | 3.50 | 3.50 | 159.03 | 0 | 333.501843 | 0 |
| Terpinolene | 136.1252005 | 113.53 | 3.64 | 3.64 | 162.69 | 0 | 336.626127 | 0 |
| (+)-α-Pinene | 136.1252005 | 115.24 | 2.87 | 2.87 | 151 | 0 | 303.035852 | 0 |
| 3-Canene | 136.1252005 | 115.59 | 2.87 | 2.87 | 151.08 | 0 | 303.035852 | 0 |
| (+)-β-Pinene | 136.1252005 | 115.59 | 2.93 | 2.93 | 149.18 | 0 | 302.068025 | 0 |
| Myrcene | 136.1252005 | 116.96 | 3.69 | 3.69 | 172.59 | 0 | 349.873088 | 0 |
| Camphene | 136.1252005 | 111.81 | 2.93 | 2.93 | 147.95 | 0 | 302.068025 | 0 |
| β-Caryophyllene | 204.1878008 | 167.72 | 4.75 | 4.75 | 229.32 | 0 | 411.970758 | 0 |
| (+)-α-Longipinene | 204.1878008 | 171.15 | 4.12 | 4.12 | 220.78 | 0 | 381.504767 | 0 |
| (-)-Isolongifolene | 204.1878008 | 169.44 | 4.08 | 4.08 | 222.73 | 0 | 379.259056 | 0 |
| (+)-Sativene | 204.1878008 | 171.84 | 4.22 | 4.22 | 211.22 | 0 | 382.782651 | 0 |
| (+)-Longifolene | 204.1878008 | 170.47 | 4.18 | 4.18 | 217.73 | 0 | 380.53694 | 0 |
| (-)-Caryophyllene oxide | 220.1827154 | 182.81 | 4.87 | 4.87 | 269.42 | 20.23 | 421.190323 | 52.152873 |
| Butylated hydroxytoluene | 220.1827154 | 180.07 | 3.52 | 3.52 | 233.04 | 12.53 | 397.227075 | 22.68203 |
| Camphor | 152.1201151 | 122.1 | 2.08 | 2.08 | 165.56 | 17.07 | 305.688433 | 43.490521 |
| 2-Methoxy-4-vinylphenol.Kosher | 150.0680796 | 102.21 | 2.12 | 2.12 | 159.3 | 29.46 | 330.318931 | 58.183583 |
| (-)-Fenchone | 152.1201151 | 118.67 | 2.50 | 2.50 | 165.91 | 17.07 | 305.688433 | 43.490521 |
Note: Molecular volume is the 3D volume for each molecule using the current 3D coordinates. Log D is the octanol-water partition coefficient calculated taking into account the ionization states of the molecule in different pH. Molecular surface area is the total surface area for each molecule using a 2D approximation. Molecular polar surface area is the polar surface area for each molecule using a 2D approximation. Molecular SASA is the total solvent accessible surface area for each molecule using a 3D method. Molecular polar SASA is the polar solvent accessible surface area for each molecule using a 3D method.
Comparison of binding affinities (indicated by 1/Ki*1000) between DhelOBP21 and its mutant at different pH. (A, B) The binding ability of ligands with the molecular volume between 100 and 125 ų. (C, D) The binding ability of ligands with the molecular volume between 160 and 185 ų.
Comparison of shape of DhelOBP21 and its mutants. The green areas express hydrophobicity and red areas express hydrophilia of binding cavity. The red atom is oxygen atom. The blue atom is nitrogen-atom. The gray molecule in the cavity is 1-NPN.
Predicted hydrogen-bond interaction by molecular docking. The green amino acid is nonpolar. The red amino acid is polar. The dashed lines with arrows express the predicted hydrogen-bond interaction.
Discussion
Herein, we cloned and identified the DhelOBP21 from D. helophoroides, which belongs to the Minus-C insect OBPs subfamily. A phylogenetic analysis showed that DhelOBP21, along with the other two Minus-C OBPs DhelOBPs 5 (five Cys residues) and 6 (four Cys residues) formed a branch, while other classical OBPs grouped into another branch [35]. The predicted 3D structure of DhelOBP21 contains only two disulfide bridges and a hydrophobic cavity, which is similar to that reported for AmelOBP14. In OBP14, the C-terminus segment forms an external seventh helix at the interface between the protein exterior and the internal cavity. It was located near the binding site opening and allowed the cavity to close completely. A similar helix was not observed in DhelOBP21. Although the C-terminus extended into the protein core, similar to Lush, it was also shorter.
By analyzing the molecular volume and hydrophobicity of ligands, we found that if we wanted to analyze binding ability, we first need to discuss whether the molecular volume or size fits the binding cavity or not. In an analysis of the binding ability of ligands with those proteins, when the molecular volume of ligands was in the range of 160-185 ų, the ligands had poorer binding ability. Those ligands with a molecular volume between 100 and 125 ų tend to have better binding affinity. That might occur because the volume of the former was so great that many collisions are induced between the cavity of the ligand, and the binding free energy is dramatically increased, which can hinder ligand binding in the pocket. Additionally, we suspect that if the ligands have a volume that is too small, it may have negative effects on binding. The 2-methoxy-4-vinylphenol Kosher has the smallest volume among the ligands less than 150 ų, and it has the low binding values with the wild type and mutant protein compared to other ligands under 150 ų at different pH values. We speculate that if the ligand volume is too small, the relative surface area of the interaction is also smaller and the binding ability is also poor. Therefore, the volume of proteins and ligands should be appropriate.
However, we know that a certain degree of conformational flexibility must be present to allow ligands to access the central binding pocket in most OBPs, particularly LUSH [19, 21], AgamOBP4 [14], and AmelASP2 [41]. We considered that protein plasticity could influence the binding range of ligands and the intensity of binding with specific ligands. Studies of Minus-C OBP AmelOBP14-odorant complexes have shown that the cavity volume can vary to some extent in association with ligand sizes. It could help the OBP to bind more ligands. Meanwhile, high levels of conformational flexibility may be important to tightly control responses that are specific ligands, such as pheromones [42]. Although different levels of conformational flexibility exist in OBPs, a molecular volume that is too big could result in more collisions between atoms in the ligand and the cavity when the ligand enters the binding cavity. The importance of flexibility might reflect binding with specific ligands to enhance ligand-binding abilities. Moreover, the ligands that we tested were from the woodland where D. helophoroides resides. The ligand shapes were not similar to those of MOP and PEG, which have long chains [13, 16]. This molecular volume specificity is important for protein evolution.
Hydrophobic contacts have been reported in many studies. We carried out studies aimed at understanding the influence of ligand hydrophobicity. In the DhelOBP-WT binding analysis at pH 7.4, ligands with greater hydrophobicity tended to have a stronger binding ability (Fig. 5C). This was also influenced by ligand orientation and steric hindrance. In cooperation with the conformational flexibility of a cavity, the more residues belonging to the cavity wall that interact with the ligands, the easier it is for a ligand to be retained in the cavity. Hydrogen-bond interactions have also been studied recently. Many studies have mutated residues that could form hydrogen-bonds with specific ligands to prove the function of a residue [28, 31, 43]. However, in our present study, ligands with polar atoms did not have greater binding affinities. Especially for the mutant protein S67A at pH 5.0, although increased hydrophobicity could influence binding with polar ligands, these ligands could strengthen the binding ability by enhancing the hydrophobicity of the binding cavity. CquiOBP1 and AmelOBP14 had the similar modes of action, whereas CquiOBP1 does not recognize the specific functional group of MOP but instead recognizes the length of the lipid chain that fits its hydrophobic tunnel [16]. AmelOBP14 does not form any hydrogen bonds, but binds the citralva by establishing many hydrophobic contacts with the cavity wall residues [25]. Thus, ligand hydrophobicity and the binding cavity had a greater influence than hydrogen bonding in DhelOBP21.
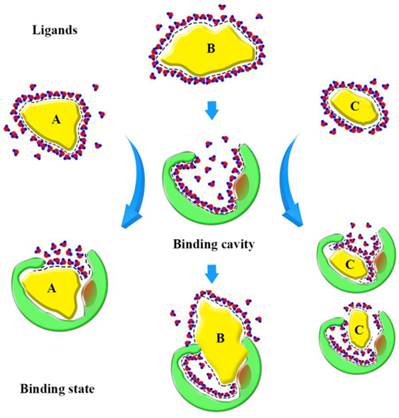
Based on our findings, we can form a model to explain the binding state (Fig. 10). Before binding, hydrophobic ligands and cavity were surrounded by water, and water forms a cage that wraps around the ligands. When ligands enter the binding cavity, hydrophobic interactions make the hydrophobic ligands and cavity draw close together. Additionally, the aqueous phase concentrates to become more structured. The cage hydrates bound the ligands in cavity. In this process, the characteristics of the ligands can influence binding. The shape of ligand A conforms to the shape of the binding cavity, and establishes remarkable hydrophobic interactions. The volume of ligand B is too big to fit the binding cavity, so more collisions between the atoms of the ligand and cavity could occur when it enters the binding cavity. Ligand C is small, so it can 'roam around' the cavity and bind with the amino acid residues in various conformations and, because of that, it can be released easily. The binding process is dynamic.
Sketch of binding state. The yellow graphics express the different ligands. The green graphics express the binding cavity. And the red circles mean the oxygen atom and the blue circles mean the hydrogen atom, they form the hydrones. The dashed lines express the hydrophobic interaction.
Notably, pH affected the binding of DhelOBP21 and the mutant protein S67A in opposite ways. The Ser67 residue may play an important role in the release of ligands. We also noticed at the N-terminal base there were three residues that exhibited different protonated states at pH 7.4 and 5.0 (Fig. S4). We speculate this may affect comformational changes in the N-terminus. Furthermore, studies of 3D structure of THP12 reported that its structural stability is highly pH-dependent and circular dichroism studies revealed a loss of helicity upon changing the pH from 7.5 to 3.0 [44]. The binding affinities of mutant S67A with most ligands showed remarkable improvement at pH 5.0, while the more hydrophobic cavity was beneficial for ligand binding. Additionally, 3D structure of DhelOBP21 only contained five helices, unlike most other insect OBPs that contain of six α-helices. We speculate that one reason for this difference is that template 1C3Y lacks ligands, so the structure is expansive. Thus, we could infer the cavity was extended or open at a low pH. However, the release mechanism for DhelOBP21 remains unclear. Future studies will be needed to confirm these ligand-OBP interaction findings.
Supplementary Material
Figures S1-S4.
Acknowledgements
This study was supported and funded by the National Natural Science Foundation of China (31230015), Program for New Century Excellent Talents in University (NCET-11-0649) and Fundamental Research Funds for the Central Universities (2013PY046).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Arthurs SP, Tofangsazi N, Meagher RL, Cherry R. Attraction of Plecia nearctica (Diptera: Bibionidae) to Floral Lures Containing Phenylacetaldehyde. Florida Entomologist. 2012;95:199-201
2. Wang N, Wang N, Niu L, Bian S, Xiao J, Huang D. Odorant-binding protein (OBP) genes affect host specificity in a fig-pollinator mutualistic system. Insect molecular biology. 2014;23:621-31
3. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annual review of entomology. 2013;58:373-91
4. Vogt RG. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. Insect pheromone biochemistry and molecular biology. 2003:391-445
5. Vogt RG. Molecular basis of pheromone detection in insects. Comprehensive insect physiology, biochemistry, pharmacology and molecular biology. 2005;3:753-804
6. Venthur H, Mutis A, Zhou JJ, Quiroz A. Ligand binding and homology modelling of insect odorant-binding proteins. Physiological Entomology. 2014;39:183-98
7. Krieger J, von Nickisch-Rosenegk E, Mameli M, Pelosi P, Breer H. Binding proteins from the antennae of Bombyx mori. Insect biochemistry and molecular biology. 1996;26:297-307
8. Stengl M, Zufall F, Hatt H, Hildebrand JG. Olfactory receptor neurons from antennae of developing male Manduca sexta respond to components of the species-specific sex pheromone in vitro. The Journal of neuroscience. 1992;12:2523-31
9. Lagarde A, Spinelli S, Tegoni M, He X, Field L, Zhou J-J. et al. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. Journal of molecular biology. 2011;414:401-12
10. Forêt S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome research. 2006;16:1404-13
11. Zhou J-J, Huang W, Zhang G-A, Pickett JA, Field LM. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004;327:117-29
12. Tsitsanou K, Thireou T, Drakou C, Koussis K, Keramioti M, Leonidas D. et al. Anopheles gambiae odorant binding protein crystal complex with the synthetic repellent DEET: implications for structure-based design of novel mosquito repellents. Cellular and Molecular Life Sciences. 2012;69:283-97
13. Wogulis M, Morgan T, Ishida Y, Leal WS, Wilson DK. The crystal structure of an odorant binding protein from Anopheles gambiae: evidence for a common ligand release mechanism. Biochemical and biophysical research communications. 2006;339:157-64
14. Davrazou F, Dong E, Murphy EJ, Johnson HT, Jones DN. New insights into the mechanism of odorant detection by the malaria-transmitting mosquito Anopheles gambiae. Journal of Biological Chemistry. 2011;286:34175-83
15. Leite NR, Krogh R, Xu W, Ishida Y, Iulek J, Leal WS. et al. Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive “Lid”. PLoS One. 2009;4:e8006
16. Mao Y, Xu X, Xu W, Ishida Y, Leal WS, Ames JB. et al. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proceedings of the National Academy of Sciences. 2010;107:19102-7
17. Tsitsanou KE, Drakou CE, Thireou T, Gruber AV, Kythreoti G, Azem A. et al. Crystal and solution studies of the “Plus-C” odorant-binding protein 48 from Anopheles gambiae control of binding specificity through three-dimensional domain swapping. Journal of Biological Chemistry. 2013;288:33427-38
18. Zhou J-J, Robertson G, He X, Dufour S, Hooper AM, Pickett JA. et al. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. Journal of molecular biology. 2009;389:529-45
19. Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nature Structural & Molecular Biology. 2003;10:694-700
20. Mohanty S, Zubkov S, Gronenborn AM. The solution NMR structure of Antheraea polyphemus PBP provides new insight into pheromone recognition by pheromone-binding proteins. Journal of molecular biology. 2004;337:443-51
21. Thode AB, Kruse SW, Nix JC, Jones DN. The role of multiple hydrogen-bonding groups in specific alcohol binding sites in proteins: insights from structural studies of LUSH. Journal of molecular biology. 2008;376:1360-76
22. Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chemistry & biology. 2000;7:143-51
23. Damberger FF, Ishida Y, Leal WS, Wüthrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. Journal of molecular biology. 2007;373:811-9
24. Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289-93
25. Spinelli S, Lagarde A, Iovinella I, Legrand P, Tegoni M, Pelosi P. et al. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect biochemistry and molecular biology. 2012;42:41-50
26. Li Z-Q, Zhang S, Luo J-Y, Cui J-J, Ma Y, Dong S-L. Two Minus-C odorant binding proteins from Helicoverpa armigera display higher ligand binding affinity at acidic pH than neutral pH. Journal of insect physiology. 2013;59:263-72
27. Rusconi B, Maranhao A, Fuhrer J, Krotee P, Choi S, Grun F. et al. Mapping the Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) using modeling techniques, site directed mutagenesis, circular dichroism and ligand binding assays. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2012;1824:947-53
28. Zhuang X, Wang Q, Wang B, Zhong T, Cao Y, Li K. et al. Prediction of the key binding site of odorant-binding protein of Holotrichia oblita Faldermann (Coleoptera: Scarabaeida). Insect molecular biology. 2014;23:381-90
29. Lu Y, Li H, Zhuang S, Zhang D, Zhang Q, Zhou J. et al. Olfactory biosensor using odorant-binding proteins from honeybee: Ligands of floral odors and pheromones detection by electrochemical impedance. Sensors and Actuators B: Chemical. 2014;193:420-7
30. Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E. et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5:e9471
31. Ahmed T, Zhang T-t, Wang Z-y, He K-l, Bai S-x. Three Amino Acid Residues Bind Corn Odorants to McinOBP1 in the Polyembryonic Endoparasitoid of Macrocentrus cingulum Brischke. PloS one. 2014;9:e93501
32. Wei J-R, Yang Z-Q, Poland TM, Du J-W. Parasitism and olfactory responses of Dastarcus helophoroides (Coleoptera: Bothrideridae) to different Cerambycid hosts. BioControl. 2009;54:733-42
33. Yang Z-Q, Wang X-Y, Zhang Y-N. Recent advances in biological control of important native and invasive forest pests in China. Biological Control. 2014;68:117-28
34. Xiao G. Forest insects of China. China Forestry Publishing House. 1991
35. Wang J, Li D-Z, Min S-F, Mi F, Zhou S-S, Wang M-Q. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2014;11:1-8
36. Boratyn GM, Schaffer A, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12
37. Chemical Computing Group Inc. Molecular Operating Environment (MOE), 2013.08. Chemical Computing Group Inc, 1010 Sherbooke St West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2015
38. Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. Journal of medicinal chemistry. 2003;46:499-511
39. Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A, Cambillau C. et al. Revisiting the specificity of mamestra brassicaeand Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. Journal of Biological Chemistry. 2001;276:20078-84
40. Lautenschlager C, Leal WS, Clardy J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: implications for pheromone recognition. Structure. 2007;15:1148-54
41. Lescop E, Briand L, Pernollet J-C, Guittet E. Structural basis of the broad specificity of a general odorant-binding protein from honeybee. Biochemistry. 2009;48:2431-41
42. Ziemba BP, Murphy EJ, Edlin HT, Jones DN. A novel mechanism of ligand binding and release in the odorant binding protein 20 from the malaria mosquito Anopheles gambiae. Protein Science. 2013;22:11-21
43. Yi X, Zhang Y, Wang P, Qi J, Hu M, Zhong G. Ligands Binding and Molecular Simulation: the Potential Investigation of a Biosensor Based on an Insect Odorant Binding Protein. International journal of biological sciences. 2015;11:75
44. Rothemund S, Liou Y-C, Davies PL, Krause E, Sönnichsen FD. A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Structure. 1999;7:1325-32
Author contact
![]() Corresponding author: Address: College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, P.R.China. Tel.: (0086) 13627126839 Fax: (0086) -27-87280920. E-mail: mqwanghzau.edu.cn (M.-Q Wang)
Corresponding author: Address: College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, P.R.China. Tel.: (0086) 13627126839 Fax: (0086) -27-87280920. E-mail: mqwanghzau.edu.cn (M.-Q Wang)

 Global reach, higher impact
Global reach, higher impact