10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(2):257-269. doi:10.7150/ijbs.13538 This issue Cite
Research Paper
Wheat Transcription Factor TaAREB3 Participates in Drought and Freezing Tolerances in Arabidopsis
The National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Received 2015-8-12; Accepted 2015-10-28; Published 2016-1-1
Abstract
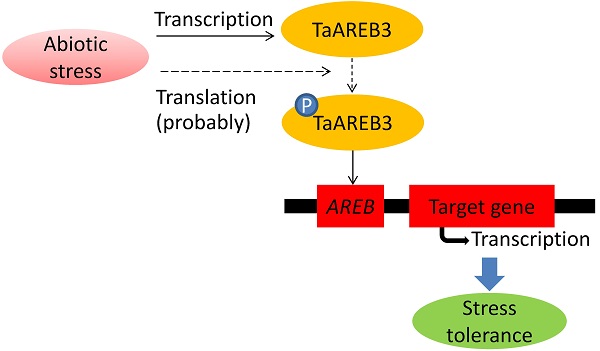
AREB (ABA response element binding) proteins in plants play direct regulatory roles in response to multiple stresses, but their functions in wheat (Triticum aestivum L.) are not clear. In the present study, TaAREB3, a new member of the AREB transcription factor family, was isolated from wheat. Sequence analysis showed that the TaAREB3 protein is composed of three parts, a conserved N-terminal, a variable M region, and a conserved C-terminal with a bZIP domain. It belongs to the group A subfamily of bZIP transcription factors. TaAREB3 was constitutively expressed in stems, leaves, florets, anthers, pistils, seeds, and most highly, in roots. TaAREB3 gene expression was induced with abscisic acid (ABA) and low temperature stress, and its protein was localized in the nucleus when transiently expressed in tobacco epidermal cells and stably expressed in transgenic Arabidopsis. TaAREB3 protein has transcriptional activation activity, and can bind to the ABRE cis-element in vitro. Overexpression of TaAREB3 in Arabidopsis not only enhanced ABA sensitivity, but also strengthened drought and freezing tolerances. TaAREB3 also activated RD29A, RD29B, COR15A, and COR47 by binding to their promoter regions in transgenic Arabidopsis. These results demonstrated that TaAREB3 plays an important role in drought and freezing tolerances in Arabidopsis.
Keywords: Abscisic acid, drought tolerance, freezing tolerance, gene expression
Introduction
Abiotic stresses, such as drought and freezing, limit world growth in wheat production [1, 2]. When encountering unfavorable environmental conditions, plants produce elevated levels of abscisic acid (ABA), which controls guard cell aperture and activates expression of many stress-related genes involved in signal transduction pathways [3, 4]. The ABA-mediated signal network in Arabidopsis has been well studied. The ABA receptor PYR/PYL/RCAR binds ABA, and the bound complex binds to, and inhibits, protein phosphatase activity of PP2Cs; inhibition by PP2C on SnRK2 kinase activity is released, in turn allowing accumulation of activated SnRK2 kinases, whose direct targets include SLOW ANION CHANNEL 1, an anion channel that controls guard cell aperture, and downstream transcription factors. Transcription factors activate expression of target genes [5-7].
AREBs, also named ABFs (ABRE binding factors), which belong to the bZIP transcription factor family, are regulated by ABA and have important roles in plant responses to abiotic stress [8-10]. Numerous ABA and/or stress-regulated genes contain a (C/T)ACGTGGC consensus sequence, known as the ABA responsive element (ABRE), in their promoter regions [11-13]. This cis-element was originally discovered in the RAB16 and Em genes in rice and wheat, respectively [14, 15]. The wheat Em gene contains an ACGT core sequence (CACGTG) known as a “G-box” [16]. Many AREB genes, such as AREB1/ABF2 and ABF3, which belong to the bZIP family, were found by yeast one hybrid experiments using the ABRE motif as bait [17, 18].
Basic region/leucine zipper (bZIP) proteins are classified by their DNA-binding domains [19]. They have a basic region that binds DNA and a leucine zipper dimerization motif [20]. In Arabidopsis, bZIP proteins can be placed in ten groups based on sequence similarity of the basic region and additional conserved motifs [8, 21]. AREB transcription factors belong to group A [18, 22].
The regulatory effects of AREB transcription factors on target genes are mainly induced by abiotic stresses, and thereby help plants to adjust to new environments [23, 24]. The target genes mostly encode late embryogenesis abundant proteins (LEA), osmotin, proline or betaine [25]. LEA gene families encompass numerous genes, such as RD (responsive to dehydration), COR (cold regulated), ERD (early responsive to dehydration) and RAB (responsive to ABA) [26, 27]. Their protein products are rich in hydrophilic amino acids that can protect protein integrity and cell structure from stress damage [28, 29]. Osmotic adjustment substances, such as proline and betaine, also have important roles in stress tolerance [30].
Many studies have reported that ABA and AREB play vital roles in abiotic stress tolerance in Arabidopsis [31, 32], but how they work in wheat remains largely unknown [33-36]. In this study, we isolated a novel AREB transcription factor, TaAREB3, from wheat. Functional analysis showed that TaAREB3 can bind to promoters of RD29A, RD29B, COR15A and COR47, and activate their expression under stress conditions. Transformed Arabidopsis overexpressing TaAREB3 showed enhanced freezing and drought tolerances.
Materials and methods
Plant materials and growth conditions
Hanxuan 10, a drought tolerant wheat cultivar was used for isolation of TaAREB3. Eight diploid species, including three A genome (T. urartu) accessions, two B genome (Aegilops speltoides) accessions and three D genome (Ae. tauschii) accessions were used to study gene origins.
Arabidopsis thaliana (Columbia ecotype) seeds were sown on MS medium with 3% (w/v) sucrose and 0.8% (w/v) agar. After a 2-day treatment at 4ºC the plates were transferred to a light-fitted incubator at 22ºC under long days (23 h light/1 h darkness). One week later, the seedlings were transferred to soil and cultured in a greenhouse at 22ºC (light) and 18ºC (darkness) under long day conditions (16 h light/8 h darkness).
Expression pattern of TaAREB3 in wheat
To detect TaAREB3 responses to abiotic stresses, two-week-old wheat seedlings were treated at low temperature (4ºC), or sprayed with 50 μM ABA solution. Plant tissues (stems, leaves, florets, anthers, pistils, seeds and roots) were sampled for expression analysis at 0, 1, 3, 6, 12, 24, 48, and 72 h after treatment.
Isolation of full-length cDNA and phylogenetic tree construction of TaAREB3
Using sequences of Arabidopsis AREB family members as references, a complete wheat AREB cDNA (named as TaAREB3) sequence information was obtained from the wheat A and D genome databases by tBLASTn [37, 38]. The full-length cDNA of the TaAREB3 gene was then cloned from wheat total RNA by reverse transcription polymerase chain reaction (RT-PCR). Through amino acid sequence analyses using protein BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) various bZIP genes were obtained from plant species, such as Arabidopsis, rice, barley, and Brachypodium distachyon. Phylogenetic analysis was performed by the maximum likelihood method using Molecular Evolutionary Genetics Analysis (MEGA) 5 software.
Genetic characterization of TaAREB3
To analyze the genomic origin of TaAREB3 genome-specific sequences of TaAREB3 were amplified using the wheat diploid progenitor species. DNA fragments were ligated into a pEASY-Blunt cloning vector, and the vectors were transformed into E. coli; plasmids were extracted and sequenced with a DNA Analyzer ABI 3730. Diversity of the sequencing results was analyzed with the MegAlign program in DNAStar software.
Subcellular localization of TaAREB3
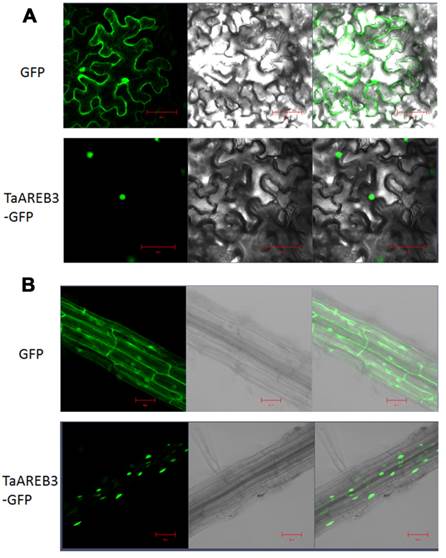
The full-length cDNA of TaAREB3 was fused into the modified vector pCAMBIA1300 with a 35S promoter and GFP tag [39]. The fusion vector and control (GFP) were transformed into Agrobacterium tumefaciens strain GV3101, which was then transformed into tobacco and Arabidopsis to obtain transient expression of TaAREB3 and stable transgenic lines, respectively. GFP fluorescence was recorded with a camera fitted to a confocal laser scanning microscope (Zeiss LSM700).
Phenotypic analyses of transgenic plants
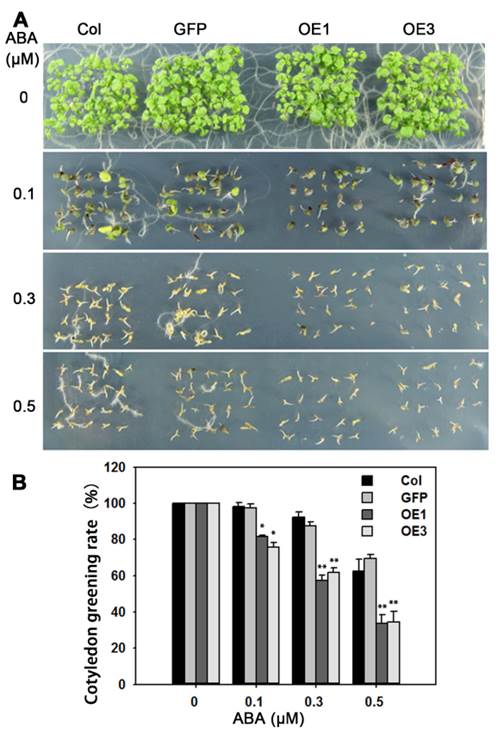
Post-germination seedling greening (that is, numbers of seedlings with green cotyledons) levels were scored for wild type and transgenics at one week after germination on MS media containing 0, 0.1, 0.3, 0.5 μM ABA [40]. Freezing tolerance assays were performed as described by Shi [41]. Briefly, two-week-old seedlings grown at 21ºC on 0.8% agar plates were treated at -12ºC for 12 h after a 4 d, 4ºC treatment (cold acclimation, CA), or at -10ºC for 12 h with no cold acclimation (NA). After the freezing treatments and a 2-d recovery at 21ºC, seedling survival rates were determined. Ion leakage was assayed as previously described [42]. Briefly, 1 g of two-week-old seedling tissue was immersed in 10 ml water for 30 min with gentle shaking, and conductivity of the solution was recorded. Conductivity was again measured after the solution was boiled for 10 min and cooled to room temperature. Ion leakage was calculated as the proportion of conductivity before boiling relative to that after boiling. Three-week-old seedlings of wild type (WT) and transgenic plants deprived of water for 2 weeks were analyzed for drought tolerance as described [43].
Transcriptional activation activity of TaAREB3
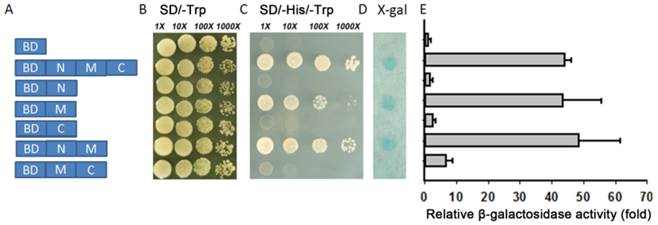
For transcriptional activation analysis full-length or partial sequences of TaARBE3 cDNA were fused in-frame with the GAL4 DNA binding domain in the pGBKT7 vector. The constructs were transformed into yeast strain AH109 containing the His3 and LacZ reporter genes following the Yeastmaker™ Yeast Transformation System 2 User Manual. Transformed yeast cells were cultured on synthetic-defined (SD) plates containing SD/-Trp, and SD/-His/-Trp media and then subjected to LacZ staining. O-nitrophenyl-β-d-galactopyranoside (ONPG) was used for quantitative assays of β-galactosidase activity.
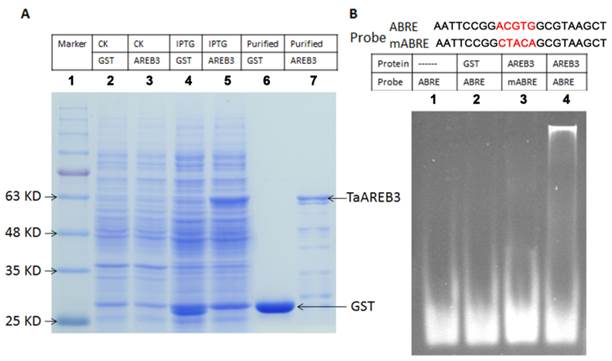
Purification of recombinant protein and electrophoretic mobility shift assays (EMSA)
The cDNA of TaAREB3 was fused to pGEX-4T1 with GST tag, and its protein was expressed in an E. coli BL21 cell line. Protein expression and purification were carried out following the standard protocol [44]. Optimal conditions for fusion were created by adding 0.2 mM isopropyl-β-d-thiogalactoside (IPTG), and incubating the E. coli at 37ºC for 6 h. The recombinant protein was purified by Glutathione Sepharose 4B (GE Healthcare). Two reverse complementary oligonucleotides were synthesized and annealed to one strand as probe. A mixture of a 0.5 μg DNA fragment and 20 μg purified protein was incubated in 30 mM Tris-HCl buffer (pH 8.0) at room temperature for 30 min, and then loaded onto a 10% native polyacrylamide gel [45]. After electrophoresis in 0.5×TBE buffer at 120 V for about 1 h, the gel was stained with ethidium bromide for visualization of DNA bands.
Expression of stress-responsive genes
WT and transgenic Arabidopsis were grown on MS plates for two weeks before seedlings were treated with 50 μM ABA for 0, 1, 3, and 6 h. Total RNAs were extracted and cDNA templates for real-time PCR were obtained as previously described [46]. Seven stress responsive genes (RD29A, RD29B, COR15A, COR47, RD22, RAB18 and P5CS) were detected by real-time PCR performed in triplicate with an ABI 7900 system using the SYBR Green PCR master mix kit according to the manufacturer's instructions. Primers for the abiotic stress responsive genes are listed in Supplementary table 2.
Chromatin immunoprecipitation (ChIP) analyses
Two-week-old seedlings of WT and three transgenic lines (two with pCAMBIA1300-TaAREB3-GFP and one with the empty vector pCAMBIA1300-GFP) were used for ChIP following the published procedure [47]. Anti-GFP antibody (Abcam, AB290) and Salmon Sperm DNA/Protein A Agarose (Millipore, 16-157) were used to precipitate the chromatin complex. ChIP products were dissolved in 500 μL TE and 5 μL was used for each real-time PCR. The primer sequences are listed in Supplementary table 3. Three independent experiments, each with three biological replicates were performed; each biological replicate had three technical replicates.
Results
Isolation and structural analysis of TaAREB3
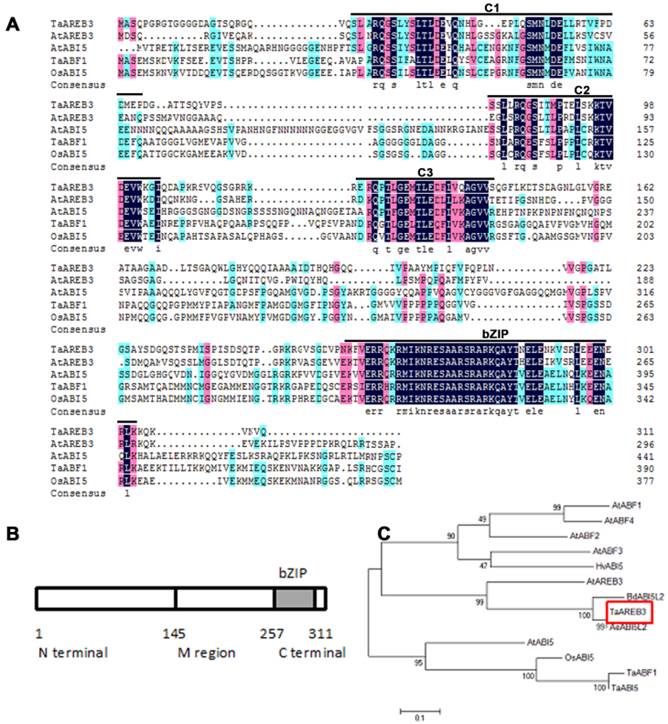
The full-length cDNA sequence of TaAREB3 gene was isolated and sequence analysis revealed that it contained a 936 bp sequence encoding a polypeptide of 311 amino acids with a predicted molecular mass of 34 kDa. The TaAREB3 protein contained a conserved N-terminal of 145 amino acids (a.a.) with Ser/Thr-like protein kinase phosphorylation sites, a variable M region (146-257 a.a.) and a conserved C-terminal with a bZIP domain (258-311 a.a.) (Figure 1A, 1B). Alignment and phylogenetic tree analysis revealed that TaAREB3 is most similar to the AREB subgroup member AtAREB3 that belongs to the bZIP family A group (Figure 1C).
Genetic characteristics of TaAREB3
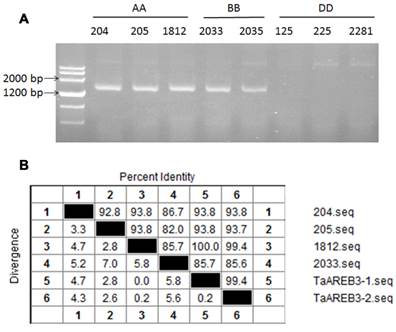
Eight accessions of wild diploid relatives of wheat species were used to identify the genomic origin of TaAREB3. As shown in Figure 2A, TaAREB3 gene was amplified only in accessions carrying A or B genomes. We then sequenced DNA fragments and found two variants in wheat cv. Hanxuan 10, namely, TaAREB3-1 and TaAREB3-2. TaAREB3-1 had 100% identity with the DNA fragment in T. uratu accession 1812 (Figure 2B), whereas TaAREB3-2 differed by 9 SNPs. TaAREB3-1, hereafter written as TaAREB3, was chosen for further functional analyses.
Amino acid sequences of AREB-related proteins and phylogenic relationships of selected AREB-related proteins. (A) Comparison of AREB-related proteins with high sequence similarity to TaAREB3. Amino acids shaded by color are conserved, with black indicating the highest similarity, red less, and blue least. Consensus sequence bases are shown below the sequences. The positions of the C1 to C3 conserved domains and bZIP domains are indicated by lines above the protein sequences. The C1, C2 and C3 domains contain potential phosphorylation sites [48]. The highly conserved bZIP domain suggests that all members of this subfamily recognize similar cis elements. (B) Structure of TaAREB3 protein. The TaAREB3 protein has three parts. (C) Phylogeny of AREB-related proteins. Alignments were made in Clustal W using default parameters.
Genetic characteristics of TaAREB3. (A) Analysis of TaAREB3 in 8 diploid accessions of hexapolid wheat progenitors. Accessions 204, 205 and 1812 are T. urartu (AA). 2033 and 2035 are Ae. speltoides (BB); and 125, 225 and 2281 are Ae. tauschii (DD). (B) Sequence diversity of TaAREB3 in the different accessions. Numbers 1 to 4 represent the TaAREB3 sequences in 204, 205, 1812, and 2033, respectively, whereas 5 and 6 represent TaAREB3-1 and TaAREB3-2 in Hanxuan 10.
Expression patterns of the TaAREB3 gene
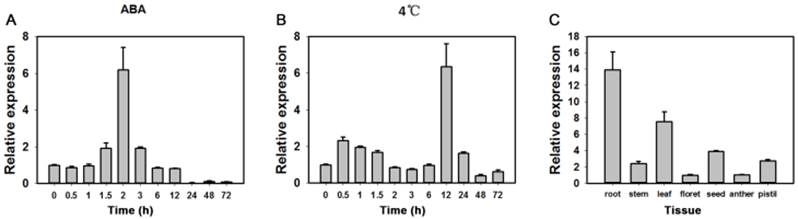
In tests for potential functions real-time PCR were performed to identify expression patterns of the TaAREB3 gene in 2-week-old wheat seedlings under various stress conditions. As shown in Figure 3A and 3B, TaAREB3 expression was induced by ABA and low temperature. Transcription levels of TaAREB3 in seedlings began to increase after exposure to ABA for 1.5 h and reached their highest levels at 2 h, when levels were 6-fold that of the non-treated control. After 12 h, expression declined to a level much lower than the control (Figure 3A). TaAREB3 expression was also induced under low temperature conditions, and mRNA accumulation reached its highest level of 6-fold that of the control treatment at 12 h and then decreased over the following 24 h (Figure 3B). These results suggested that TaAREB3 was induced by ABA and low temperature. Expression of TaAREB3 was identified by qRT-PCR in a range of tissues (Figure 3C), but was highest in roots, suggesting that its most important functions might occur in roots.
Subcellular localization of TaAREB3 protein
We examined the subcellular localization of TaAREB3-GFP in tobacco leaves and stable transgenic Arabidopsis plants with a fluorescence confocal microscope. Fluorescence was mainly detected in the nuclei in transient assays of epidermal leaf cells of tobacco (Figure 4A), and in the nuclei of root cells from T3 transgenic Arabidopsis lines (Figure 4B). In controls using the empty transformation vector carrying the GFP gene, GFP was distributed throughout the cell.
Expression patterns of TaAREB3. (A) Expression patterns of TaAREB3 following ABA treatment. Two-week-old wheat seedlings were sprayed with 50 μM ABA solution. Whole plants subjected to the treatment were sampled at 0, 1, 3, 6, 12, 24, 48, and 72 h. (B) Expression patterns of TaAREB3 following 4ºC treatment. Two-week-old wheat seedlings were treated at low temperature (4ºC). Whole plants subjected to the treatment were sampled at 0, 1, 3, 6, 12, 24, 48, and 72 h. (C) Expression patterns of TaAREB3 in various wheat tissues. Tubulin was used as the control. Three independent biological experiments, each with three technical replicates were performed. Error bars refer to 2× SE.
Subcellular localization of TaAREB3 protein. (A) GFP and TaAREB3-GFP localization in transiently transfected epidermal cells of tobacco leaves. The vector harboring GFP or TaAREB3-GFP was separately introduced into Agrobacterium, and transferred into tobacco leaves. Fluorescence was detected under a laser scanning confocal microscope. The left ones are green fluorescence images, middle ones are bright field images, and the right ones are merged images. Bar = 20 μm. (B) GFP and TaAREB3-GFP localization in a root of a stable transgenic Arabidopsis plant. The vector harboring GFP or TaAREB3-GFP was separately introduced into Agrobacterium, and transferred into Arabidopsis. The roots of transformed plants were observed under a laser scanning confocal microscope. The images from left to right are the same that in (A). Bar = 100 μm.
TaAREB3 protein specifically binds to the ABRE cis-element in vitro
The promoters of ABA and/or stress regulated genes share a consensus sequence known as the ABA-responsive element (ABRE) and the AREB proteins bind with ABRE cis-elements [13, 14]. Electrophoretic mobility shift assays were performed to test whether TaAREB3 binds to ABRE cis-acting elements in vitro. Expression and purification of protein were carried out using E. coli BL21 cultures harboring a pGEX-4T1 vector with the TaAREB3 cDNA, with GST used as a control (Figure 5A). Probes for ABRE and mutated ABRE (mABRE) are shown in Figure 5B. Purified TaAREB3 bound to ABRE was identified by much slower migration compared to the free probe (Figure 5B, lane 4), but TaAREB3 could not bind to mABRE (Figure 5B, lane 3), and GST could not bind to ABRE (Figure 5B, lane 2). Thus, TaAREB3 protein binds to the ABRE cis-element in vitro.
Transcriptional activation of TaAREB3
The yeast system was used to determine whether TaAREB3 could act as a transcriptional activator. Full-length TaAREB3 and different fragments were fused to the DNA-binding domain of GAL4 (pGBKT7 plasmid) to identify its ability to activate transcription of the upstream GAL4 sequence-driven His and LacZ reporter genes (Figure 6A). Yeast cells containing only pGBKT7 were used as the negative control. They all grew well on SD/-Trp, but only yeasts that carried M region fragments grew on the SD/-His/-Trp medium and showed LacZ staining (Figure 6B, 6C, 6D). This provided evidence that transcriptional activation activity was controlled by the M region. Yeasts with both the M and C terminal regions failed to grow on the SD/-His/-Trp medium and lacked LacZ staining, indicating that the C terminal may have an inhibitory effect on the M region's transcriptional activation capability. A relative quantitative assay of β-galactosidase activity using ONPG as substrate provided similar results (Figure 6E).
Overexpression of TaAREB3 in Arabidopsis affected ABA sensitivity
We transformed Arabidopsis with TaAREB3 and obtained more than 20 transgenic lines. The expression levels of TaAREB3 in 6 lines are shown in Supplementary figure 1. Lines OE1 and OE3 with different expression levels of TaAREB3 were selected for functional studies. Because TaAREB3 is induced by ABA we tested whether overexpression of TaAREB3 in Arabidopsis affects ABA sensitivity. We scored cotyledon greening at one week after completion of germination. At 0.3 μM ABA, about 60% of OE1 and OE3 germinated seedlings displayed green cotyledons compared to about 90% for WT (Figure 7A). This showed that overexpression of TaAREB3 led to severe ABA sensitivity and that TaAREB3 was involved in ABA signal transduction.
Purification of TaAREB3 protein and EMSA. (A) Expression and purification of TaAREB3 protein. GST and TaAREB3 protein are indicated with arrows. Lane 1 is a size marker. Lane 2 contains the entire E. coli (which has the pGEX-4T1 vector) protein suspension before IPTG induction. Lane 3 contains entire E. coli (which has the pGEX-4T1 vector fused with TaAREB3) protein suspension before IPTG induction. Lane 4 contains entire E. coli (which has the pGEX-4T1 vector) protein suspension after IPTG induction. Lane 5 contains entire E. coli (which has pGEX-4T1 vector fused with TaAREB3) protein suspension after IPTG induction. The TaAREB3 band is indicated in lanes 5 and 7. Lane 6 contains purified GST protein. Lane 7 contains purified TaAREB3 fused protein. (B) EMSA proves that TaAREB3 can bind ABRE in vitro. Probe sequences are shown above lanes. The mutant sequence difference is shown in red script. Lane 1 contains only the ABRE probe. Lane 2 contains the ABRE probe + GST protein. Lane 3 contains mABRE + TaAREB3 protein. Lane 4 contains ABRE + TaAREB3 protein.
Transcriptional activation activity. (A) Different parts of TaAREB3 were fused with the GAL4 DNA binding domain in a pGBKT7 vector. Transcriptional activation activities in subfigures B, C, D and E correspond to each of the seven combinations in A. (B) Vectors transformed into yeast strain AH109 containing the His3 and LacZ reporter genes all grew very well in SD/-Trp medium. (C) Only yeast cultures with vectors containing the M domain, but not the M + C domains, grew very well in SD/-Trp/-His medium. (D) LacZ staining. (E) Quantitative analysis of β-galactosidase activity using ONPG as substrate. X-axis indicates the relative fold of β-galactosidase activity, compared to to the empty vector. Three independent biological experiments, each with three technical replicates were performed. Error bars refer to 2× SE.
Overexpression of TaAREB3 in Arabidopsis affects ABA sensitivity. (A) Comparisons of cotyledon greening on MS, or MS supplemented with different concentrations of ABA between WT, GFP and two TaARE3-OE lines. (B) Statistical analyses of cotyledon greening rates. Three independent experiments were performed, and at least 50 seeds were counted in each experiment. Error bars refer to 2× SE. * P < 0.05, ** P < 0.01 (Student's t-test).
Overexpression of TaAREB3 confers freezing tolerance
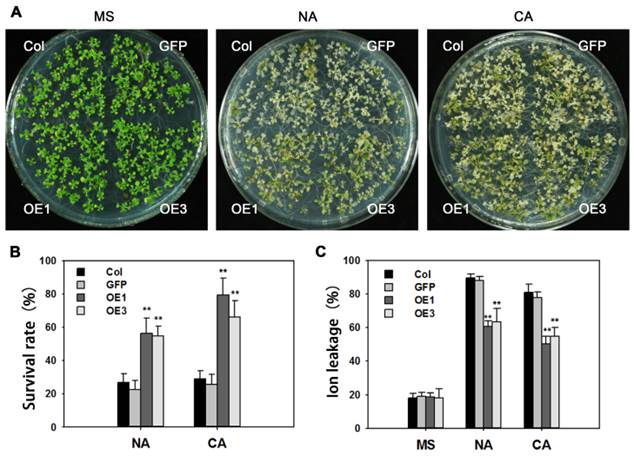
Many reports indicate that AREB proteins participate in freezing tolerance. After treatment of -10ºC for 12 h, about 80% of non-acclimated WT seedlings were dead, whereas survival in transgenic lines was around 50%. Following cold acclimation at 4ºC for 4 days, and -12ºC for 12 h, survival rates of cold-acclimated TaAREB3-OE seedlings were 45% higher than that for WT seedlings (Figure 8A and 8B). Ion leakages were measured as indicators of membrane stability. When exposed to freezing stress, TaAREB3-OE Arabidopsis seedlings showed about 30% ion less leakage than WT plants under both non-acclimated and acclimated conditions (Figure 8C). These results indicate that overexpression of TaAREB3 confers plant freezing tolerance.
TaAREB3 overexpression in Arabidopsis strengthens drought tolerance
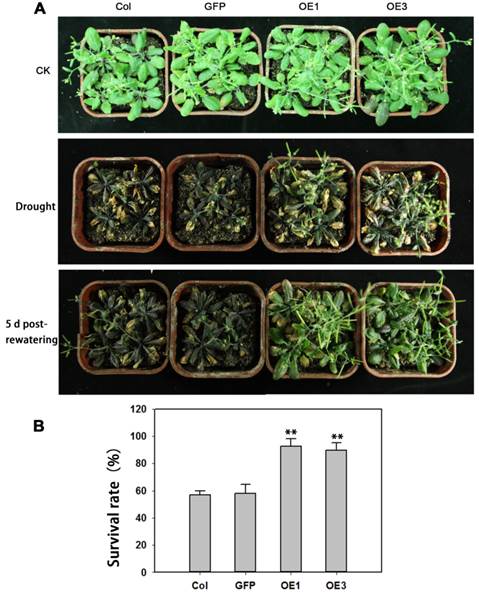
The performance of soil-grown TaAREB3 transgenic Arabidopsis seedlings was investigated under water deprivation conditions. No obvious morphological differences between the WT and transgenic lines were observed under normal conditions. After 3 weeks of normal growth, the plants were deprived of water for 2 weeks and then normal watering was resumed. WT and GFP plants displayed severe wilting phenotypes with some dead rosette leaves, but TaAREB3 transgenic lines displayed less wilting and better recovery (Figure 9A). After re-watering for 5 days, about 90% of transgenic plants survived, compared to about 60% for the controls (Figure 9B).
TaAREB3 regulated expression of stress-responsive genes
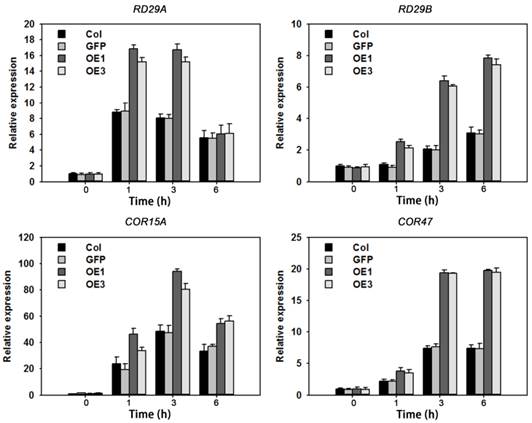
The above results suggested that overexpression of TaAREB3 caused enhanced ABA sensitivity and tolerance to drought and freezing. To further understand the molecular mechanisms we assayed expression levels of seven known abiotic stress-responsive genes [30, 49-52] in WT and TaAREB3 transgenic lines under ABA treatment and normal conditions. Transcripts of four genes (RD29A, RD29B, COR15A and COR47) were similar under normal conditions, but with ABA treatment, transcript levels were significantly higher in transgenic plants than in WT (Figure 10). Transcript levels of the other three genes (RD22, RAB18 and P5CS) were not significantly changed (data not shown).
Overexpression of TaAREB3 in Arabidopsis confers freezing tolerance. (A) Freezing tolerance phenotypes of TaAREB3-OE plants. The left panel shows two-week-old seedlings pre-treatment, and the other two show two-week-old seedlings treated at -10ºC for 12 h (non-acclimated, NA) or -12ºC for 12 h after 4ºC treatment for 4 d (cold acclimated, CA), and photographed after 2 d of recovery. (B) Survival rates of the seedlings in (A). Three independent experiments were performed and at least 50 seedlings were counted in each experiment. Error bars refer to 2× SE. ** P <0.01 (Student's t-test). (C) Ion leakage assays of the seedlings in (A). Three independent experiments were performed. Error bars refer to 2× SE. ** P <0.01 (Student's t-test).
Overexpression of TaAREB3 strengthens drought tolerance. (A) Drought tolerance in Arabidopsis wild type (Col), GFP and TaAREB3-OE lines grown in soil. CK, 5-week-old plants under well-watered. Drought, 3-week-old well-watered plants were deprived of watering for 2 weeks. The bottom panel of photograph was taken 5 days after re-watering. (B) Survival rates from drought tolerance experiments. Three independent experiments were performed and at least 30 seedlings were counted in each experiment. Error bars refer to 2× SE. ** P < 0.01 (Student's t-test).
TaAREB3 regulated expression of stress-responsive genes. Time courses of relative transcript levels of RD29A, RD29B, COR15A and COR47 in WT and a GFP control, and TaAREB3-OE lines treated with 50 μM ABA. Actin2 was used as the control. Three independent biological experiments were carried out, each with three technical replicates. Error bars refer to 2× SE.
TaAREB3 binds to stress-responsive gene promoters in vivo
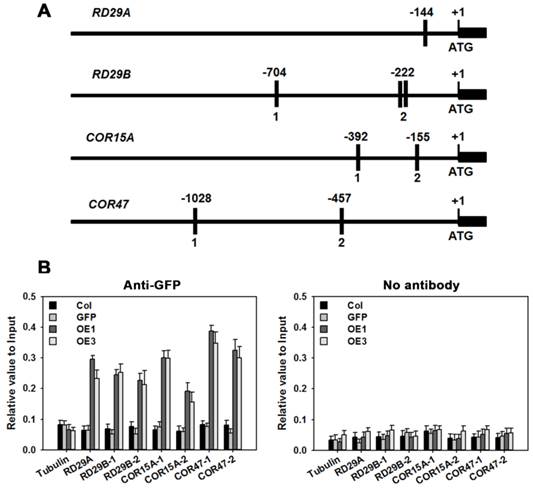
Based on information at the plantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), 1, 3, 2 and 2 copies of the ABRE cis-element were identified in the promoter regions of genes RD29A, RD29B, COR15A and COR47, respectively (Figure 11A). ChIP was performed to determine whether TaAREB3 could bind directly to the promoter regions in vivo. The transgenic TaAREB3-GFP lines and WT and GFP controls were compared. Anti-GFP was used for immunoprecipitation of protein-DNA complexes. Real-time PCR was used to detect DNA fragment abundance relative to the Input, i.e. all protein-DNA complexes saved before adding antibody. TaAREB3-GFP bound to the RD29A, RD29B, COR15A and COR47 promoter regions containing ABRE cis-elements, but did not bind to the Tubulin promoter region lacking ABRE cis-elements. In the absence of the GFP antibody, we could not detect TaAREB3-GFP bond to DNA fragments. These results indicated that TaAREB3 could bind the ABRE cis-elements in the promoter regions of the downstream genes and activate expression in transgenic Arabidopsis under stress conditions, thus leading to enhanced stress tolerance.
RD29A, RD29B, COR15A, COR47 are direct target genes of TaAREB3. (A) Bars indicate putative TaAREB3 binding sites. A 1.5 kb upstream sequence is shown. The ATG translation start site is at position +1. Primers used for ChIP PCR are listed in Supplementary table 3. (B) ChIP assays of TaAREB3-GFP binding to target gene promoters. Three independent experiments were performed, each with three biological replicates. Each biological replicate had three technical replicates. The results of independent experiments were similar. The data presented were from one experiment. Error bars refer to 2× SE.
Discussion
TaAREB3 is a novel member of the AREB family
We identified a novel wheat AREB transcription factor, TaAREB3. According to its structural features, the protein belongs to the bZIP family A group. In addition to its bZIP domain, TaAREB3 protein contains conserved C1, C2 and C3 three domains containing potential phosphorylation sites in the N terminals. The middle region of the protein is highly variable and therefore might contribute to functional diversity. Compared with other bZIP family A group members the C terminal of TaAREB3 lacks a C4 domain (where is also a potential phosphorylation site) [48].
The bZIP family A group in Arabidopsis, has 13 members, including the well studied AtbZIP39/ABI5, AtbZIP36/ABF2/AREB1, AtbZIP38/ABF4/AREB2, AtbZIP66/AREB3, AtbZIP40/GBF4, AtbZIP35/ABF1 and AtbZIP37/ABF3 [8]. Although these genes have been shown to take part in plant stress response, different genes may function in different environments. For example, overexpression of AtABF3 or AtABF4 both enhance drought tolerance [9]; OsABI5 overexpression enhances salinity tolerance [53]; AtAREB1/ABF2 is required for winter seedling survival and its overexpression confers multiple stress tolerances [31, 54]; and TaABL1 (ABI-like) improves multiple abiotic stress tolerances in transgenic plants [35]. Our results showed that overexpression of TaAREB3 can enhance plant tolerances to drought and freezing. Why and how different AREBs respond to different stresses need further study.
Different expression patterns of TaAREB3 and AtABI5
TaAREB3, also named as TaABI5L2 (ABA Insensitive 5 like 2) [38]. ABI5, which was reported as a bZIP family A group member was induced by ABA, cold and NaCl in Arabidopsis seedlings mainly immediately after germination [55]. In this study TaAREB3 was strongly induced by ABA and low temperature in young seedlings. Thus AtABI5 and TaAREB3 expression levels and response times differ under different abiotic stress conditions. AtABI5 expression was highest in mature seeds [56] suggesting that ABI5 controls ABRE-mediated transcription mainly in seeds [4]. Although TaAREB3 activity was detected in a range of tissues, its expression was highest in roots. This suggests that TaAREB3 has a different expression pattern from AtABI5. We therefore compared the promoter regions of these two genes, and found there was very low similarity (data not shown), perhaps explaining the different expression patterns.
Target gene differences between TaAREB3 and other AREBs
We compared TaAREB3 and other AREB-regulated abiotic stress responsive genes. For example, ABI5 mainly activates several late embryogenesis-abundant genes, including AtEm1 and AtEm6 [48], and Tamarix hispida ThABF1 directly regulates expression of ThbZIP1 [57]. Expression levels of stress-responsive genes RD29B and RAB18 were up-regulated in TaABL1 transgenic lines under both stress and normal conditions [35]. Wabi5 functions as a transcriptional activator and positively regulates the expression of the Wdhn13, Wrab18, and Wrab19 genes under drought stress [58]. We found that four stress-responsive genes (RD29A, RD29B, COR15A, COR47) were more highly expressed in TaAREB3 transgenic plants than in controls under ABA treatment. This suggests that although AREB protein can bind to ABRE, binding to different downstream target genes may be specific. Under normal conditions expression of the four genes was the same in control and transgenic plants suggesting that overexpression of TaAREB3 was not sufficient to active expression of downstream target genes. More TaAREB3 target genes need to be studied by ChIP sequencing.
TaAREB3 may be phosphorylated and function under stress
Although transgenic Arabidopsis plants had TaAREB3 protein under normal conditions, WT and transgenic plants displayed the same phenotypes and same expression levels of TaAREB3 target genes. This suggests that TaAREB3 must be activated before it functions. Previous studies indicated that ABA and stress can induce both transcriptional and post-translational regulation of AREBs [59, 60]. In-gel kinase assays showed that the activation of AREB proteins requires phosphorylation by protein kinase SnRK2 in an ABA-dependent manner [18]. Phosphorylation was necessary for AREB to induce downstream gene expression [61, 62]. TaAREB3 has conserved C1, C2, and C3 domains in the N terminal; these contain phosphorylation sites and therefore may function only after being phosphorylated by SnRK2 under stress conditions. Nevertheless, further study is required to determine how this protein is post-translationally regulated.
In conclusion, we characterized a novel wheat AREB transcription factor TaAREB3, which can be induced by ABA and low temperature. In heterologous Arabidopsis TaAREB3 can bind to the promoters of RD29A, RD29B, COR15A, and COR47 to initiate activity. Overexpression of TaAREB3 in Arabidopsis not only enhanced ABA sensitivity, but also increased drought and freezing tolerances. These results contribute to our understanding of the TaAREB3 gene and provide a candidate gene for improvement of stress tolerance in wheat.
Supplementary Material
Supplementary table 1: Primers used for vector construction; Supplementary table 2: Primers used for detecting gene expression by real-time PCR and semi-quantitative PCR; Supplementary table 3: Primers used for ChIP assays; Supplementary Figure 1: Semi-quantitative PCR detecting different expression levels in Arabidopsis lines transformed with TaAREB3.
Acknowledgements
We thank Professor Robert A. McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for revising the manuscript. This study was supported by the National High-tech R&D Program (2011AA100501), National Natural Science Foundation of China (31461143024) and Agricultural Science and Technology Innovation Program (ASTIP).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fuller MP, Fuller AM, Kaniouras S, Christophers J, Fredericks T. The freezing characteristics of wheat at ear emergence. European Journal of Agronomy. 2007;26:435-41
2. Martin B, Ruiz-Torres NA. Effects of water-deficit stress on photosynthesis, its components and component limitations, and on water use efficiency in wheat (Triticum aestivum L.). Plant Physiology. 1992;100:733-9
3. Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Reports. 2013;32:959-70
4. Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of Plant Research. 2011;124:509-25
5. Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068-71
6. Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A. et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064-8
7. Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino-Powell KM. et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21259-64
8. Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T. et al. bZIP transcription factors in Arabidopsis. Trends in Plant Science. 2002;7:106-11
9. Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343-57
10. Zhang LN, Zhang LC, Xia C, Zhao GY, Liu J, Jia JZ. et al. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiologia Plantarum. 2015;153:538-54
11. Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251-64
12. Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. The Journal of Biological Chemistry. 2000;275:1723-30
13. Busk PK, Pages M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425-35
14. Guiltinan MJ, Marcotte WR Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267-71
15. Mundy J, Yamaguchi-Shinozaki K, Chua NH. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1406-10
16. Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:7089-93
17. Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M. et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470-88
18. Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11632-7
19. Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:105-68
20. Latchman DS. Transcription factors: An overview. The International Journal of Biochemistry & Cell Biology. 1997;29:1305-12
21. Correa LGG, Riano-Pachon DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One. 2008:3
22. Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4782-7
23. Koornneef M, Leon-Kloosterziel KM, Schwartz SH, Zeevaart JAD. The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiology and Biochemistry. 1998;36:83-9
24. Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology. 2000;3:217-23
25. Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247-73
26. Welin BV, Olson A, Nylander M, Palva ET. Characterization and differential expression of dhn/lea/rab-like genes during cold acclimation and drought stress in Arabidopsis thaliana. Plant Molecular Biology. 1994;26:131-44
27. Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008:9
28. Chinnusamy V, Zhu J, Zhu JK. Salt stress signaling and mechanisms of plant salt tolerance. Genetic Engineering. 2006;27:141-77
29. Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin CT, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13404-9
30. Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchishinozaki K. et al. Correlation between the induction of a gene for delta(1)-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic-stress. Plant Journal. 1995;7:751-60
31. Sharma PD, Singh N, Ahuja PS, Reddy TV. Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter. Molecular Biology Reports. 2011;38:5147-59
32. Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J. et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant Journal. 2010;61:672-85
33. Johnson RR, Shin M, Shen JQ. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Molecular Biology. 2008;68:93-103
34. Harris LJ, Martinez SA, Keyser BR, Dyer WE, Johnson RR. Functional analysis of TaABF1 during abscisic acid and gibberellin signalling in aleurone cells of cereal grains. Seed Science Research. 2013;23:89-98
35. Xu DB, Gao SQ, Ma YZ, Xu ZS, Zhao CP, Tang YM. et al. ABI-like transcription factor gene TaABL1 from wheat improves multiple abiotic stress tolerances in transgenic plants. Functional & Integrative Genomics. 2014;14:717-30
36. Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology. 2002;130:837-46
37. Jia J, Zhao S, Kong X, Li Y, Zhao G, He W. et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91-5
38. Ling HQ, Zhao SC, Liu DC, Wang JY, Sun H, Zhang C. et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87-90
39. Gong ZH, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803-14
40. Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant Journal. 2010;63:749-65
41. Shi YT, Tian SW, Hou LY, Huang XZ, Zhang XY, Guo HW. et al. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and Type-A ARR genes in Arabidopsis. Plant Cell. 2012;24:2578-95
42. Lee H, Guo Y, Ohta M, Xiong LM, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO Journal. 2002;21:2692-702
43. Mao XG, Chen SS, Li A, Zhai CC, Jing RL. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One. 2014:9
44. Chong L. Molecular cloning - A laboratory manual, 3rd edition. Science. 2001;292:446 -
45. Hu L, Liang W, Yin C, Cui X, Zong J, Wang X. et al. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell. 2011;23:515-33
46. Mao XG, Zhang HY, Qian XY, Li A, Zhao GY, Jing RL. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. Journal of Experimental Botany. 2012;63:2933-46
47. Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols. 2008;3:1018-25
48. Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F. et al. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell. 2002;14:1391-403
49. Nakayama K, Okawa K, Kakizaki T, Honma T, Itoh H, Inaba T. Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiology. 2007;144:513-23
50. Thalhammer A, Hundertmark M, Popova AV, Seckler R, Hincha DK. Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochimica et Biophysica Acta. 2010;1798:1812-20
51. Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular & General Genetics. 1993;236:331-40
52. Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14570-5
53. Zou MJ, Guan YC, Ren HB, Zhang F, Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Molecular Biology. 2008;66:675-83
54. Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant Journal. 2004;40:75-87
55. Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiology. 2002;129:1533-43
56. Kim SY, Ma JZ, Perret P, Li ZS, Thomas TL. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiology. 2002;130:688-97
57. Ji XY, Liu GF, Liu YJ, Nie XG, Zheng L, Wang YC. The regulatory network of ThbZIP1 in response to abscisic acid treatment. Frontiers in Plant Science. 2015:6
58. Kobayashi F, Maeta E, Terashima A, Takumi S. Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiologia Plantarum. 2008;134:74-86
59. Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K. et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1988-93
60. Chen YT, Liu HX, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant Journal. 2013;75:965-76
61. Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T. et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant Journal. 2005;44:939-49
62. Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiologia Plantarum. 2013;147:15-27
Author contact
![]() Corresponding author: Ruilian Jing, email: jingruiliancn
Corresponding author: Ruilian Jing, email: jingruiliancn

 Global reach, higher impact
Global reach, higher impact