10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(2):246-256. doi:10.7150/ijbs.13805 This issue Cite
Research Paper
A Group of Novel Serum Diagnostic Biomarkers for Multidrug-Resistant Tuberculosis by iTRAQ-2D LC-MS/MS and Solexa Sequencing
1. Institute of Cell Biology, Zhejiang University, Hangzhou 310058, P.R. China
2. Department of Respiratory Medicine, The Sixth Hospital of Shaoxing, Shaoxing 312000, P.R. China
3. Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou 310013, P.R. China
4. Department of Tuberculosis, The First Hospital of Jiaxing, Jiaxing 314001, P.R. China
Received 2015-9-10; Accepted 2015-11-11; Published 2016-1-1
Abstract
The epidemic of pulmonary tuberculosis (TB), especially multidrug-resistance tuberculosis (MDR-TB) presented a major challenge for TB treatment today. We performed iTRAQ labeling coupled with two-dimensional liquid chromatography-tandem mass spectrometry (2D LC-MS/MS) and Solexa sequencing among MDR-TB patients, drug-sensitive tuberculosis (DS-TB) patients, and healthy controls. A total of 50 differentially expressed proteins and 43 differentially expressed miRNAs (fold change >1.50 or <0.60, P<0.05) were identified in the MDR-TB patients compared to both DS-TB patients and healthy controls. We found that 22.00% of differentially expressed proteins and 32.56% of differentially expressed miRNAs were related, and could construct a network mainly in complement and coagulation cascades. Significant differences in CD44 antigen (CD44), coagulation factor XI (F11), kininogen-1 (KNG1), miR-4433b-5p, miR-424-5p, and miR-199b-5p were found among MDR-TB patients, DS-TB patients and healthy controls (P<0.05) by enzyme-linked immunosorbent assay (ELISA) and SYBR green qRT-PCR validation. A strong negative correlation, consistent with the target gene prediction, was found between miR-199b-5p and KNG1 (r=-0.232, P=0.017). Moreover, we established the MDR-TB diagnostic model based on five biomarkers (CD44, KNG1, miR-4433b-5p, miR-424-5p, and miR-199b-5p). Our study proposes potential biomarkers for MDR-TB diagnosis, and also provides a new experimental basis to understand the pathogenesis of MDR-TB.
Keywords: multidrug-resistance tuberculosis, proteomic, transcriptomic, serum, biomarkers
Introduction
Tuberculosis (TB) is caused by infection with Mycobacterium tuberculosis (Mtb). The efforts to control TB have been impaired by the emergence of multidrug-resistant (MDR) strains and HIV epidemic. According to the World Health Organization (WHO) report, there were 9 million newly diagnosed TB cases and 1.5 million death cases worldwide in 2013 [1]. Meanwhile, there were 480,000 newly diagnosed multidrug-resistance tuberculosis (MDR-TB) cases and 210,000 death cases [2]. Globally, 3.5% of newly diagnosed TB cases and 20.5% of previously treated TB cases were estimated to have had MDR-TB. The MDR-TB rate in China has been estimated to be 11.6% for new and 35.9% for previously treated TB cases [3], which is significantly higher than global average. Only 48% of MDR-TB patients have been reported to be successfully treated [2], while 90% of drug-sensitive tuberculosis (DS-TB) patients were cured. Therefore, MDR-TB has become the most difficult problem for TB prevention and treatment, leading to the widespread of TB.
Epidemiological reports declared that irrational prescription, inadequate administration of drug, and intermittent drug therapy are the most common reasons for causing MDR-TB. The key to control MDR-TB is early diagnosis [4, 5]. Drug susceptibility testing (DST) is the golden standard for MDR-TB diagnosis. However, DST takes 6-8 weeks to obtain results. Microscopic-observation drug susceptibility (MODS) testing [6] have shortened the time needed for phenotypic assays to between 1 and 2 weeks, however, it might over-diagnose MDR-TB. The results of fluorophage method matched 100% [7] with that of DST, but the verified sample size was too small (only 7 cases per group). Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique [8, 9] could only detect specific mutations, and is not suitable for other clinical causes. Current routine methods are not very effective or rapid. Therefore, there is an urgent need to obtain novel biomarkers suitable for MDR-TB diagnosis.
MDR strains can cause different immune response than drug-sensitive (DS) strains [10, 11]. A small sample size experiment found that different strains of Mtb can cause differential miRNAs expression in human macrophages [12]. In this study, we investigated serum biomarkers from MDR-TB patients using iTRAQ labeling coupled with two-dimensional liquid chromatography-tandem mass spectrometry (2D LC-MS/MS) and Solexa sequencing. Integrative analysis and biological verification were performed before model establishment. Our study proposes potential biomarkers for MDR-TB diagnosis, and also provides a new experimental basis to understand the pathogenesis of MDR-TB.
Material and Methods
Patients and control subjects
This study was approved by the Ethics Committee of the Faculty of Medicine (Zhejiang University, China). Written informed consents were obtained from all subjects before blood sampling. A total of 42 subjects with MDR-TB (33 males, 9 females), aged 23-76 years (mean age 44.88 ± 14.56 years), were recruited from the Sixth Hospital of Shaoxing and the First Hospital of Jiaxing during April 2013 and May 2015. In addition, 60 DS-TB subjects (39 males, 21 females), aged 18-65 years (mean age 40.48 ± 16.46 years), were recruited from the Sixth Hospital of Shaoxing at the same period. The healthy control group with no history of TB or other immune diseases comprised 60 healthy subjects (42 males, 18 females), aged 24-73 years (mean age 42.57 ± 13.17 years), were recruited from the Zhejiang Hospital (Table S1). Blood was drawn into regular bottles in the morning from each patient before the anti-TB therapy. Similarly, fasting blood samples were drawn from healthy controls. The samples were stored at - 80°C for further analysis. Data including age, gender and clinical examination findings of TB patients and healthy controls were collated into databases.
iTRAQ-2D LC-MS/MS
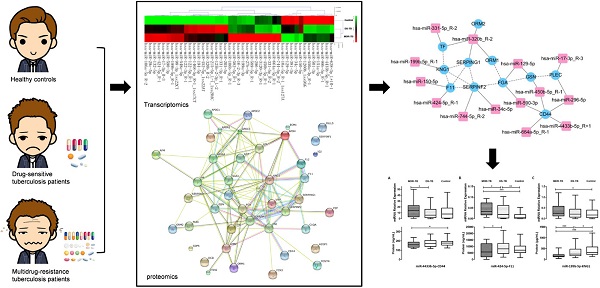
The workflow of our study is shown in Fig. 1. In order to increase the precision and accuracy of the data in proteomics study [13], equal amount of 10 different samples were mixed to produce a sample group. In addition, 10 samples were randomly divided into two pools as biological replicates. Then, six iTRAQ labeled sample pools were generated (healthy controls group, DS-TB group, and MDR-TB group; each for two subgroups).
High-abundance serum proteins such as albumin, IgG, and haptoglobin were removed by using the Human 14 Multiple Affinity Removal System (Agilent Technologies, Santa Clara, CA, USA). Then, the proteins were concentrated and desalted [14]. A total of 100 μg protein from each group was soaked in ice-cold acetone, and then centrifuged. After that, the samples were digested overnight with trypsin at 37°C. Finally, iTRAQ reagents (Applied Biosystems, Foster city, CA, USA) were labeled for six groups: healthy control group, iTRAQ reagent 113, 117; DS-TB group, iTRAQ reagent 114, 118; MDR-TB group, iTRAQ reagent 116, 121. The six sample groups were mixed, desalted, and dried.
The iTRAQ labeled peptides were separated by polysulfoethyl column (2.1×100 mm, 5 μm, 200Ǻ; Nest Group, Inc., Southborough, MA, USA) of strong cation exchange (SCX) chromatography [15]. A total of ten SCX components were collected, concentrated, and dissolved. Samples were subsequently loaded onto the ZORBAX 300SB-C18 column (5 μm, 200Ǻ, 0.1 × 150mm, Microm, Auburn, CA, USA). The components produced by SCX chromatography were subjected to MS analysis twice. The ratio of the peak area of the iTRAQ reporter ion reflected the relative abundance of the peptide and protein [16, 17]. Protein identification and quantification were performed using the ProteinPilotTM software (Applied Biosystems, version 4.2). The ProGroup algorithm was used to identify peptides. MS/MS data were searched against the Human International Protein Index database (version 3.87) as described previously [14, 16, 17]. In order to reduce false positive results, a strict cutoff for protein identification was applied with the unused ProtScore > 1.3 and at least one peptide with a 95% confidence limit [18, 19]. The expression ratio higher than 1.50-fold or lower than 0.60-fold were considered significant.
The workflow for serum biomarkers of multidrug-resistant tuberculosis, drug-sensitive tuberculosis, and healthy controls using iTRAQ-2D LC-MS/MS and Solexa sequencing technology. MARS, multiple affinity removal system; SCX, strong cation exchange.
Solexa sequencing
Total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) following the manufacturer's procedure. The total RNA quantity and purity were analyzed by the Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number >7.0. Approximately 1 μg of total RNA were used to prepare small RNA library according to the protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA) [20]. The total RNA of 10 healthy controls, 10 DS-TB patients, and 10 MDR-TB patients were purified directly for Solexa sequencing analysis. Then, we performed the single-end sequencing (36 bp) on Illumina Hiseq2500 following the recommended protocol.
The raw reads were subjected to the Illumina pipeline filter (Solexa 0.3), and then the dataset was further processed with an in-house program, ACGT101-miR (LC Sciences, Houston, Texas, USA) to remove adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, unique sequences with length in 18~26 nucleotide were mapped to specific species precursors in miRBase 20.0 by BLAST search to identify known miRNAs and novel 3p- and 5p- derived miRNAs. Length variation at both 3' and 5' ends and one mismatch inside of the sequence were allowed in the alignment. The unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs. The unique sequences mapping to the other arm of known specific species precursor hairpin opposite to the annotated mature miRNA-containing arm were considered to be novel 5p- or 3p- derived miRNA candidates. The remaining sequences were mapped to other selected species precursors (with the exclusion of specific species) in miRBase 20.0 by BLAST search, and the mapped pre-miRNAs were further BLASTed against the specific species genomes to determine their genomic locations. The above two we defined as known miRNAs. The unmapped sequences were BLASTed against the specific genomes, and the hairpin RNA structures containing sequences were predicated from the flank 80 nt sequences using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Bioinformatics analysis
The differentially expressed protein-protein network was analyzed by STRING software (http://string-db.org/). The norm data of differentially expressed miRNAs were transformed and hierarchical cluster analysis was performed using the TIGR MultiExperiment Viewer 4.0 (MeV 4.0) software program (http://www.tm4.org/mev.html). To predict the genes targeted by differentially expressed miRNAs, two computational target prediction algorithms (TargetScan 50 and miRanda 3.3a) were used to identify miRNA binding sites. Finally, the data predicted by both algorithms were combined and the overlaps were calculated. The cellular component, molecular function, and biological process were analyzed through Gene Ontology (GO) database (http://geneontology.org/). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping was performed by KEGG Mapper (http://www.genome.jp/kegg/mapper.html), and the enrichment analysis was performed by Blast2GO PRO software (https://www.blast2go.com/, version 2.8). Interaction network of differentially expressed miRNAs and proteins (related genes) were performed by Cytoscape software (http://www.cytoscape.org/, version 3.2.1) [21].
Enzyme-linked immunosorbent assay (ELISA) methods
Human CD44 ELISA kit (Abcam, Cambridge, MA, USA; the detection limit was 0.113 ng/mL; SwissProt: P16070), human factor XI (F11) ELISA kit (Abcam, Cambridge, MA, USA; the detection limit was 1.5 ng/mL; SwissProt: P03951), and human kininogen-1 (KNG1) ELISA kit (Abnova Co., Taipei, Taiwan; the detection limit was 0.1 μg/mL; SwissProt: P01042), were used to detect protein levels in serum. Serum samples were diluted with dilution factors of 1:40, 1:1,200, and 1:200 for CD44, F11, and KNG1, respectively. The protein concentrations were measured according to the manufacturer's instructions.
SYBR green qRT-PCR assay
RNA molecules was isolated from the serum using miRcute miRNA Isolation kit (TIANGEN Biotech, Beijing, China) according to the manufacturer's instructions. And then polyadenylated by poly(A) polymerase and subsequently converted into cDNA by reverse transcriptase with oligodT priming (miRcute miRNA First-strand cDNA Kit, TIANGEN Biotech, Beijing, China) according to the manufacturer's instructions. SYBR green qRT-PCR assay (miRcute miRNA qPCR Detection Kit, TIANGEN Biotech, Beijing, China) was used for miRNA quantification in serum samples. Forward primer of has-miR-4433b-5p, has-miR-424-5p, has-miR-199b-5p and has-U6 were designed by TIANGEN Biotech. The total volume of reaction mixture was 20 μl, including 10 μl 2*miRcute miRNA premix (with SYBR&ROX), 0.4 μl forward primer (10 μM), 0.4 μl reverse primer (10 μM), 2 μl cDNA, 1.6 μl 50*ROX reference dye, and 5.6 μl ddH2O. Then, cooled the plate and centrifuged plate briefly in Mini Plate Spinner. The PCRs were carried out with incubation at 94°C for 2 min followed by 45 cycles of 94°C for 20 s and 60°C for 34 s using an ABI PRISM 7300 detection system (Applied Biosystems, Foster City, CA, USA). Each sample was run in duplicate for two times.
Statistical analysis
For qRT-PCR data, the expression level of miRNA was normalized to miR-U6 that was stable in serum samples [22]. The relative expression levels of each target miRNAs (Log2 relative level) were calculated according to the difference in CT values between the target miRNAs and U6 (ΔCT). The expression level of miRNA was calculated using the ΔΔCT method [23]. The negative controls for templates were ddH2O.
The parametric data were tested using the chi-square test for the composition ratios, t-tests for means of two groups, and one-way analysis of variance (ANOVA) for means of three or more groups. Nonparametric analysis was performed using the Mann-Whitney U-test and the Kruskal-Wallis H-test. Spearman correlation method was performed to determine association between two different parameters. P values <0.05 were considered statistically significant by the SPSS software (Chicago, IL, version 16.0). Receiver operating characteristic (ROC) curve were calculated by MedCalc Software (Belgium, Version 12.4.2.0) and the diagnostic score of MDR-TB patients was set as 1, whereas that of DS-TB patients and healthy controls was set as 0. Selected biomarkers were transferred to Biomarker Pattern Software (BPS, Ciphergen Biosystems) to construct the decision tree of MDR-TB [24]. Accuracy was defined as the proportion of correct state classifications.
Results
Clinical data analysis
Clinical data from 42 MDR-TB patients, 131 DS-TB patients and 150 healthy controls were collected and analyzed. There were significant differences between MDR-TB patients and healthy controls (P<0.05). In addition, significant differences of albumin, albumin/globulin ratio, and fibrinogen levels were also found between MDR-TB patients and DS-TB patients (P<0.05) (Table 1).
Proteomic and transcriptomic results
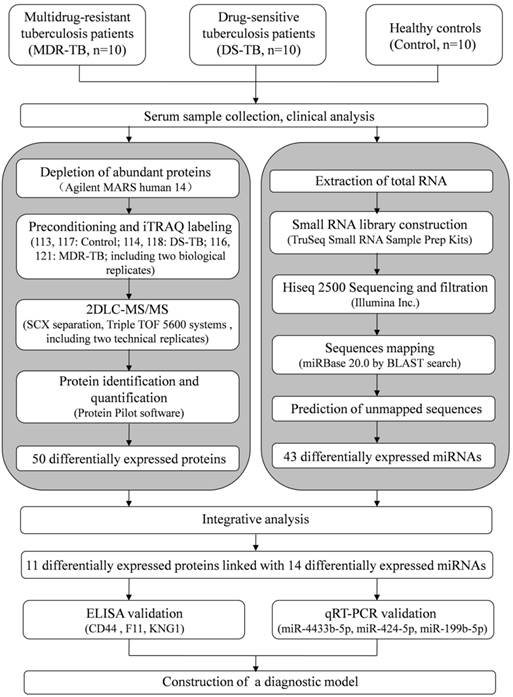
MDR-TB patients, DS-TB patients and healthy controls were screened using iTRAQ-2D LC-MS/MS. The experiment was repeated twice and detected 160 proteins. Further screening revealed 50 differentially expressed proteins in MDR-TB patients compared with both DS-TB patients and healthy controls, including 16 up-regulated proteins (>1.50-fold, P<0.05) and 34 down-regulated proteins (<0.60-fold, P<0.05) (Table S2). GO enrichment analysis of differentially expressed proteins revealed that most of the proteins were involved in the biological regulation (29, 11.11%), metabolic process (29, 11.11%), and single-organism process (29, 11.11%) (Fig. 2A). In addition, the proteins were located in the extracellular region (26, 26.26%), cell (20, 20.20%) and the organelles (19, 19.19%) (Fig. 2B) and possessed binding (28, 37.84%), catalytic activity (12, 16.22%), and enzyme regulator activity (12, 16.22%) (Fig. 2C). The KEGG pathway mapping revealed some significant pathways: complement and coagulation cascades (7 proteins) and extracellular matrix (ECM)-receptor interaction (3 proteins) (Fig. 2D). Protein-protein interactions were noted among all the proteins (Fig. 2E).
We detected 202,848, 239,654 and 102,624 valid reads in MDR-TB patients, DS-TB patients and healthy controls, respectively by Solexa sequencing (Table S3), and predicted 668 new miRNAs. In all, 43 differentially expressed miRNAs including 29 up-regulated miRNAs (>1.50-fold, P<0.05) and 14 down-regulated miRNAs (<0.60-fold, P<0.05) were identified (Table S4). GO annotations to genes revealed significant biological processes (DNA-dependent, regulation of transcription, and multicellular organismal development), cellular components (nucleus and cytoplasm), and molecular functions (protein binding) (Fig. 3A). KEGG enrichment analysis also revealed significant differences in metabolic pathways, pathways in cancer, cytokine-cytokine receptor interaction, and MAPK signaling pathway (Fig. 3B). Cluster analysis revealed many clustering patterns (Fig. 3C).
Clinical data of pulmonary tuberculosis patients and healthy controls.
| P value a | ||||||
|---|---|---|---|---|---|---|
| MDR-TB (N = 42) | DS-TB (N = 131) | Controls (N = 150) | MDR-TB vs DS-TB | MDR-TB vs Controls | DS-TB vs Controls | |
| Total protein (g/L) | 67.64 ± 8.39 | 70.58 ± 6.14 | 73.99 ± 3.52 | 0.067 | <0.001*** | <0.001*** |
| Albumin (g/L) | 36.86 ± 5.31 | 41.02 ± 4.82 | 46.26 ± 2.52 | <0.001*** | <0.001*** | <0.001*** |
| Globulin (g/L) | 30.78 ± 7.84 | 29.40 ± 5.49 | 27.73 ± 3.29 | 0.339 | 0.002** | 0.002** |
| A/G | 1.26 ± 0.31 | 1.43 ± 0.30 | 1.70 ± 0.25 | 0.023* | <0.001*** | <0.001*** |
| Total cholesterol (mmol/L) | 3.84 ± 1.00 | 3.74 ± 0.85 | 5.03 ± 1.06 | 0.061 | <0.001*** | <0.001*** |
| Triglyceride (mmol/L) | 1.07 ± 0.60 | 1.03 ± 0.54 | 1.55 ± 1.50 | 0.777 | 0.165 | <0.001*** |
| HDL-C (mmol/L) | 1.14 ± 0.32 | 1.22 ± 0.39 | 1.35 ± 0.29 | 0.397 | 0.003** | 0.001** |
| LDL-C (mmol/L) | 2.51 ± 0.87 | 2.27 ± 0.62 | 2.83 ± 0.74 | 0.150 | 0.079 | <0.001*** |
| Lipoprotein (mg/L) | 229.21 ± 166.49 | 177.34 ± 140.85 | 203.06 ± 154.97 | 0.145 | 0.495 | 0.159 |
| APOA1 (g/L) | 1.08 ± 0.25 | 1.13 ± 0.26 | 1.24 ± 0.26 | 0.493 | 0.015* | <0.001*** |
| APOB (g/L) | 0.92 ± 0.25 | 0.84 ± 0.20 | 0.92 ± 0.45 | 0.102 | 0.958 | 0.077 |
| CRP (mg/L) | 34.89 ± 32.93 | 21.04 ± 30.51 | 1.26 ± 1.68 | 0.071 | <0.001*** | <0.001*** |
| Pre-albumin (g/L) | 0.14 ± 0.06 | 0.18 ± 0.07 | 0.22 ± 0.06 | 0.052 | <0.001*** | <0.001*** |
| IgG (g/L) | 15.40 ± 6.51 | 14.31 ± 3.78 | 12.73 ± 2.43 | 0.319 | <0.001*** | <0.001*** |
| IgA (g/L) | 5.72 ± 5.43 | 3.87 ± 5.07 | 2.07 ± 0.84 | 0.153 | <0.001*** | <0.001*** |
| IgM (g/L) | 1.17 ± 0.70 | 1.41 ± 0.67 | 0.99 ± 0.52 | 0.209 | 0.199 | <0.001*** |
| Complement 3 (g/L) | 1.14 ± 0.27 | 1.17 ± 0.25 | 0.71 ± 0.41 | 0.603 | <0.001*** | <0.001*** |
| Complement 4 (mg/L) | 341.06 ± 98.19 | 355.08 ± 99.88 | 138.01 ± 115.53 | 0.825 | <0.001*** | <0.001*** |
| INR | 1.06 ± 0.09 | 1.07 ± 0.10 | 1.01 ± 0.08 | 0.711 | 0.045* | <0.001*** |
| Fibrinogen (g/L) | 6.46 ± 1.81 | 4.97 ± 2.03 | 3.53 ± 0.65 | 0.007** | <0.001*** | <0.001*** |
| D-dimer (μg/L) | 1055.33 ± 569.76 | 800.96 ± 1053.96 | 115.4 ± 50.84 | 0.689 | <0.001*** | <0.001*** |
All data are presented as the mean ± SD. MDR-TB: multidrug-resistant tuberculosis; DS-TB: drug-sensitive tuberculosis; A/G: albumin/globulin ratio; HDL-C: high density lipoprotein cholesterol; LDL-C: low density lipoprotein cholesterol; APOA1: apolipoprotein A1; APOB: apolipoprotein B; CRP: C-reactive protein; INR: international normalized ratio. aP value between two groups, for t-test. * P<0.05, ** P<0.01, *** P < 0.001.
Data mining of the set of multidrug-resistant tuberculosis serum proteins biomarker candidates. (A) Biological process; (B) Cellular component; (C) Molecular function; (D) KEGG pathway mapping; (E) The network of proteins analyzed by String software.
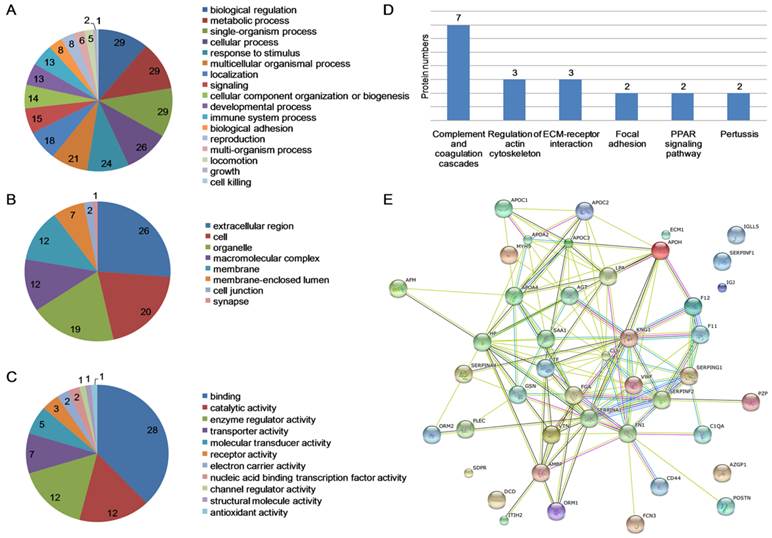
Data mining of the set of multidrug-resistant tuberculosis serum miRNAs biomarker candidates. MDR-TB: multidrug-resistant tuberculosis; DS-TB: drug-sensitive tuberculosis. (A) GO analysis; (B) KEGG analysis; (C) cluster analysis.
Biomarkers prediction and validation
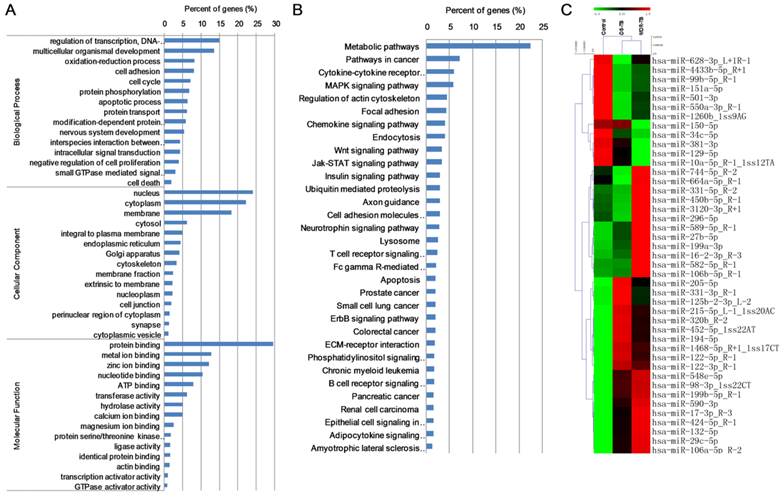
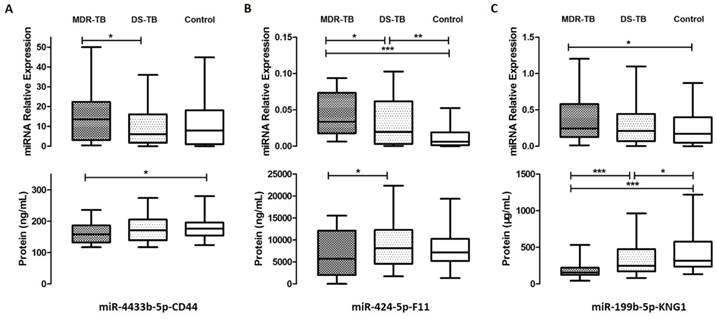
We predicted target genes of 43 differentially expressed miRNAs, combined with proteomic results of iTRAQ-2D LC-MS/MS. All of related 11 differentially expressed proteins and 14 differentially expressed miRNAs were found and the network was established mainly in complement and coagulation cascades (Fig. 4). The bioinformatics analysis is shown in Table 2. We validated all the differentially expressed miRNAs (more than 50 copies) including miR-150-5p, miR-199b-5p, miR-320b, miR-424-5p, miR-4433b-5p, and miR-450b-5p, except miR-17-3p as the only related protein plectin does not have commercial ELISA kit. However, significant differences were found only in miR-199b-5p, miR-424-5p, and miR-4433b-5p (P<0.05). According to the relevance between miRNAs and proteins (predicted genes), we obtained three miRNA-protein (predicted gene) pairs: miR-4433b-5p-CD44, miR-424-5p-F11, and miR-199b-5p-KNG1. Significant differences in miR-4433b-5p, CD44, miR-424-5p, F11, miR-199b-5p, and KNG1 were found among 42 MDR-TB patients, 60 DS-TB patients and 60 healthy controls (P<0.05) (Fig. 5). Serum miR-4433b-5p level was significantly increased in MDR-TB patients compared with DS-TB patients (expression level=2.23, P=0.033), while the CD44 level was significantly reduced (P=0.031) in MDR-TB patients compared with healthy controls. Also, serum miR-424-5p level was significantly increased (expression level=3.34, P=0.029) while the F11 level was significantly reduced (P=0.041) in MDR-TB patients compared with DS-TB patients. In addition, significantly increased miR-424-5p levels were also observed between MDR-TB patients and healthy controls (expression level=7.17, P<0.001), and DS-TB patients and healthy controls (expression level=2.15, P=0.009). However, neither miR-4433b-5p and CD44, nor miR-424-5p and F11 had correlation between miRNA and protein (predicted gene).
Significantly increased miR-199b-5p level (expression level=2.13, P=0.027) and significantly reduced KNG1 level (P<0.001) were found in MDR-TB patients compared with healthy controls. KNG1 levels also showed significant differences between DS-TB patients and MDR-TB patients (P<0.001), and DS-TB patients and healthy controls (P=0.039). Moreover, negative correlation was observed between miR-199b-5p and KNG1 (r=-0.232, P=0.017). We further observed a positive correlation between KNG1 and F11 protein (r=0.308, P<0.0001), and a negative correlation between KNG1 and miR-4433b-5p (r=-0.182, P=0.044) using the Spearman correlation analysis.
Integrative proteomic and transcriptomic analysis. FGA, F11, KNG1, SERPINF2, and SERPING1 involved in complement and coagulation cascades in KEGG pathways.
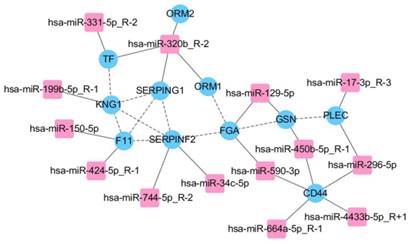
The bioinformatics analysis of 11 differentially expressed proteins and 14 differentially expressed miRNAs.
| miRNA name | Gene Annoation | Protein name | GO Function | GO name | KEGG name |
|---|---|---|---|---|---|
| hsa-miR-296-5p, hsa-miR-4433b-5p_R+1, hsa-miR-450b-5p_R-1, hsa-miR-590-3p, hsa-miR-664a-5p_R-1 | CD44 | CD44 antigen | cellular component | cytoplasm | ECM-receptor interaction |
| hsa-miR-150-5p, hsa-miR-424-5p_R-1 | F11 | Coagulation factor XI | cellular component | membrane | Complement and coagulation cascades |
| hsa-miR-129-5p, hsa-miR-590-3p | FGA | Fibrinogen alpha chain | cellular component | external side of plasma membrane | Complement and coagulation cascades |
| hsa-miR-129-5p, hsa-miR-450b-5p_R-1 | GSN | Gelsolin | molecular function | protein binding | Fc gamma R-mediated phagocytosis |
| hsa-miR-199b-5p_R-1 | KNG1 | Kininogen-1 | molecular function | zinc ion binding | Complement and coagulation cascades |
| hsa-miR-320b_R-2 | ORM1 | Alpha-1-acid glycoprotein 1 | molecular function | protein binding | - |
| hsa-miR-320b_R-2 | ORM2 | Alpha-1-acid glycoprotein 2 | cellular component | extracellular space | - |
| hsa-miR-17-3p_R-3, hsa-miR-296-5p | PLEC | Plectin | cellular component | cytoplasm | - |
| hsa-miR-34c-5p, hsa-miR-744-5p_R-2 | SERPINF2 | Alpha-2-antiplasmin | molecular function | protein binding | Complement and coagulation cascades |
| hsa-miR-320b_R-2 | SERPING1 | Plasma protease C1 inhibitor | molecular function | protein binding | Complement and coagulation cascades, Pertussis |
| hsa-miR-320b_R-2, hsa-miR-331-5p_R-2 | TF | Serotransferrin | molecular function | protein binding | Mineral absorption |
GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; ECM: extracellular matrix.
Serum levels of three miRNA-protein (predicted gene) pairs among multidrug-resistant tuberculosis, drug-sensitive tuberculosis patients, and healthy controls. MDR-TB: multidrug-resistant tuberculosis; DS-TB: drug-sensitive tuberculosis. A P-value of less than 0.05 indicates statistical significance using the Mann-Whitney U-test. *P < 0.05, **P < 0.01, ***P <0.001.
Further clinical data correlation analysis revealed negative correlations between KNG1 and IgG (r=-0.382, P=0.007), miR-4433b-5p and complement C4 (r=-0.343, P=0.021). Positive correlations were found between miR-424-5p and international normalized ratio (INR) (r=0.374, P=0.030), miR-199b-5p and high density lipoprotein cholesterol (HDL-C), D-dimer (r=0.295, P=0.040; r=0.767, P=0.016, respectively).
ROC analysis and decision tree establishment
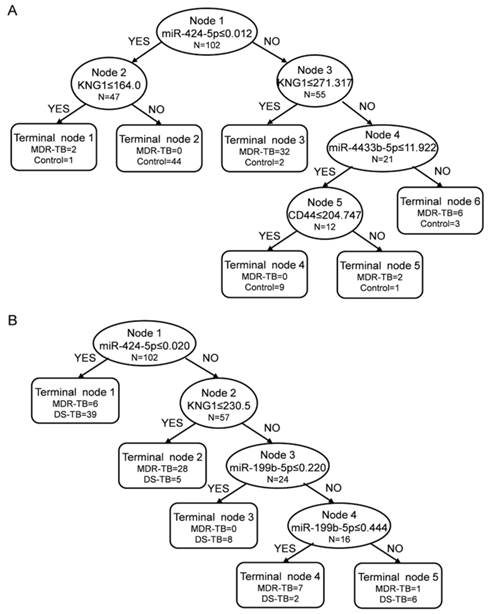
We performed ROC analysis to evaluate the sensitivity and specificity of the six biomarkers, and the areas under the curve (AUC) were 0.662 for miR-4433b-5p, 0.627 for CD44, 0.825 for miR-424-5p, 0.573 for F11, 0.663 for miR-199b-5p, and 0.870 for KNG1 in MDR-TB patients and healthy controls. The decision tree we established to identify MDR-TB patients and healthy controls possess a sensitivity of 100.00% and a specificity of 88.33% (overall accuracy 93.14%) (Fig. 6A). We used 10-fold cross validation to validate the diagnostic model. The decision tree discriminated MDR-TB patients from the healthy controls with an accuracy of 83.33% (83.33% sensitivity and 83.33% specificity).
In addition, the AUC were 0.599 for miR-4433b-5p, 0.548 for CD44, 0.493 for miR-424-5p, 0.527 for F11, 0.633 for miR-199b-5p, and 0.684 for KNG1 in MDR-TB patients and DS-TB patients. The corresponding decision tree possess a sensitivity of 83.33% and a specificity of 88.33% (overall accuracy 86.27%) (Fig. 6B). The decision tree discriminated MDR-TB patients from DS-TB patients with an accuracy of 75.49% (73.81% sensitivity and 76.67% specificity) by using 10-fold cross validation.
Discussion
MDR-TB patients are infected with strains resistant to both isoniazid and rifampicin. The highest MDR-TB rates have been found in countries of Eastern Europe and central Asia, where MDR strains threaten to become as common as pan-susceptible strains [25]. Clinical data analysis showed liver dysfunction (decreased albumin), immune system activation (increased globulin, IgG), complement system activation (increased C4, C3), and coagulation disorder (increased INR, fibrinogen) in MDR-TB patients as well as DS-TB patients (Table 1). In addition, there were significant differences of albumin, albumin/globulin ratio, and fibrinogen levels between MDR-TB patients and DS-TB patients (P<0.05). We screened proteomic and transcriptomic differences between MDR-TB patients, DS-TB patients, and healthy controls by using iTRAQ-2D LC-MS/MS and Solexa sequencing, and found 50 differentially expressed proteins and 43 differentially expressed miRNAs. Previous studies observed reduced serotransferrin [14] and fibronectin (FN1) [26] levels, and increased miR-199a-3p [27] and miR-122 [28] levels in TB patients than the healthy controls, which is consistent with our results. Moreover, differentially expressed proteins and miRNAs in our study were also involved in the pathogenesis of pulmonary TB. FN1 is an important extracellular matrix protein, which can combine with Mtb and regulate recruitment and retention of T cells [29], and may indicate pathological changes in granuloma formation of TB [26]. Haptoglobin (HP) was increased in TB patients and might indicate Traditional Chinese Medicine (TCM) syndrome [30], while 2-2 phenotype of HP was associated with high mortality rate of pulmonary TB [31]. Up-regulation of miR-132 was found after Mtb infection, which can limit macrophage response to IFN-γ by altering p300 protein [32]. The miR-125b was also increased after Mtb infection, allowing Mtb to subvert host immunity by translational repression and/or degradation of TNF mRNA [33].
Decision trees in the diagnostic model for the multidrug-resistant tuberculosis by the Biomarker Patterns Software. The diagnostic model shows the tree structure and sample distribution of the set. MDR-TB: multidrug-resistant tuberculosis; DS-TB: drug-sensitive tuberculosis. (A) diagnostic model for MDR-TB patients and healthy controls; (B) diagnostic model for MDR-TB patients and DS-TB patients.
In the present study, 22.00% (11 out of 50) of the proteins and 32.56% (14 out of 43) of the miRNAs correlated with each other (Fig. 4), suggesting that transcriptional regulation and post-transcriptional regulation play important roles in the pathogenesis of MDR-TB. Both the proteomic and transcriptomic data are important in deciphering the molecular processes involved in MDR-TB pathogenesis. The integrative transcriptomic and proteomic data not only highlighted a set of miRNAs/proteins that were possibly involved in the diagnosis of MDR-TB but also revealed molecular characterizations associated with the pathogenesis of MDR-TB in the hosts.
Integrative analysis highlighted the complement and coagulation cascades (Table 2). In our study, complement C3, C4, and fibrinogen increased in MDR-TB patients compared with healthy controls (P<0.001, P<0.001, and P<0.001, respectively). Moreover, fibrinogen level in MDR-TB patients was even higher than that of DS-TB patients (P=0.007) (Table 1), indicating complement and coagulation cascades disorder in MDR-TB patients. Zhu et al. [34] found reduced neutrophil peptides 1, 2 and 3 levels in MDR-TB patients compared with DS-TB patients, suggesting disorder of innate immune response and reduction of airway defense function. Tan et al. [10] proposed immune-suppression of Th1- and Th2-type responses in MDR-TB patients. Basile et al. [11] reported significantly increased IL-17+ cells level by MDR strains infection compared with DS strains, which might cause immunopathological damage. Therefore, we suspected that MDR-TB patients undergo abnormalities of immune system, limitations of pathogen clearance, and disorders of complement system and fibrinolysis system.
Significant differences of miR-4433b-5p, CD44, miR-424-5p, F11, miR-199b-5p, and KNG1 were found among MDR-TB patients, DS-TB patients and healthy controls (P<0.05). Ple et al. [35] reported miR-4433 as a kind of human platelet miRNAs. The predicted target-CD44 is a member of the hyaluronate receptor family of cell adhesion molecules, and plays an important role in lymphocyte trafficking [36]. Leemans et al. [37] identified CD44 as a new binding site for Mtb that mediates mycobacterial phagocytosis and protective immunity against pulmonary TB. Previous studies have found that miR-424 regulate transcriptional factor NFI-A, thereby controlling human monocyte/macrophage differentiation [38], and may lead to a disorder of T cells and B cells proportion [39]. Consistent with our results, increased levels of miR-424 have been found in active TB [39], Crohn's disease [40], and Alzheimer's disease [41]. F11 may be involved in leukocyte trafficking, either directly or, via inflammatory pathways, indirectly [42]. Furthermore, activation and/or consumption of F11 during infection or inflammation have been documented in animal models and human patients. Our results manifested that MDR-TB patients undergo immune system activation, and coagulation system disorders.
Consistent with the target gene prediction, our study also found a negative correlation between miR-199b-5p and KNG1 (r=-0.232, P=0.017). Qi et al. [27] and Zhang et al. [28] also evaluated the target gene prediction, but failed to validate predicted genes/proteins. The miR-199b-5p is also a kind of human platelet miRNA [35] while KNG1 is essential for blood coagulation. Analyses of the urine proteome of patients with chronic pancreatitis [43] and interstitial cystitis [44] showed reduced KNG1 levels. It has been suggested that KNG1 is proteolyzed by neutrophils at the inflammation sites. Lower level of KNG1 was found in the fibrosis/cirrhosis patients compared with the non-fibrosis hepatitis C patients [45], indicating that KNG1 was directly related to the severity of impaired liver function. Therefore, we suspected that the increased miR-199b-5p level in MDR-TB patients inhibited KNG1 target gene expression through post-transcriptional regulation, and then reduced KNG1 protein level, resulting in blood coagulation and fibrinolysis system imbalance. Moreover, MDR-TB patients suffered a more serious liver damage than DS-TB patients.
Our study also found positive correlation between KNG1 and F11 (r=0.308, P<0.0001) which is consistent with a genome-wide association study indicating that both KNG1 and F11 gene can regulate F11 protein level [46].
We established a diagnostic model for MDR-TB patients and healthy controls with a sensitivity of 100.00% and a specificity of 88.33% (overall accuracy 93.14%) (Fig. 6A). We also established a diagnostic model for MDR-TB patients and DS-TB patients with a sensitivity of 83.33% and a specificity of 88.33% (overall accuracy 86.27%) (Fig. 6B). The diagnostic model established by multiple markers increased sensitivity and specificity compared with a single marker model, leading to an earlier diagnosis.
In conclusion, we screened and identified proteomic and transcriptomic biomarkers for MDR-TB patients, and focused on complement and coagulation cascade by integrative analysis. By ELISA and SYBR green qRT-PCR validation, we established the MDR-TB diagnostic model with CD44, KNG1, miR-4433b-5p, miR-424-5p, and miR-199b-5p biomarkers. Our study proposes potential biomarkers for MDR-TB diagnose, and allows a better understanding of the pathogenesis of MDR-TB and DS-TB.
Supplementary Material
Supplementary Tables.
Abbreviations
TB: tuberculosis; MDR-TB: multidrug-resistance tuberculosis; DS-TB: drug-sensitive tuberculosis; Mtb: Mycobacterium tuberculosis; WHO: World Health Organization; DST: drug susceptibility testing; MODS: microscopic-observation drug susceptibility; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; ELISA: enzyme-linked immunosorbent assay; 2D LC-MS/MS: two-dimensional liquid chromatography-tandem mass spectrometry; SCX: strong cation exchange; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; ECM: extracellular matrix; ANOVA: one-way analysis of variance; ROC: receiver operating characteristic; AUC: areas under the curve; CD44: CD44 antigen; F11: coagulation factor XI; KNG1: kininogen-1; FN1: fibronectin; HP: haptoglobin; A/G: albumin/globulin ratio; HDL-C: high density lipoprotein cholesterol; LDL-C: low density lipoprotein cholesterol; APOA1: apolipoprotein A1; APOB: apolipoprotein B; CRP: C-reactive protein; INR: international normalized ratio; TCM: Traditional Chinese Medicine.
Acknowledgements
We thank the pulmonary DS-TB patients and MDR-TB patients enrolled in our study and the study coordinators and staff at the hospitals for their contributions to this research. This work was supported by grants from National Natural Science Foundation of China (No.81573709, No.81273882), the National Basic Research Program of China (No.2014CB543002) and the National Special Sci-Tech Projects (No.2012ZX10005001-006).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global tuberculosis report 2014. World Health Organization. http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf?ua=1
2. Multidrug-resistant TB (MDR-TB): 2014 Update. World Health Organization. http://www.who.int/tb/challenges/mdr/mdr_tb_factsheet.pdf?ua=1
3. Technical Guidance Group of the Fifth National TB Epidemiological Survey, The Office of the Fifth National TB Epidemiological Survey. The fifth national tuberculosis epidemiological survey in 2010. Chin J Antituber. 2012;34(8):485-508
4. Iseman MD. Evolution of drug-resistant tuberculosis: a tale of two species. Proc Natl Acad Sci U S A. 1994;91(7):2428-2429
5. Liang L, Wu Q, Gao L. et al. Factors contributing to the high prevalence of multidrug-resistant tuberculosis: a study from China. Thorax. 2012;67(7):632-638
6. Moore DA, Mendoza D, Gilman RH. et al. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J Clin Microbiol. 2004;42(10):4432-4437
7. Jain P, Hartman TE, Eisenberg N. et al. phi(2)GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J Clin Microbiol. 2012;50(4):1362-1369
8. Ahmad S, Jaber AA, Mokaddas E. Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis (Edinb). 2007;87(2):123-129
9. Pulimood AB, Peter S, Rook GW. et al. In situ PCR for Mycobacterium tuberculosis in endoscopic mucosal biopsy specimens of intestinal tuberculosis and Crohn disease. Am J Clin Pathol. 2008;129(6):846-851
10. Tan Q, Xie WP, Min R. et al. Characterization of Th1- and Th2-type immune response in human multidrug-resistant tuberculosis. Eur J Clin Microbiol Infect Dis. 2012;31(6):1233-1242
11. Basile JI, Geffner LJ, Romero MM. et al. Outbreaks of mycobacterium tuberculosis MDR strains induce high IL-17 T-cell response in patients with MDR tuberculosis that is closely associated with high antigen load. J Infect Dis. 2011;204(7):1054-1064
12. Zheng L, Leung E, Lee N. et al. Differential MicroRNA Expression in Human Macrophages with Mycobacterium tuberculosis Infection of Beijing/W and Non-Beijing/W Strain Types. PLoS One. 2015;10(6):e0126018
13. Karp NA, Huber W, Sadowski PG. et al. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010;9(9):1885-1897
14. Xu DD, Deng DF, Li X. et al. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2014;14(2-3):322-331
15. Chen G, Zhang Y, Jin X. et al. Urinary proteomics analysis for renal injury in hypertensive disorders of pregnancy with iTRAQ labeling and LC-MS/MS. Proteomics Clin Appl. 2011;5(5-6):300-310
16. Wang YS, Cao R, Jin H. et al. Altered protein expression in serum from endometrial hyperplasia and carcinoma patients. J Hematol Oncol. 2011;4:15
17. Jin GZ, Li Y, Cong WM. et al. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res. 2011;10(8):3418-3428
18. Xiao Z, Li G, Chen Y. et al. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J Histochem Cytochem. 2010;58(6):517-527
19. Ye H, Sun L, Huang X. et al. A proteomic approach for plasma biomarker discovery with 8-plex iTRAQ labeling and SCX-LC-MS/MS. Mol Cell Biochem. 2010;343(1-2):91-99
20. Tan Y, Pan T, Ye Y. et al. Serum microRNAs as potential biomarkers of primary biliary cirrhosis. PLoS One. 2014;9(10):e111424
21. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504
22. Yang JS, Li BJ, Lu HW. et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36(4):3035-3042
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-408
24. Liu J, Li Y, Wei L. et al. Screening and identification of potential biomarkers and establishment of the diagnostic serum proteomic model for the Traditional Chinese Medicine Syndromes of tuberculosis. J Ethnopharmacol. 2014;155(2):1322-1331
25. Seung KJ, Keshavjee S, Rich ML. Multidrug-Resistant Tuberculosis and Extensively Drug-Resistant Tuberculosis. Cold Spring Harb Perspect Med. 2015;5(9):a017863
26. Wang C, Li YY, Li X. et al. Serum complement C4b, fibronectin, and prolidase are associated with the pathological changes of pulmonary tuberculosis. BMC Infect Dis. 2014;14:52
27. Qi Y, Cui L, Ge Y. et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. 2012;12:384
28. Zhang X, Guo J, Fan S. et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One. 2013;8(12):e81076
29. Rojas RE, Thomas JJ, Gehring AJ. et al. Phosphatidylinositol mannoside from Mycobacterium tuberculosis binds alpha5beta1 integrin (VLA-5) on CD4+ T cells and induces adhesion to fibronectin. J Immunol. 2006;177(5):2959-2968
30. Jiang TT, Wang C, Wei LL. et al. Serum protein gamma-glutamyl hydrolase, Ig gamma-3 chain C region, and haptoglobin are associated with the syndromes of pulmonary tuberculosis in traditional Chinese medicine. BMC Complement Altern Med. 2015;15:243
31. Kasvosve I, Gomo ZA, Mvundura E. et al. Haptoglobin polymorphism and mortality in patients with tuberculosis. Int J Tuberc Lung Dis. 2000;4(8):771-775
32. Ni B, Rajaram MV, Lafuse WP. et al. Mycobacterium tuberculosis decreases human macrophage IFN-gamma responsiveness through miR-132 and miR-26a. J Immunol. 2014;193(9):4537-4547
33. Rajaram MV, Ni B, Morris JD. et al. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011;108(42):17408-17413
34. Zhu LM, Liu CH, Chen P. et al. Multidrug-resistant tuberculosis is associated with low plasma concentrations of human neutrophil peptides 1-3. Int J Tuberc Lung Dis. 2011;15(3):369-374
35. Ple H, Landry P, Benham A. et al. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746
36. DeGrendele HC, Estess P, Siegelman MH. Requirement for CD44 in activated T cell extravasation into an inflammatory site. Science. 1997;278(5338):672-675
37. Leemans JC, Florquin S, Heikens M. et al. CD44 is a macrophage binding site for Mycobacterium tuberculosis that mediates macrophage recruitment and protective immunity against tuberculosis. J Clin Invest. 2003;111(5):681-689
38. Rosa A, Ballarino M, Sorrentino A. et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104(50):19849-19854
39. Wang C, Yang S, Sun G. et al. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS One. 2011;6(10):e25832
40. Lin J, Welker NC, Zhao Z. et al. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27(4):602-608
41. Wang WX, Huang Q, Hu Y. et al. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121(2):193-205
42. Tucker EI, Gailani D, Hurst S. et al. Survival advantage of coagulation factor XI-deficient mice during peritoneal sepsis. J Infect Dis. 2008;198(2):271-274
43. Weeks ME, Hariharan D, Petronijevic L. et al. Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma. Proteomics Clin Appl. 2008;2(7-8):1047-1057
44. Canter MP, Graham CA, Heit MH. et al. Proteomic techniques identify urine proteins that differentiate patients with interstitial cystitis from asymptomatic control subjects. Am J Obstet Gynecol. 2008;198(5):553 e551-556
45. Henkel C, Schwamborn K, Zimmermann HW. et al. From proteomic multimarker profiling to interesting proteins: thymosin-beta(4) and kininogen-1 as new potential biomarkers for inflammatory hepatic lesions. J Cell Mol Med. 2011;15(10):2176-2188
46. Sabater-Lleal M, Martinez-Perez A, Buil A. et al. A genome-wide association study identifies KNG1 as a genetic determinant of plasma factor XI Level and activated partial thromboplastin time. Arterioscler Thromb Vasc Biol. 2012;32(8):2008-2016
Author contact
![]() Corresponding author: Dr. Ji-Cheng Li, Zhejiang University School of Medicine, 866 Yuhangtang Road, Hangzhou 310058, P.R. China. Tel: +86 571-88208088; Fax: +86 571-88208088. Email: lijichenedu.cn.
Corresponding author: Dr. Ji-Cheng Li, Zhejiang University School of Medicine, 866 Yuhangtang Road, Hangzhou 310058, P.R. China. Tel: +86 571-88208088; Fax: +86 571-88208088. Email: lijichenedu.cn.

 Global reach, higher impact
Global reach, higher impact