10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(5):505-517. doi:10.7150/ijbs.14212 This issue Cite
Research Paper
Transcriptional Reactivation of OTX2, RX1 and SIX3 during Reprogramming Contributes to the Generation of RPE Cells from Human iPSCs
1. Stem Cell Research Center, Department of Cell Biology, School of Basic Medical Sciences, Peking University, Haidian District, Beijing, 100191, China
2. Department of Histology and Embryology, Institute of Chinese Medicine, Hunan University of Chinese Medicine, Science Garden District of Hanpu, Changsha, Hunan, 410208, China
3. Peking University Eye Center, Peking University Third Hospital, Beijing, 100191, China
4. Lund Stem Cell Center, University Hospital, Lund University, Lund, 22242, Sweden
5. Beijing Cellonis Biotechnologies Co.Ltd, Zhongguancun Bio-Medicine Park, Beijing, 100191, China
6. Department of Biology, College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, Zhejiang, 321004, China.
* These authors contributed equally to this work.
Received 2015-10-24; Accepted 2016-1-16; Published 2016-2-20
Abstract
Directed differentiation of human induced pluripotent stem cells (iPSCs) into retinal pigmented epithelium (RPE) holds great promise in cell replacement therapy for patients suffering from degenerative eye diseases, including age-related macular degeneration (AMD). In this study, we generated iPSCs from human dermal fibroblasts (HDFs) by electroporation with episomal plasmid vectors encoding OCT4, SOX2, KLF4, L-MYC together with p53 suppression. Intriguingly, cell reprogramming resulted in a metastable transcriptional activation and selective demethylation of neural and retinal specification-associated genes, such as OTX2, RX1 and SIX3. In contrast, RPE progenitor genes were transcriptionally silent in HDFs and descendant iPSCs. Overexpression of OCT4 and SOX2 directly stimulated the expression of OTX2, RX1 and SIX3 in HDFs and iPSCs. Luciferase and chromatin immunoprecipitation (ChIP) assays further identified an OCT4- and two SOX2-binding sites located in the proximal promoter of OTX2. Histone acetylation and methylation on the local promoter also participated in the reactivation of OTX2. The transcriptional conversion of RX1 and SIX3 genes partially attributed to DNA demethylation. Subsequently, iPSCs were induced into the RPE cells displaying the characteristics of polygonal shapes and pigments, and expressing typical RPE cell markers. Taken together, our results establish readily efficient and safe protocols to produce iPSCs and iPSC-derived RPE cells, and underline that the reactivation of anterior neural transcription factor OTX2, eye field transcription factor RX1 and SIX3 in iPSCs is a feature of pluripotency acquisition and predetermines the potential of RPE differentiation.
Keywords: induced pluripotent stem cells, retinal pigment epithelium, reprogramming, epigenetic modification
Introduction
Retinal pigment epithelium (RPE) is a monolayer of polarized and terminally differentiated pigmented cells lying between the photoreceptor cells layer and the choroid. Apart from absorbing stray light, RPE cells are crucial for the integrity and proper visual function of the outer retina [1]. Aged-related macular degeneration (AMD), triggered mainly by the recession and death of RPE, is the leading cause of blindness and visual impairment in the elderly worldwide. Currently, there is no effective treatment for the more prevalent wet form of AMD [2]. Transplantation of RPE and other cells is an emerging field of AMD therapy. Various cell types of different origin have been examined for their potential application in treating AMD, including immortalized RPE cell lines, RPE isolated from donors, RPE derived from embryonic stem cells (ESCs), and other non-RPE cell lines [3-7].
The recent discovery of induced pluripotent stem cells (iPSCs) that generated by reprogramming adult cells with defined factors (c-MYC, OCT4, SOX2, KLF4) might provide another promising source of immune-matched autologous cells for RPE cell replacement [8]. iPSCs can differentiate into RPE cells that are highly similar to native RPE and induced RPE derived from ESCs [9-12]. Transplantation of iPSC-derived RPE has been shown to improve visual function in a rat model of AMD [13]. Nevertheless, the application of iPSCs in regenerative medicine encounters several safety issues. For instance, c-MYC is a well-characterized proto-oncogene with great tumorigenic potential. Recent study has generated human iPSCs replacing c-MYC with transformation-deficient L-MYC to reduce the risk of tumorigenicity. L-MYC protein has shorter amino acid sequences than c-MYC in the N-terminus, along with significantly lower transformation property [14]. In addition, iPSCs generated using integrative viruses usually lead to genomic instability and rapid immunological rejection by the recipient after transplantation. Moreover, iPSCs usually suffer from incomplete reprogramming and retain a residual memory of somatic donor cells in transcriptional and epigenetic pattern. Thus, incompletely reprogrammed cells cannot fully replicate the pluripotent features of ESCs and still display heterogeneous propensity for lineage specification [15]. Therefore, the methods for reprogramming should also be refined to avoid serious risks, and achieve complete conversion of somatic cells to pluripotency state.
In this study, we generated iPSCs from human dermal fibroblasts (HDFs) using viral free non-integrating episomal vectors and further expanded the iPSC clones in defined culture conditions. Under feeder-free and xeno-free conditions, the generated iPSCs were sequentially differentiated to neural and then to RPE fate by chemical compounds. The iPSC-derived pigmented cells exhibit characteristics of authentic RPE cells, including proper pigmentation, morphology, and specific marker expression. The eye field is a presumptive eye tissue existing at the neural plate stage prior to optic vesicle formation. In vertebrates, several transcription factors such as OTX2, LHX2, SIX3, RX1 or SIX6 are required in the eye field [16]. Notably, we found that OTX2, RX1 and SIX3 were epigenetically silenced in parental donor cells and underwent transcriptional reactivation during cellular reprogramming. We further showed that OCT4 and SOX2 are direct regulators of these genes. Our finding implicates that the selective reactivation of OTX2, RX1 and SIX3 during reprogramming, is a prerequisite for rendering the RPE differentiation potential to iPSCs. Taken together, our results optimized the protocols of iPSCs acquisition and RPE cells induction with minimizing potential risks in therapeutic application, and demonstrated that OTX2, RX1 and SIX3 are selectively reactivated by OCT4 and SOX2 during reprogramming. These findings are helpful for evaluating the reprogramming efficiency and RPE propensity of reprogrammed cells.
Materials and Methods
Cell culture
Human dermal fibroblasts (HDFs) from aborted fetuses were purchased from Peking Union Medical College Cell Resource Center. HDF cells were cultured in standard culture media containing DMEM medium supplemented with 10% fetal bovine serum (Hyclone, USA). ARPE-19 cells (ATCC, USA) were established in DMEM/F12 medium containing 20% fetal bovine serum, sodium bicarbonate, and L-glutamine. H9 human ES cells and iPS cells were maintained in feeder-free Essential 8 Medium (E8, Life Technology) on matrigel (BD Biosciences). ESCs and iPSCs were passaged every 3-5 days by 0.5 mM EDTA (Life Technology).
Generation of iPSCs from HDFs
Yamanaka episomal plasmids obtained from Addgene were used in experiments: pCXLE-hOCT3/4-shp53 (#27077), pCXLE-hSK (#27078), pCXLE-hUL (#27080) and pCXLE-EGFP (#27082). 5 × 105 HDFs were counted and resuspended in nucleofector solution supplied in the Amaxa Nucleofector kit (Lonza). Episomal plasmids were added to the cell suspensions at 10 μg each per reaction. Cell suspensions were transfected using program U-023 on a Amaxa Nucleofector device. Immediately after transfection, cells were resuspended in fibroblast medium and transferred to a 60 mm tissue culture dish coated with matrigel. The next day, the culture medium was changed to Essential 6 medium (E6, Life technology) supplemented with 100 ng/mL bFGF (Peprotech) and 1μM hydrocortisone (Sigma). After 3-5 day when the cell confluence at 60-70%, E6 medium was supplemented with bFGF and 100 μM sodium butyrate (Sigma). By day 25-30 post transfection, colonies emerged with distinct ESC-like features of a compact and flat appearance and EGFP-negative, an indicative of exogenous gene disappearance. A single colony was picked and passaged with E8 medium. iPSCs more than 10 passages were used for differentiation into RPE cells.
RNA isolation, RT-PCR and quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen). cDNAs were synthesized using the M-MLV reverse transcription kit (Promega) following the manufacturer's instructions. PCR was performed with ExTaq (Takara). Quantitative PCR was performed using SYBR Green Real-time PCR Master Mix (Promega) and analyzed with the Mx3000P real-time PCR system (Agilent Technologies, Santa Clara, USA). The relative expression levels were normalized to those of GAPDH based on the Delta Ct Method. Primers for RT-PCR and real-time PCR are listed in Table S1.
Characterization of iPSCs
Alkaline phosphatase (ALP) staining was performed using the alkaline phosphatase kit (ZSGB-BIO) according to the manufacturer's instructions. To test the in vivo pluripotency, teratoma formation was performed. In brief, iPSCs were suspended at 1 × 107 cells/ml in DMEM/F12. 100 μl of cell suspension (1 × 106 cells) were injected subcutaneously into the posterior limbs of 4-w-old BALB/c SCID mice. After two months, teratomas were dissected and fixed in 4% paraformaldehyde. Paraffin-embedded tissue was sliced and stained with hematoxylin and eosin. Chromosomal G-band analyses were performed in the Cytogenetics Laboratory at Peking University.
In vitro differentiation of iPSCs
For retinal differentiation, iPS colonies were treated with Y-27632 (Tocris, 10 μM) for 1 hour, and dissociated into clumps with 0.5 mM EDTA. The produced clumps were transferred to low attachment culture dishes in embryoid bodies (EBs) medium containing DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.1 mM non-essential amino acids, 2 mM L-glutamine, and 20% KSR (Knock Out Replacement, Invitrogen). After three days of EBs formation, the medium was switched to differentiation medium (GMEM, 0.1 mM non-essential amino acids, 1 mM pyruvate, and 0.1 mM 2-mercaptoethanol) containing 20% KSR for another three days, then in 15% KSR-containing differentiation medium for 9 days, and finally in 10% KSR-containing differentiation medium. CKI-7 (Sigma, 5 μM) and SB-431542 (Sigma, 5 μM) were added to the medium for 21 days during suspension culture. The medium was changed every 2-3 days. Formed cell aggregates were then replated on matrigel-coated dishes. In adherent cultures, cells were incubated in 10% KSR-containing differentiation medium. Approximately after 10 weeks, these pigmented clusters were isolated using a needle and spread onto matrigel-coated plates for expansion.
Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde. Permeabilization was performed with 0.2% Triton X-100. Primary antibodies included NANOG, MITF, RPE65, ZO-1 (Abcam), OCT4, TRA-1-81 (Santa Cruz), SOX2, SSEA4, PAX6 (Chemicon). Secondary antibodies used were Alexa Fluor 488 or 594 goat anti-mouse/rabbit IgG antibody (ZSGB-BIO) diluted at 1:500. Cell nuclei were counterstained with Hoechst 33342. Labeled cells were imaged with a laser-scanning confocal microscope (Leica).
RNA interference (RNAi)
To inhibit OCT4 or SOX2 expression in the H9 and iPSCs, RNA interference-mediated silencing was performed using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer's instructions. The SOX2 siRNAs (sense, 5′-GGAAUGGACCUUGUAUAGAUCtt-3′; anti-sense, 5′-UCUAUACAAGGUCCAUUCCCC-3′) and the OCT4 siRNAs (sense, 5'-GGAUUAAGUUCUUCAUUCAtt-3'; anti-sense, 5'-UGAAUGAAGAACUUAAUCCCA-3') were synthesized by GenePharma (Shanghai, China). The cells were harvested 48 h after transfection. Western blot assay was used to evaluate the RNAi effect.
Western blot analysis
Primary antibody were used, including OCT4 (Santa Cruz), SOX2 (Chemicon) and ACTIN (Santa Cruz). Images were captured by the Odyssey Fluorescent Western Scanning System (LI-COR, Lincoln, USA). Data were analyzed with the Odyssey Imaging System.
Methylation analysis
Primers for all the genes were designed to cover the region with the most CpG sites. The selected amplicon was located in the promoter region of the gene. The mass spectra were collected using a MassARRAY Compact MALDI-TOF (SEQUENOM, BioMiao Biological Technology, Beijing) and the spectra's methylation ratios were generated by the EpiTYPER software.
Luciferase assay
A pGL3-basic plasmid containing the OTX2 promoter region (ATG-1008 bp) and mutant SOX2 and OCT4 sites were synthesized by Genenode (Beijing, China). Transfections were carried out in 293T cells using Neofect DNA transfection reagent (Neofect Biotech, China). Cells that were transfected with the expression vectors pBK-SOX2, pBK-OCT4, and the reporter vectors were harvested 48 h after transfection. Reporter activities were measured by using the dual-luciferase reporter assay system (Promega). Each assay was performed in duplicate, and all results are shown as mean ± SD for at least three independent assays.
Chromatin immunoprecipitation (ChIP) assays
For ChIP experiments, chromatin was cross-linked with 1% formaldehyde for 10 min at room temperature. Cells were washed twice in ice-cold PBS, resuspended in the SDS-lysis buffer, and sonicated until the cross-linked chromatin was sheared to an average DNA fragment length of 100-800 bp. Normal IgG was used as a negative control, and antibodies against SOX2, OCT4, acetyl-histone H3K9 and trimethyl-histone H3K9 (Abcam) used for each immunoprecipitation. The precipitated DNA was amplified by real-time PCR for fragments of the OTX2 promoter. The primers are 5'- TTAAAATCTCTGCCATGGAAA-3' and 5'-AGAACAAAAACCCGTGCCTT-3'. Each value represents the average of at least 3 independent experiments.
Phagocytosis assay
Cells were incubated in medium containing Cy3-conjugated 1 μm polystyrene microspheres (Invitrogen) at a concentration of 1 × 108 beads/mL at 37°C for 2 and 5 h respectively. The fluorescence signal was observed with a laser-scanning confocal microscope.
Statistical analysis
All data were expressed as the mean ± SD from at least three independent experiments. The Student's two-tailed t-test with significance set at p<0.05 was used to determine statistical significance for each assay. Error bars indicate standard deviation.
Results
Generation and characterization of iPSCs from human dermal fibroblasts
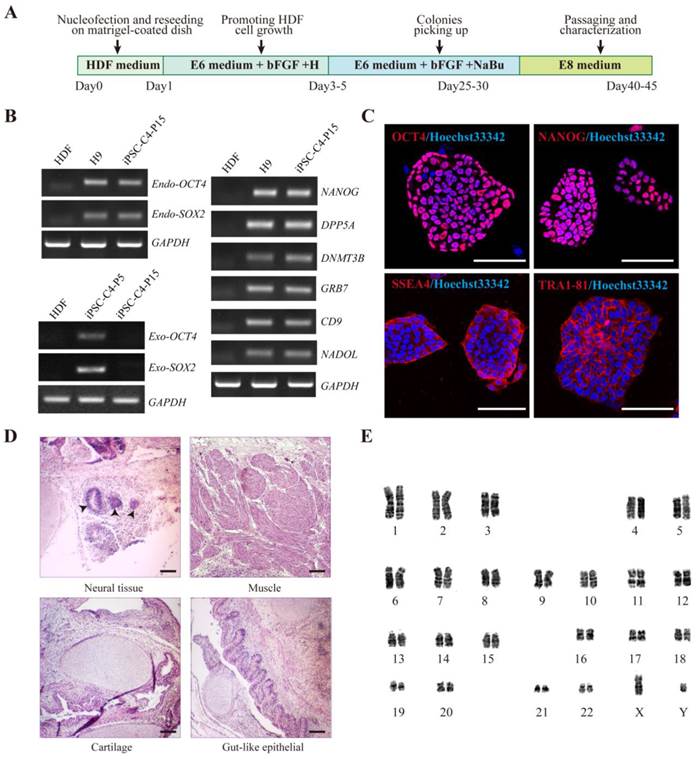
To generate integration-free iPSCs, the Yamanaka episomal plasmids (encoding OCT4, SOX2, LIN28, L-MYC, KLF4 and p53 shRNA) were electro-transfected into human dermal fibroblasts (HDFs) using the Amaxa Nucleofector system. After transfection, the cells were cultured on matrigel-coated dishes in supplemented E6 medium to improve cell proliferation and reprogramming efficiency at indicated time points (Fig. 1A). The transfection efficiency was monitored by cotransfection with pCXLE-EGFP plasmid. At 24 hours after electroporation, EGFP fluorescence was observed in approximately 36 ± 16% cells. Approximately 15 days later, cell aggregates indicative of transformed cells emerged from 5×105 original cells. With continuous culture, the size of cell clumps increased gradually. About day 25 after electroporation, several of these colonies began to exhibit characteristic hESC-like compact and flat features and stained positive for alkaline phosphatase (Fig. S1), Individual cells within the colonies displayed typical morphology similar to that of human ES cells, with a high nucleus-to-cytoplasm ratio and conspicuous nucleoli. Additionally, the number of EGFP-positive cells gradually diminished with cell division, indicating the silence of exogenous genes from the iPSCs. From days 25 to 30, a single colony was picked and propagated in feeder-free conditions. Of the original 5×105 HDFs, we routinely observed a yield of 5-10 iPS colonies.
The pluripotency of HDF-derived iPSCs was further evaluated. RT-PCR analysis revealed that HDF-derived iPSCs (hiPSCs) expressed endogenous OCT4, SOX2 as well as other hESC markers, such as NANOG, DAPP5, DNMT3B, GRB7, CD9 and NADOL. In contrast, the expression of exogenous transduced reprogramming factors was silenced (Fig. 1B). Immunofluorescence staining confirmed the positive expression of typical pluripotent markers such as OCT4, NANOG, SSEA4, and TRA-1-81 in HDF-derived iPSCs (Fig. 1C). Bisulfite sequencing further revealed that the cytosine guanine dinucleotides (CpG) in both OCT4 and NANOG promoters were highly demethylated in these iPS cells, similar to human H9 ESCs and in contrast to the original HDFs (Fig. S2).
To further assess the pluripotency of HDF-derived iPSCs, in vivo teratoma formation assay was performed by injecting iPSCs into subcutaneous tissue of BALB/c SCID mice. After two months, these iPSCs formed teratomas and histological examination showed that the teratomas were composed of cells characteristic of all three germ layers (Fig. 1D), which were confirmed by in vitro analysis. Meanwhile, karyotype analysis after passage 15 showed that the iPSCs maintained normal karyotypes (Fig. 1E). Taken together, HDFs were successfully and efficiently reprogrammed into non-integrating iPSCs.
Derivation of iPSCs from HDFs using an episomal electroporation system. A. Time schedule of iPSCs generation. B. RT-PCR analysis of endogenous, exogenous transgenes, and ES cell-marker genes in iPS cell clone4 after passage 15 (iPSC-C4-P15). Primers used for Endo-OCT4 and Endo-SOX2 specifically detect the transcripts from the endogenous genes, but not from the transgenic transcripts. Parental HDFs were used as negative control. H9 and iPSCs-C4-P5 were used as positive controls respectively. C. Immunofluorescence analysis of iPS cell pluripotent markers, OCT4, NANOG, SSEA4 and TRA-1-81(Red). Nuclei were counterstained with Hoechst 33342 (blue). Shown were the merge images. Scale bar = 25 μm. D. Hematoxylin and eosin staining of teratomas sections of iPSCs-C4-P15 injected subcutaneously into immunocompromised SCID mice for two months. Shown were neural tissue (ectoderm, tailless arrow), gut-like epithelium (endoderm), striated muscle and cartilage (mesoderm). Scale bar = 100 μm. E. Karyotyping analysis showing normal karyotyping of iPSCs after passage 15.
Comparative gene expression profiling in HDFs, iPSCs and ESCs
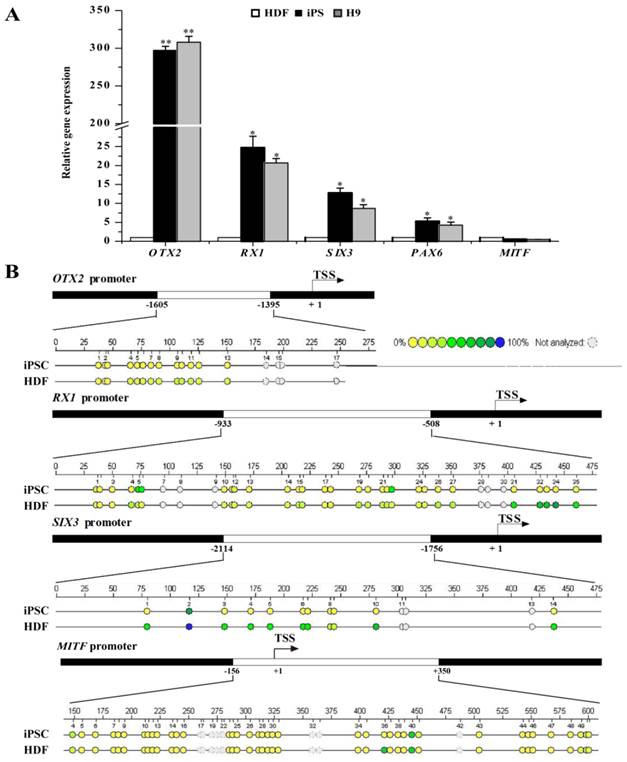
Fully reprogrammed cells should resemble ESCs in expression profiling of genes including pluripotent genes as well as RPE differentiation-associated genes. The transcriptional and epigenetic status of RPE differentiation-associated genes may bias the differentiation potential of iPSCs. Thus, we further investigated the expression profiling of RPE differentiation-associated genes in HDFs, HDF-derived iPSCs, and human H9 ESCs. As shown in Fig. 2A, iPSCs and ESCs showed similar expression pattern in RPE differentiation-associated genes. The mRNA level of OTX2, a transcription factor essential for early neural specification was relatively abundant in iPSCs and ESCs. In contrast, its expression was extremely low in donor HDFs. After reprogramming, its expression was dramatically elevated by approximately 300-fold. Meanwhile, the expression of RX1 and SIX3, which are involved in retinal development in iPSCs and ESCs, was 25- and 13-fold higher than donor HDFs, respectively. Nevertheless, the expression of RPE progenitor markers such as MITF still remained a relative silent status after reprogramming. Consistent with the augmented expression levels, the promoters of RX1 and SIX3 genes showed fewer methylated CpG dinucleotides in iPSCs, but were densely methylated in HDFs (Fig. 2B). Comparatively, the OTX2 promoter was hypomethylated in HDFs, and nearly demethylated in iPSCs. In contrast, the promoter of MITF showed a nearly equivalent level of methylated CpG in iPSCs compared with in HDFs (Fig. 2B). These evidences indicate that the process of reprogramming led to the conversion of retinal developmental genes from a condensed methylation state in HDFs to an accessible demethylated one in pluripotent stem cells in a supervised hierarchical pattern. Seemingly, DNA demethylation of genes associated with neural and retinal development during reprogramming may be a determinative event for rendering retinal differentiation potential to reprogrammed cells.
RPE differentiation-associated gene expression in HDFs, iPSCs, and ESCs. A. Real-time PCR analysis of RPE differentiation-associated genes in HDFs, iPSCs, and ESCs. HDF cells were used as control (* p < 0.05, ** p < 0.01 vs the control). B. Bisulfite genomic sequencing of the promoter regions of OTX2, RX1, SIX3 and MITF in iPSCs and HDFs. Different colors indicate the level of methylated CpGs.
Regulatory roles of OCT4 and SOX2 on expression of OTX2, RX1, and SIX3 genes
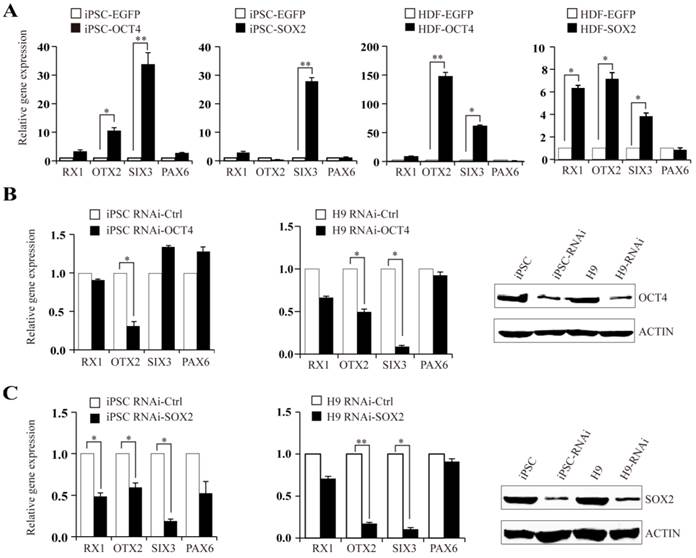
OTX2, RX1, and SIX3 genes were shown to have moderate activation in iPSCs and ESCs. We further investigated the regulatory role of OCT4 and SOX2 on OTX2, RX1 and SIX3 expression. HDFs and iPSCs transiently transfected with OCT4 or SOX2 plasmids were analyzed using real-time PCR for the expression of OTX2, RX1 and SIX3 genes. In HDF-derived iPSCs, OCT4 augmented the expression of OTX2 and SIX3 genes by 12- and 35-fold, respectively, with little change in RX1 and PAX6 levels. Transfection of HDFs with OCT4 plasmids increased OTX2 and SIX3 transcript levels by approximately 150- and 60-fold, respectively, whereas the RX1 transcript level was elevated 10-fold (Fig. 3A). In comparison, SOX2 overexpression triggered a significant increase of SIX3 and RX1 in both HDF and iPSC cells, while only slightly affecting the expression of PAX6. Accordingly, knockdown of OCT4 or SOX2 expression in iPSCs by RNAi reduced the expression of their respective targeted genes. Similar results were observed in human H9 ESCs (Fig. 3B and C). These results suggest that the metastable activation of OTX2, RX1 and SIX3 genes in pluripotent stem cells is regulated by OCT4-SOX2 axis, directly or indirectly, as part of the program maintaining pluripotency. Considering the retained methylation state of RX1 and SIX3 promoters in untreated HDFs, it seems that OCT4 and SOX2 alone can directly alter the silent epigenetic state of these genes into an active or "poised yet repressed" one.
OCT4 and SOX2 regulate expression of OTX2, RX1, and SIX3 genes. A. Expression levels of RX1, OTX2, SIX3, and PAX6 following overexpressing of OCT4 or SOX2 plasmids in iPSCs and HDFs. iPSCs and HDF transfected with EGFP plasmid as control. B and C. iPSCs and H9 cells, were treated with OCT4 or SOX2 siRNA respectively. Real-time PCR analysis for RX1, OTX2, SIX3 and PAX6 expression after RNAi transfection. The efficiency of RNAi was monitored by Western blot. (* p < 0.05, ** p < 0.01 vs the control).
The requirement of OCT and SOX motif for activation of OTX2 promoter. A. The putative OCT and SOX binding sites at approximately 500 ~ 600 bp upstream of OTX2 ATG were shown. Conserved sequences among human, mouse, and rat species are in the box. B. Luciferase assay in 293T cells was performed to evaluate the transcriptional activity of the OTX2 promoter by cotransfecting SOX2 or OCT4 expressing plasmids (pBK-SOX2 or pBK-OCT4). Wild-type OTX2 promoter-luciferase construct (wt pGL); mt SOX-pGL, OTX2 promoter-luciferase construct with mutant SOX consensus sites; mt OCT-pGL, OTX2 promoter-luciferase construct with mutant OCT consensus sites. Data are presented as fold activity over basal promoter activity. (* p < 0.05, ** p < 0.01 vs the control). C. ChIP assay was carried out to detect OCT4 and SOX2 binding on the OTX2 promoter in iPSCs. Input represented 10% of the total input chromatin, and IgG served as a negative control. D. The immunoprecipitated DNA fragments were amplified by real-time PCR for the OTX2 promoter region containing the putative OCT-SOX binding sites in HDFs and iPSCs. Each bar represents the mean ± SD for triplicate experiments.
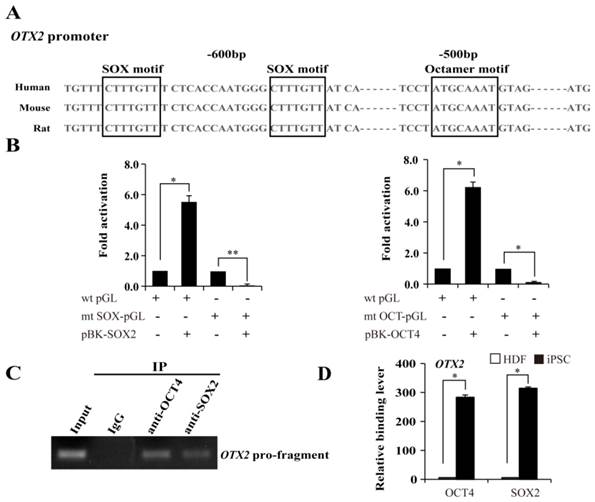
Identification of OCT4 and SOX2 binding sites on the proximal OTX2 promoter
Based on the above mentioned, we then used the luciferase assay to examine the response patterns of the sequences upstream of OTX2 to the overexpression of SOX2 and OCT4 in 293T cells. OCT4 specifically interacts with the octamer motif ''ATGCTAAT'' through the POU domain. SOX2, an HMG domain-containing transcription factor, preferentially binds to the consensus SOX motif “CATTGTT” or “CTTTGTT”. A tight transcriptional cooperation between OCT4 and SOX2 is important for governing pluripotency. Promoter analysis suggests that a typical OCT4 octamer motif exists and along with two SOX “CTTTGTT” elements located upstream of the ATG translation start site in the OTX2 promoter (Fig. 4A). Subsequently, we constructed a wild-type luciferase reporter containing 1008 bp upstream of the ATG site, and mutants destroying the OCT4 or SOX2 binding sites in the OTX2 promoter. All the constructs were transfected into 293T cells. As shown in Fig. 4B, heterologous expression of OCT4 or SOX2 evoked a 5.5- and 6.2-fold increase in the wild-type reporter activity in 293T cells, respectively. Meanwhile, the mutant constructs exhibited variable levels of luciferase activity decreased to 4.5 and 5.9%, respectively. To further evaluate recruitment of OCT4 and SOX2 on the OTX2 promoter, ChIP assays were conducted in iPSCs. DNA fragments containing the putative OCT-SOX composite elements of OTX2 were enriched with specific OCT4 and SOX2 antibodies and detected by PCR (Fig. 4C). The control antibody, confirming the specificity of binding, did not detect enrichment. As shown in Fig. 4D, no significant enrichment for OCT4 and SOX2 were observed on the OTX2 promoter regions in HDFs. After reprogramming, the OTX2 promoter was significantly occupied by OCT4 and SOX2. Taken together, our results strongly suggest that OTX2 expression in pluripotent stem cells is dependent on the recruitment of OCT4 and SOX2 onto the defined sites of the proximal OTX2 promoter.
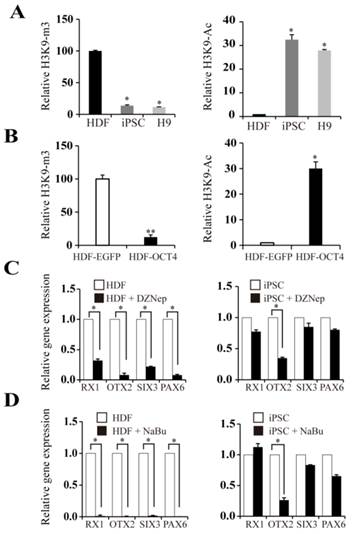
Histone modifications on the OTX2 promoter
Given the hypomethylation status of the OTX2 promoter in HDFs, we speculated that histone modification may be involved in the reactivation of OTX2. H3K9 acetylation is generally a transcriptionally permissive modification. H3K9 trimethylation is a stable repressive modification and opposes the transcription-activating acetylation of the same H3K9. We found that the OTX2 promoter in HDFs harbored high levels of H3K9 trimethylation (Fig. 5A). When cells were reprogrammed into the pluripotent state, the degree of H3K9 acetylation on the OTX2 promoter was significantly enhanced, while the enrichment of H3K9 trimethylation was decreased. Moreover, transfection of HDFs with OCT4 plasmids directly triggered the acetylation and demethylation of H3K9 (Fig. 5B). Intriguingly, DZNep, a global histone methylation inhibitor, conversely led to the decrease of OTX2 as well as RX1 and SIX3 in HDFs and iPSCs (Fig. 5C). Sodium butyrate, a histone deacetylase inhibitor, also inhibited the expression of OTX2, RX1 and SIX3 genes (Fig. 5D). Therefore, these results indicate that the local alteration of histone modifications on the OTX2 promoter, irrespective of the global changes, contributes to the reactivation of OTX2.
Histone modifications on the OTX2 promoter. A. ChIP assays were performed with antibodies against H3K9-ac and H3K9-m3 in HDFs, iPSCs, and H9. Immunoprecipitated DNA fragments were amplified by real-time PCR for the promoter region of OTX2. HDFs served as a control. B. ChIP assays were conducted with antibodies against H3K9-ac, H3K9-m3 in HDFs and HDFs-overexpressed OCT4. Immunoprecipitated DNA fragments were amplified by real-time PCR for the promoter region of OTX2. HDFs served as a control. C. Real-time PCR analyses for expression of RX1, OTX2, SIX3, PAX6 in HDF and iPSCs treated with DZNep for 48 hours respectively. D. Expression levels of RX1, OTX2, SIX3, and PAX6 genes in HDF and iPSCs treated with sodium butyrate (NaBu) for 48 hours respectively were assessed by real-time PCR. Each bar represents mean ± SD from three independent experiments, each in triplicate. (* p < 0.05, ** p < 0.01 vs the control).
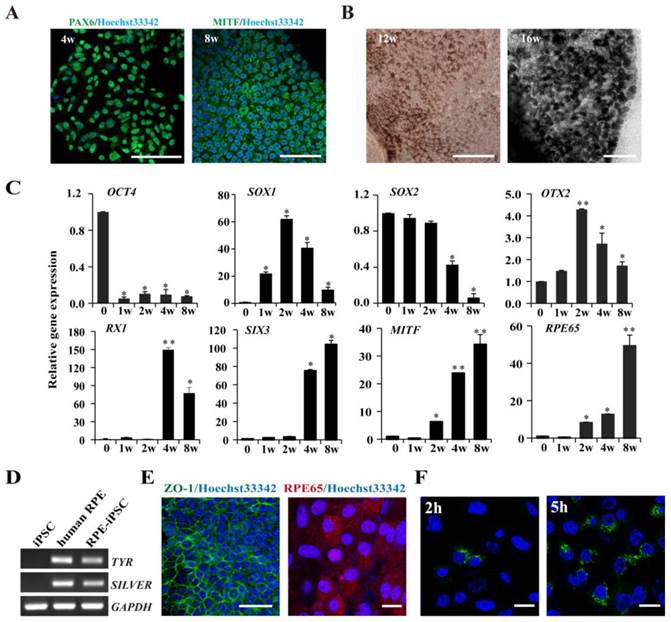
Differentiation of hiPSCs into RPE cells
We next set out to determine the RPE differentiation capability of HDF-derived iPSCs with metastable reactivation of OTX2, RX1 and SIX3 genes. A stepwise protocol with defined small molecules was used to induce RPE differentiation. As shown in Fig. S3A, dissociated iPSCs were cultured in a suspension culture for 21 days in the presence of CKI-7 and SB-431542 which block Wnt and Nodal signaling respectively. Under the suspension and serum-free conditions (SFEB/CS), floating iPSCs spontaneously formed EBs give rise to neuroectodermal cells. Subsequently, EBs were plated onto a matrigel-coated dish, which supports adhesion of cell aggregates and subsequent retinal differentiation. Approximately 6 weeks after the start of induction, some polygonal morphology of RPE-like cells can be observed (Fig. S3B). Later then, 33.1±4.8% of the RPE-like cells developed pigmented areas at week 8 after initiation of induction. Immunofluorescence staining showed that the cultured cells expressed neural retinal progenitor marker (PAX6) at week 4 and RPE progenitor marker (MITF) at week 8 (Fig. 6A). These pigmented clusters were isolated manually and spread onto plates for expansion. Continued culture resulted in the formation of a confluent cell monolayer that displayed increased pigmentation and the typical cobblestone shape during week 12 to 16 (Fig. 6B). Within differentiated colonies, 55.7% ± 3.5% of the cells were pigmented.
Time-course analysis of gene expression revealed that HDF-derived hiPSCs underwent a stepwise differentiation through neural, retinal, and RPE fates under defined culture conditions. Specific markers that reflect the states of pluripotency (OCT4), early neural specification (OTX2), general neural transcription factors (SOX1, SOX2), retinal specification (RX1, SIX3), RPE progenitor (MITF) and mature RPE (RPE65) were examined by real-time PCR. As shown in Fig. 6C, the expression of undifferentiated ESC marker OCT4 rapidly decreased at the early stage during suspension culture. At the middle stage of suspension culture (2 weeks), the expression of the early neural transcription factors OTX2 reached the peak but gradually decreased during the following progression of retinal differentiation. The definitive neural transcription factor SOX1 reached the peak at week 2 and another neural specification gene SOX2 prolong expression until 4 weeks. The expression of retinal specification transcription factors, RX1 and SIX3, which play crucial roles in retinal specification, appeared after 1 week of differentiation and remained at elevated levels throughout the late stage of differentiation. Simultaneously, MITF, a transcription factor required for RPE differentiation, was detected at 2 weeks after induction and significantly augmented after 4 and 8 weeks. Another RPE marker RPE65, involved in the conversion of all-trans-retinol to 11-cis-retinal, exhibited a similar expression pattern to that of MITF and was strongly expressed in differentiated pigmented cells at 4 and 8 weeks. RT-PCR further confirmed that HDF-derived iPSCs were successfully differentiated into RPE cells, as demonstrated by the expression of the mature RPE markers and the functional genes (TYR and SILVER) involved in pigment synthesis (Fig. 6D). Immunofluorescence staining with specific antibodies showed that the pigmented RPE cells expressed RPE65, a mature RPE marker at 16 weeks. Additionally, the RPE-like cells were positively stained with anti-ZO-1 antibody, implicating that pigment cells formed tight junctions with a polygonal shape (Fig. 6E). As known, phagocytosis of photoreceptor outer segment (POS) is a physiological function of RPE cells in vivo. Then we conducted a latex bead phagocytosis assay and indeed detected phagocytosis activity in iPSCs-derived RPE cells (Fig. 6F). Taken together, these data indicate that fully reprogrammed iPSCs from HDF were induced to differentiate into pigmented RPE cells under nonxenogeneic conditions.
RPE cells derived from iPSCs and characterization of iPSC-derived RPE Cells. A. Immunofluorescence images demonstrating the expression of differentiation of, neural retina progenitors, PAX6 at week 4; RPE progenitor marker, MITF at week 8. B. Phase-contrast images were shown for pigmented, hexagonal RPE-like cells at week 12 and week 16 (Scale bar = 100 μm). C. Real-time PCR analysis at sequential time points for expression of key genes in early neural and RPE development. Undifferentiated iPS cells were used as negative control. (* p < 0.05, ** p < 0.01 vs the control). D. Characterization of mature RPE genes in iPSC-derived RPE-like cells derived from iPSCs using RT-PCR, undifferentiated iPSCs and human RPE cells (ARPE-19) were used as negative and positive controls respectively. E. Immunostaining showing that the pigmented cells expressed tight junction protein ZO-1 (Scale bar = 50 μm) and the mature RPE marker, RPE 65 after induction for 16 weeks (Scale bar = 25 μm). F. IPSC-induced RPE cells phagocytose latex beads. Nuclei were counterstained with Hoechst 33342 (Scale bar = 25 μm).
Discussion
In this study, we efficaciously generated human iPSCs from fibroblasts with a modified reprogramming recipe, replacing c-MYC with transformation-deficient L-MYC to reduce the risk of tumorigenicity. In serum-free, feeder-free and xeno-free conditions, we used viral free non-integrating episomal vectors to generate iPSCs that are less immunogenic and tumorigenic. Besides, we established an induction procedure for derivation of RPE cells from iPSCs with small molecules in serum-free, xeno-free culture conditions. It minimized the risks of pathogen cross-transfer, contamination from xenogeneic components and zoonosis. Our results demonstrated the validity of the modified protocols in cell reprogramming and RPE induction.
Besides, we found that OTX2, RX1 and SIX3 are selectively reactivated after reprogramming. OTX2, RX1 and SIX3 genes all encode crucial transcriptional factors that are required for forebrain development and retinal cell fate determination. Genetic mutation or malfunction of these genes results in serious malformations of forebrain and ocular organs, as well as erroneous retinal pigment epithelium specification [16]. Developmentally, RPE cells are derived from neuroectodermal cells. OTX2 as one of the genes in anterior neuroectoderm was expressed earliest [17, 18]. In the mouse retina, Otx2 ablation leads to downregulation of Otx2 target genes that are associated with RPE functions [19]. Gene expression analysis has identified that Otx2 directly regulates most RPE-specific genes [20]. SIX3 is essential for development of the optic vesicles by repressing Wnt signal pathway [21]. Overexpression of RX1 in Xenopus embryos results in an optic vesicle derived RPE cells [22]. Besides, during the stepwise RPE development, these transcriptional factors constitute a hierarchical gene regulatory network. For instance, OTX2 has been proven to activate RX1 [23]. Herein, we elucidated that SOX2 together with OCT4 exert an important regulatory effect on the moderate reactivation of OTX2, RX1 and SIX3 genes. SOX2, a SRY-like HMG box transcription factor which is critical for maintenance of pluripotent state of ESCs, also plays a pivotal role in the eye and forebrain development. In previous studies, PAX6, RX1 and SIX3 genes have been identified as SOX2 transcriptional targets in forebrain development [24, 25]. In our study, SOX2 mainly facilitated the reactivation of OTX2 and RX1 during reprogramming, while OCT4 predominantly stimulated the expression of OTX2 and SIX3. We also identified an OCT4- and two SOX2-binding motifs located in the proximal promoter of OTX2 gene. Luciferase assays on wild-type and mutant upstream untranslated regions (UTRs) suggest that the intact OCT4- and SOX2-binding motifs were indispensable to activate the OTX2 promoter activity. Taken our finding together with previous reports, the metastable reactivation of OTX2, RX1 and SIX3 genes in iPSCs may attribute to the establishment of OCT4-SOX2 regulatory axis during cell reprogramming.
In this study, the HDF-derived iPSCs underwent a selective transcriptional and epigenetic reactivation on genes required for early RPE differentiation during reprogramming, eventually resembling ESCs in gene expression profiling. The transcriptional conversion of RX1 and SIX3 genes from silent to active status partially attributed to DNA demethylation. Nevertheless, the focal alteration of histone modifications including methylation and acetylation rather than DNA demethylation, accounted for the reactivation of OTX2. In comparison, the RPE progenitor genes such as MITF retained a transcriptional silent status. It seems that the epigenetic choreography of RPE differentiation is composed by sequential phases and hierarchical mode of DNA demethylation on the promoters of neural, retinal, and RPE genes. Indeed, data from ESCs implicated that genes associated with early development are arranged within a bivalent chromatin structure. It is considered as a "poised yet repressed" status, which can be promptly activated upon differentiation. In this way, pluripotent stem cells maintain a delicate balance between self-renewal and susceptibility to differentiation, and are highly sensitive to inductive signals [26]. Obviously, the retinal propensity of HDF-derived iPSCs was predetermined as characterized by the metastable reactivation of OTX2, RX1 and SIX3 genes. Therefore, evaluating the transcriptional and epigenetic characteristics of these genes may be a simple and reliable approach to predict the early retinal propensity of iPSCs from different sources. Besides, in reprogramming to pluripotency, the binding of OCT4 or SOX2 to designated sites of downstream genes can evoke a focal DNA demethylation and histone modification to generate an open/primed chromatin state [26]. It was documented that OCT4 can recruit the histone lysine demethylase to catalyze histone demethylation and nucleosome depletion [27]. In our study, histone acetylation and methylation on the local promoter contributed to the reactivation of OTX2. Collectively, the reactivation of OTX2, RX1 and SIX3 in iPSCs should be an event associated with the OCT4-SOX2 regulatory axis and coupled focal epigenetic modification.
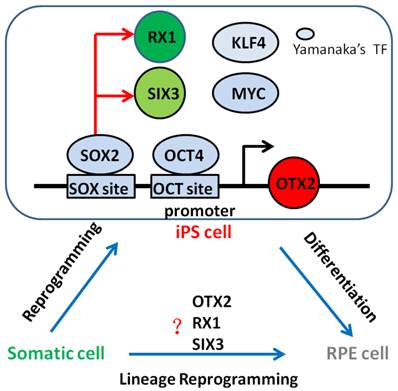
Since the landmark discovery of generating pluripotent cells from somatic cells, referred to as reprogramming in 2006 [8], researchers from all over the world insist on researching whether one cell type could be induced directly from another determined cell type without going through a pluripotent stage, only by overexpressing transcription factors or microRNAs. Meanwhile, many cell types has been direct reprogrammed successfully such as pancreatic beta cells [28], neurons [29], hepatocyte-like cells [30, 31], or hematopoietic progenitor cells [32]. In this study, we showed OTX2, RX1 and SIX3 genes that associated with early neural and retinal differentiation reactivation and demethylation during cell reprogramming. This may be the reason that iPSCs can be differentiate into RPE cells. Subsequently, we successfully obtained derived RPE cells from iPSCs under chemically defined conditions, paving the way for clinical use of RPE in AMD treatment. Thus, our data suggest the intriguing possibility that combinations of neural and retinal specific transcription factors which are high expression level in iPSCs might be able to convert from fibroblasts into RPE cells bypassing a pluripotent state, what is called lineage reprogramming. Future studies will be necessary to determine whether RPE cells could be generate directly from other determined cell type simply by overexpression such as OTX2, RX1 and SIX3 genes (Fig. 7).
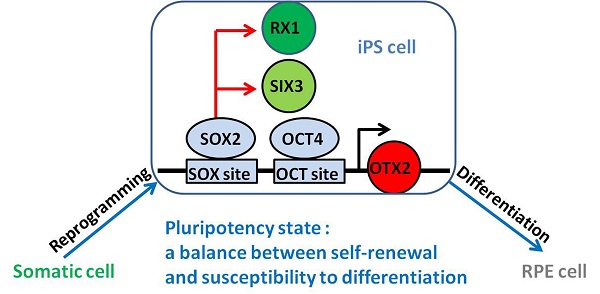
Neural and retinal specification-associated genes, OTX2, RX1 and SIX3 are very significant for RPE development. In induced pluripotent stem cells, they were reactivated by reprogramming transcription factors SOX2 and OCT4 respectively. The reactivated of these genes in iPSC cells may give rise to the differentiation of pluripotent stem cells into RPE cells. Thus, we have the hypothesis that somatic cells can direct convert into RPE cells by overexpression these genes. Yamanaka's TF: Yamanaka's reprogramming transcription factor.
Supplementary Material
Supplementary Tables and Figures.
Acknowledgements
This work was supported by Grant 2011CB510200 from the National Basic Research Program of China (973 program), the National Natural Science Foundation of China (81303004, 31470082), and Zhejiang Provincial Natural Science Foundation and Public Innovation Program (LY14C120001, 2014C37126).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Longbottom R, Fruttiger M, Douglas RH, Martinez-Barbera JP, Greenwood J, Moss SE. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18728-33
2. Heller JP, Martin KR. Enhancing RPE Cell-Based Therapy Outcomes for AMD: The Role of Bruch's Membrane. Translational vision science & technology. 2014;3:11
3. Seiler MJ, Aramant RB. Cell replacement and visual restoration by retinal sheet transplants. Progress in retinal and eye research. 2012;31:661-87
4. Idelson M, Alper R, Obolensky A, Ben-Shushan E, Hemo I, Yachimovich-Cohen N. et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell stem cell. 2009;5:396-408
5. Westenskow P, Sedillo Z, Barnett A, Friedlander M. Efficient derivation of retinal pigment epithelium cells from stem cells. Journal of visualized experiments: JoVE. 2015
6. Li Y, Atmaca-Sonmez P, Schanie CL, Ildstad ST, Kaplan HJ, Enzmann V. Endogenous bone marrow derived cells express retinal pigment epithelium cell markers and migrate to focal areas of RPE damage. Investigative ophthalmology & visual science. 2007;48:4321-7
7. Harris JR, Fisher R, Jorgensen M, Kaushal S, Scott EW. CD133 progenitor cells from the bone marrow contribute to retinal pigment epithelium repair. Stem cells. 2009;27:457-66
8. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-76
9. Borooah S, Phillips MJ, Bilican B, Wright AF, Wilmut I, Chandran S. et al. Using human induced pluripotent stem cells to treat retinal disease. Progress in retinal and eye research. 2013;37:163-81
10. Alvarez Palomo AB, McLenachan S, Chen FK, Da Cruz L, Dilley RJ, Requena J. et al. Prospects for clinical use of reprogrammed cells for autologous treatment of macular degeneration. Fibrogenesis & tissue repair. 2015;8:9
11. Carr AJ, Vugler AA, Hikita ST, Lawrence JM, Gias C, Chen LL. et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PloS one. 2009;4:e8152
12. Sugita S. Retinal regeneration with iPS cells-Clinical trials for retinal degenerative disorders. Nihon Rinsho Men'eki Gakkai kaishi= Japanese journal of clinical immunology. 2014;38:79-85
13. Kamao H, Mandai M, Okamoto S, Sakai N, Suga A, Sugita S. et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem cell reports. 2014;2:205-18
14. Lee SB, Seo D, Choi D, Park KY, Holczbauer A, Marquardt JU. et al. Contribution of hepatic lineage stage-specific donor memory to the differential potential of induced mouse pluripotent stem cells. Stem cells. 2012;30:997-1007
15. Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K. et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15(3):295-309
16. Beby F, Lamonerie T. The homeobox gene Otx2 in development and disease. Experimental eye research. 2013;111:9-16
17. Kurokawa D, Ohmura T, Sakurai Y, Inoue K, Suda Y, Aizawa S. Otx2 expression in anterior neuroectoderm and forebrain/midbrain is directed by more than six enhancers. Developmental biology. 2014;387:203-13
18. Haruta M, Sasai Y, Kawasaki H, Amemiya K, Ooto S, Kitada M. et al. In vitro and in vivo characterization of pigment epithelial cells differentiated from primate embryonic stem cells. Investigative ophthalmology & visual science. 2004;45:1020-5
19. Housset M, Samuel A, Ettaiche M, Bemelmans A, Beby F, Billon N. et al. Loss of Otx2 in the adult retina disrupts retinal pigment epithelium function, causing photoreceptor degeneration. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:9890-904
20. Samuel A, Housset M, Fant B, Lamonerie T. Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PloS one. 2014;9:e89110
21. Sinn R, Wittbrodt J. An eye on eye development. Mechanisms of development. 2013;130:347-58
22. Muranishi Y, Terada K, Furukawa T. An essential role for Rax in retina and neuroendocrine system development. Development, growth & differentiation. 2012;54:341-8
23. Danno H, Michiue T, Hitachi K, Yukita A, Ishiura S, Asashima M. Molecular links among the causative genes for ocular malformation: Otx2 and Sox2 coregulate Rax expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5408-13
24. Lee B, Song H, Rizzoti K, Son Y, Yoon J, Baek K. et al. Genomic code for Sox2 binding uncovers its regulatory role in Six3 activation in the forebrain. Developmental biology. 2013;381:491-501
25. Shafa M, Krawetz R, Rancourt DE. Returning to the stem state: epigenetics of recapitulating pre-differentiation chromatin structure. BioEssays: news and reviews in molecular, cellular and developmental biology. 2010;32:791-9
26. Shakya A, Callister C, Goren A, Yosef N, Garg N, Khoddami V. et al. Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Molecular and cellular biology. 2015;35:1014-25
27. Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627-32
28. Huang P, He Z, Ji S, Sun H, Xiang D, Liu C. et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386-9
29. Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390-3
30. Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell reports. 2014;9:1871-84
31. Shakya A, Callister C, Goren A, Yosef N, Garg N, Khoddami V. et al. Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Molecular and cellular biology. 2015;35:1014-25
32. Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035-41
Author contact
![]() Corresponding authors: Li Shen and Tao Li; Email: shenliedu.cn (LS); litaocn (TL)
Corresponding authors: Li Shen and Tao Li; Email: shenliedu.cn (LS); litaocn (TL)

 Global reach, higher impact
Global reach, higher impact