10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(5):607-616. doi:10.7150/ijbs.14802 This issue Cite
Research Paper
Ecological Trade-offs between Migration and Reproduction Are Mediated by the Nutrition-Sensitive Insulin-Signaling Pathway
1. College of Life Sciences, China Jiliang University, Hangzhou, China, 310018;
2. Division of Biological Sciences, The University of Montana, Missoula, Montana 59812, USA;
3. Department of Entomology, Washington State University, Pullman, Washington 99164, USA.
Received 2015-12-25; Accepted 2016-2-23; Published 2016-3-30
Abstract
Crowding and changes in food availability are two critical environmental conditions that impact an animal's trajectory toward either migration or reproduction. Many insects facing this challenge have evolved wing polyphenisms. When conditions favor reproduction, wing polyphenic species produce adults that either have no wings or short, non-functional wings. Facultative wing growth reflects a physiological and evolutionary trade-off between migration and reproduction, triggered by environmental conditions. How environmental cues are transduced to produce these alternative forms, and their associated ecological shift from migration to reproduction, remains an important unsolved problem in evolutionary ecology. The brown planthopper, a wing polymorphic insect exhibiting strong trade-offs in investment between migration and reproduction, is one of the most serious rice pests in Asia. In this study, we investigated the function of four genes in the insulin-signaling pathway known to couple nutrition with growth, PI3 Kinase (PI3K), PDK1, Akt (Protein Kinase B), and the forkhead gene FOXO. Using a combination of RNA interference and pharmacological inhibitor treatment, we show that all four genes contribute to tissue level regulation of wing polymorphic development in this insect. As predicted, silencing of the NlPI3K, NlAkt and NlPDK1 through dsRNA and with the pharmacological inhibitor Perifosine resulted in short-winged brown planthoppers, whereas knockdown of NlFOXO resulted in long-winged planthoppers. Morphometric analyses confirm that phenotypes from our manipulations mimic what would be found in nature, i.e., major parameters such as bristle number, wing area and body weight are not significantly different from non-experimental animals. Taken together, these data implicate the insulin-signaling pathway in the transduction of environmental factors into condition-dependent patterns of wing growth in insects.
Keywords: Brown planthopper, wing, polyphenism, evolutionary tradeoff, insulin signaling pathway, PI3K, Akt, FOXO.
Introduction
The ability of an organism to rapidly respond to changing environmental conditions has significant consequences for its survival, reproduction, and fitness [1]. Polyphenism, the developmental capacity to couple coordinated expression of alternative suites of morphological, physiological, and behavioral traits with circumstance, is an effective solution to this problem, as it permits organisms to facultatively invest in the production of costly traits only when conditions are appropriate. This rapid response to condition is commonly seen in insects such as aphids and crickets, who face a pronounced allocation tradeoff between wings and wing muscles, on the one hand, and reproduction on the other [2-10]. These insects switch between fully winged forms capable of migratory flight, and flightless forms that instead allocate resources to reproduction [11, 12].
Although it has long been evident that polyphenic insects coupled wing growth with exposure to specific environmental cues, such as increased crowding and/or deteriorating nutrition, the physiological and genetic mechanisms responsible for the alternate patterns of wing growth are less well understood. In both aphids and crickets, cue-induced differences in circulating levels of whole animal physiological signals such as juvenile hormone (JH) appear to provide the first link between external conditions and patterns of tissue growth [6, 13]. However, these hormone signals must act on specific tissues to enact morph-specific patterns of growth, and the details of these interactions are almost entirely unknown.
The brown planthopper Nilaparvata lugens Stål is a serious insect pest across Asia. As with aphids and crickets, wing growth in this species is polyphenic. Both males and females are capable of developing into either a migratory long-winged form or a reproductive short-winged form [14-16]. Populations of N. lugens experiencing crowding and low food availability produce high ratios of long winged individuals, which will take flight, migrate, and colonize new fields[17]. This occurs when the nutritional value of the rice plant decreases as the rice ages, and as greater numbers of brown planthoppers crowd the plants[17]. Newly colonizing populations that experience less crowding and high food availability and food quality have a greater proportion of short winged individuals, which have an increased capacity for reproduction. Thus, a diversity of environmental factors such as temperature [18], developmental stage of the host plant [17], and population density [19, 20], have been shown to affect the development of wings in brown planthoppers.
But what mediates the condition-dependent growth of wings in brown planthoppers? As in other species of wing polymorphic insects [6, 13], topical application of JH analogs and Precocene II (which decreases JH titers through its effects on the corpora allata [14, 21]) induces short-winged and long-winged brown planthoppers, respectively [19, 21-23]. These same studies also identified a sensitive period, between the 3rd and 4th instar, when planthoppers are sensitive to environmental conditions and when JH levels appear to regulate wing growth [19, 20]. These results suggest that JH signaling functions upstream in the regulation of brown planthopper wing polymorphism, and that circulating concentrations of JH mediate tissue-specific differential expression of genes, initiating the switch between winged and wingless trajectories of development. What remains unknown is how downstream tissues, such as wings, affected by this polyphenism modulate their growth, and whether additional signals, such as insulin and insulin-like growth factors, interact with JH to couple tissue growth with condition.
The insulin signaling (IS) pathway is known to translate environmental cues such as nutrition and stress into the regulation of the growth of animal body parts in a condition-dependent manner. It is highly conserved across animal species both in pathway members and in function. When this pathway is activated through the binding of insulin/insulin-like peptides/insulin growth factors to the insulin receptor, the resulting signal transduction cascade promotes cellular growth and proliferation [24-28]. It does this by turning on transcription factors in the nucleus that promote growth, but also through the action of the serine kinase PKB/Akt, which inhibits the forkhead box-containing O subfamily protein FOXO; FOXO is a key growth inhibitor [24, 25, 29-31]. Thus, depending on the constitutive sensitivity of a tissue, the concentration of insulin/ILPs/IGFs will determine the amount of downstream activation of the insulin signaling pathway, and, in this way, regulate growth under different environmental conditions [24, 31-36]. Importantly, the IS pathway regulates the nutrition-sensitive scaling of body parts with overall body size (allometry), as well as the nutrition-sensitive growth of exaggerated sexually selected structures and caste-specific (e.g. soldier) traits in social insects [30-32, 35, 37, 38]. Because of its general role as a tissue-specific modulator of trait growth, and its involvement in other insect polyphenisms, the IS pathway is an excellent candidate for regulating the trade-off in investment between migration and reproduction found in wing polymorphic insects. Recently, Xu et al. [39] published a study in planthoppers that showed that two insulin receptors determine alternative wing morphs in N. lugens.
In this study we tested the hypothesis that downstream components of the IS pathway mediate condition-dependent growth of wings between migratory and reproductive morphs of the brown planthopper. In contrast to Xu et al.[39], we investigated the functional importance of PI3K, PDK1, Akt, and FOXO on the growth of wings in laboratory populations of this insect.
Results
Disruption of PI3K/Akt/FOXO signaling by RNAi or chemical inhibitors changed wing-morph ratio
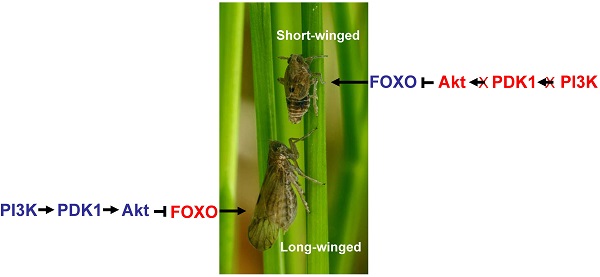
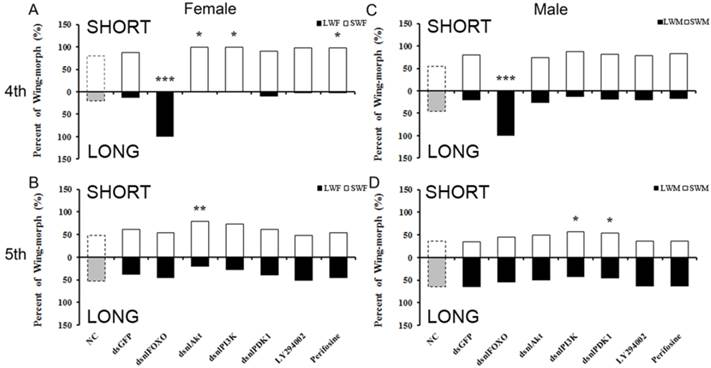
Short- and long-winged forms of the brown planthopper, and the damage they typically inflict on rice plants, are shown in Fig. 1. To test for the functional roles of NlFOXO, NlAkt, NlPI3K and NlPDK1 in the polyphenic regulation of brown planthopper wing growth. Phylogenetic analysis showed that these four genes are conserved across the species (Fig. S1-S4). We injected in vitro transcribed double stranded RNA (dsRNA) against our target genes, thus decreasing mRNA levels and disrupting signal transduction through the IS pathway. Because signaling through this pathway stimulates cell proliferation and tissue growth, we predicted that disruption of NlPI3K, NlAkt, and NlPDK1 at the 4th instar nymph stage would increase the proportion of short-winged animals, while disruption of NlFOXO at this same stage would increase the proportion of long-winged adults.
qRT-PCR was used to measure the mRNA level of NlPI3K, NlAkt, NlPDK1 and NlFOXO in knockdown and control animals. Injection of dsRNA was effective at reducing relative transcript abundance, and therefore at silencing activity of our target genes. Relative to levels in wing tissues of control animals, transcript abundances in wings of knockdown animals were 6.3%, 3.9%, 10.0%, and 4.5% respectively, 3 days after injection at the 4th instar nymphal stage (Supplemental Fig. 5). Transcript abundances were also reduced after injection at the 5th instar nymphal stage, although the development of the wing pad is less sensitive to this reduction (Supplemental Fig. 5).
As predicted, knockdown of NlPI3K and NlAkt during the 4th instar nymph stage and NlPDK1 and NlPI3K during the 5th instar nymph stage increased the proportion of short-winged adults (Fig. 2). This is also consistent with what Xu et al. (2015) found for knockdown of N. lugens Insulin Receptor 1 (NlInR1) which resulted in short-winged adults. However, knockdown of NlFOXO increased the proportion of long-winged adults (Fig. 2), as did knockdown of NlInR2[39].
Our dsRNA injection experiment also showed differential sensitivity between male and female brown planthoppers. Specifically, females were more sensitive than males to the PI3K/Akt/FOXO signaling disruption (Fig. 2). Injection of dsNlAkt, dsNlPI3K or Perifosine led to significant wing-morph ratio changes in females but not in males (Fig. 2 A, C). Injection of dsRNA during the 5th instar nymphal stage did not change the wing form as it does at the 4th instar nymph, the differential sensitivity between males and females was not apparent (Fig. 2 C,D).
To further study the role of PI3K/Akt/FOXO signaling in the brown planthopper wing-morph polyphenism, we used two chemical inhibitors, Perifosine (MedChem Express, USA), an inhibitor of Akt, and LY294002 (MedChem Express, USA), an inhibitor of PI-3K. The phenotype induced by injection of Perifosine mimicked that of NlPI3K, NlAkt or NlPDK1 dsRNA, i.e., the ratio of short-winged female adults increased, while ratio of short-winged males did not change significantly compared to the dsGFP control animals (Fig. 2B). As with the dsRNA knockdowns, the effects were apparent in 4th but not 5th instar nymphal stages, and were stronger in females than in males (Fig. 2).
The brown planthopper Niparvata lugens Stål and rice. Shown here the short-winged and long-winged form. Rice seedlings before and after brown planthopper infestation are also shown.

Wing-morph ratios changed by disruption of PI3K/Akt/FOXO signaling through injection of dsRNA or inhibitors. A, C, 4th instar nymph, B, D, 5th instar nymph. A: NC(n=82), dsGFP(n= 50), dsnlFOXO(n=49), dsnlAkt(n=32), dsnlPI3K(n=36), dsnlPDK1(n=49), LY294002(n=50), Perifosine(n= 58); B: NC(n=42), dsGFP(n=84), dsnlFOXO(n=77), dsnlAkt(n=113 ), dsnlPI3K(n=74), dsnlPDK1(n= 66), LY294002(n=71), Perifosine(n=50). C: NC(n=61), dsGFP(n=39), dsnlFOXO(n=37), dsnlAkt(n= 23), dsnlPI3K(n=23), dsnlPDK1(n=37), LY294002(n=53), Perifosine(n=51). D: NC(n=53), dsGFP(n= 58), dsnlFOXO(n=68), dsnlAkt(n=103), dsnlPI3K(n=54), dsnlPDK1(n=48), LY294002(n=52), Perifosine(n=52). *P<0.05,**P<0.01. LWF: Long-Winged Female, SWF: Short-Winged Female, LWM: Long-Winged Male, SWM: Short-Winged Male.
RNAi mediated double knockout/inhibition of PI3K/Akt/FOXO Signaling by dsRNA and chemical inhibitors
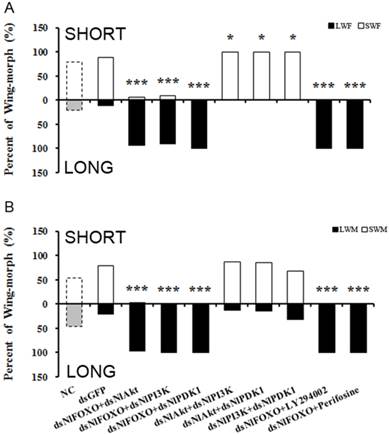
To further study the role of the insulin-signaling pathway in brown planthopper wing polyphenism, we used double gene knockdowns and then observed the resulting wing-morph ratios in the adults. As shown in Fig. 3A, knockout of NlFOXO or NlPI3K separately at the 4th instar nymph stage led to 100% long-winged females, while knockout of NlFOXO and NlPI3K led to a slight reduction to 93% long-winged females (Fig. 3A). Similarly, knockout of NlFOXO and NlAkt led to 92.2% long-winged females (Fig. 3A).
These double silencing experiments reveal some weak negative regulation of NlFOXO by its upstream regulators NlAkt and NlPI3K. However, silencing of the NlFOXO and NlPDK1 through RNAi or through pharmacological inhibition did not change the wing-morph ratio significantly.
Double knockout of NlPDK1 with either NlAkt or NlPI3K increased the percentage of female short-winged forms to 100% (Fig. 3). The female and male also showed different sensitivity, as only the double knockout of NlFOXO plus NlAkt had 2.4% short-winged forms (Fig. 3B). These results support our previous results and are also consistent with the model that FOXO acts downstream of the Akt, PI3K and PDK1. The expression levels of NlPI3K, NlAkt, NlPDK1 and NlFOXO after dsRNA injection were also measured and all showed significant reduction (Supplemental Fig. S5 B,C).
Double knockdown/inhibition of the PI3K/Akt/FOXO Signaling pathway. A, Female. NC(n=82), dsGFP(n=50), dsNlFOXO+dsNlAkt(n=63), dsNlFOXO+dsNlPI3K(n=42), dsNlFOXO+ds NlPDK1 (n=37), dsNlAkt+dsNlPI3K(n=50), dsNlAkt+dsNlPDK1(n=40), dsPI3K+dsPDK1(n=47), dsNlFOXO+LY294002(n=57), dsNlFOXO+Perifosine(n=43). B, Male. NC(n=61), dsGFP(n=39), dsNlFOXO+dsNlAkt(n=45), dsNlFOXO+dsNlPI3K(n=35), dsNlFOXO+dsNlPDK1 (n=59), dsNlAkt+dsNlPI3K(n=31), dsNlAkt+dsNlPDK1(n=40), dsPI3K+dsPDK1(n=49), dsNlFOXO+ LY294002(n=43), dsNlFOXO+Perifosine(n=31). *P<0.05, ***P<0.001. See Fig. 2 for LWF, SWF, LWM, SWM.
Wing morphologies of wildtype and knockdown animals
The brown planthopper wing has bristles along the wing veins that differ in density between short- and long-winged individuals (Fig. 4, Table S1). Although the overall vein and bristle distribution is similar in both wing forms, bristles of long-winged adults are more dispersed than those of short-winged adults, especially in the distal region of the wing (Fig. 4). In addition to wing length, we measured wing bristle density to test whether changes in wing morphology induced by perturbations to insulin signaling resembled naturally occurring differences observed between long and short winged forms. Specifically, bristle number and wing size were measured using NIH ImageJ for wings from both males and females of untreated control animals, dsGFP and water-injected control animals, and dsRNA knockdown and pharmacologically treated animals (NlFOXO, NlAkt, NlPDK1, NlPI3K, NlAkt + NlPDK1, NlPI3K + NlPDK1, NlAkt + NlPI3K, NlFOXO + NLAkt, NlFOXO + NlPI3K, NlFOXO + NlPDK1, Perifosense, LY294002, NlFOXO + Perifosene, NlFOXO + LY294002).
Dendrogram of Hierarchical cluster analysis (HCA) of the brown planthopper wings and representative of each wing-morph. Bristle number/wing area: LY294002 (LWF: n=2), Perifosine(LWF: n=2), dsNlFOXO+dsNlAkt(SWM: n=2), all the others(n=5). Body weight: dsGFP(LWF: n=6; LWM: n=8), NlAkt(LWM: n=6), NlPI3K(LWM: n=3), NlPDK1(LWF: n=5; LWM: n=7), LY294002(LWF: n=1), Perifosine(LWF: n=1: LWM: n=9), dsNlFOXO+dsNlAkt(SWF: n=4; SWM: n=1), dsNlFOXO+dsNlPI3K(SWF: n=4), dsNlAkt+dsNlPI3K(LWM: n=4),dsNlAkt+dsNlPDK1(LWM: n=6), all the others(n=10). Body length: dsGFP(LWF: n=6: LWM: n=8), NlAkt (LWM: n=6), NlPI3K(LWM: n=3), NlPDK1(LWF: n=5; LWM: n=7), LY294002(LWF: n=1), Perifosine(LWF: n=1, LWM: n=9), dsNlFOXO+dsNlAkt(SWF: n=4; SWM: n=1), dsNlFOXO+dsNlPI3K(SWF: n=4),dsNlAkt+dsNlPI3K (LWM: n=4), dsNlAkt+dsNlPDK1(LWM: n=6) , all the others(n=10). See Fig. 2 for LWF, SWF, LWM, SWM.
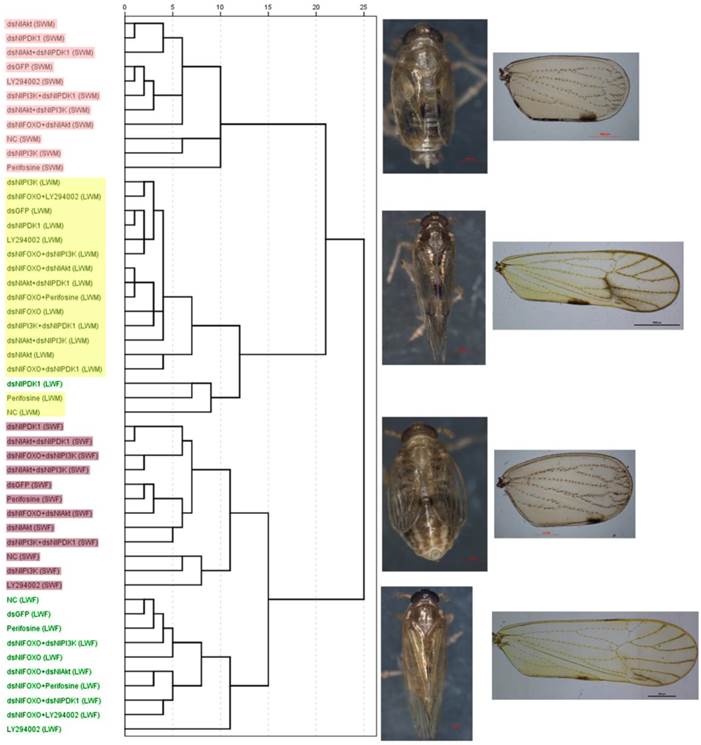
In addition, because each control and treatment population contained both long- and short-winged individuals, measurements were collected for both wing forms of each sex and treatment category, yielding a total of 50 categories. The ratio of bristle number/wing size was also calculated, and body size characteristics including body weight and body length were measured. Data were analyzed by Hierarchical Cluster Analysis (HCA) in SPSS (IBM). Results showed that all 50 categories of brown planthoppers fell neatly into 4 clusters, Long-winged Female (LWF), Long-winged Male (LWM), Short-winged Female (SWF) and Short-winged Male (SWM) (Fig. 4, Table S1). With the exception of dsNlPDK1 injected long-winged females, all wing morphologies of treated animals clustered exactly with natural long- and short-winged forms collected from the field (abbreviations in the parenthesis of the 1st column, Fig. 4, Table S1). This analysis supports the conclusion that our perturbation treatments affected the ratios of long- versus short-winged adults, but not the morphologies of the wings themselves.
Discussion
Wing size is ultimately determined by the number and size of cells. Therefore, polyphenic switching between long- and short-winged brown planthopper forms must entail changes to cell number and/or cell size. In Drosophila, signaling through the insulin pathway controls the rate of cell proliferation and cell size [29]. Specifically, increased signaling through this pathway is marked by increased levels of expression of PI3K, PDK1, and Akt, which inactivate the pathway antagonist FOXO [24, 31, 33, 40]. Overexpression of PI3K, PDK1, and/or Akt, within specific imaginal discs results in overgrowth of these structures [41-46], while over-expression of the Drosophila FOXO (dFOXO) arrests the cell cycle and reduces organ size [29, 31].
Our results are consistent with previous studies in Drosophila showing that PI3K activates PDK1, which then activates Akt, and Akt, in turn, inhibits the activity of FOXO and its downstream targets. Similar interactions among pathway elements are inferred for the brown planthopper: because NlPI3K, NlAkt and NlPDK1 negatively regulate the NlFOXO activity, the phenotypes of silencing NlFOXO are opposite to those of silencing NlPI3K, NlAkt or NlPDK1. Our results were obtained independently of Xu et al.[39] which found two separate insulin receptors regulating polyphenism in the brown planthopper. Our study focused on the downstream elements of the IS pathway and not the insulin receptor but our results confirm the findings of Xu et al.[39]. Based on our model we predicted that disruption of the insulin signaling cascade by injection of NlPI3K, NlPDK1, or NlAkt dsRNA, or by injection of chemical inhibitors of NlPI3K or NlAkt, would reduce wing growth in animals fated to become the long-winged form (Fig. 5). Because signaling through the IS pathway is expected to be largely absent in the wing primordia of animals fated to become the short-winged form, we predicted no effect of these same treatments on these animals (Fig. 5). Similarly, FOXO is predicted to be active in wing primordia of short wing, but not long wing individuals, so disruption of NlFOXO by dsRNA should increase wing growth only in those animals (Fig. 5). Our results were consistent with these predictions and with the knockdown experiments on NlInR1 and NlInR2[39]. Inactivation of NlPI3K/NlPDK1/NlAkt signaling disrupted the formation of long-winged adults, increasing the proportion of short-winged individuals, while disruption of NlFOXO enhanced wing growth in otherwise short-winged individuals, increasing the proportion of long-winged adults[39, 47].
Action model of the brown planthopper wing polymorphism regulated by the PI3K/Akt/FOXO Signaling pathway. Pathway genes marked by (*) showed marked effects on wing phenotype when perturbed, suggesting a functional role in wing growth in animals. Genes whose actions are marked by (X) had no effect on wing phenotype when perturbed, implying that they were inactive in 4th instar wings of these animals.
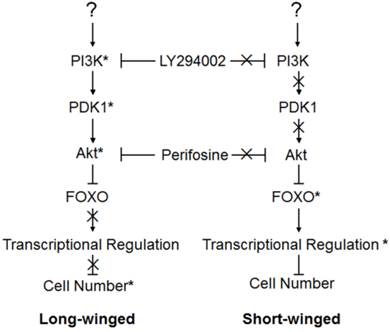
Double knockout/inhibition by dsRNA and chemical inhibitors also supported our model, and permitted us to test, to some degree, the details of the pathway interactions. For example, perturbation of FOXO, predicted to be the most downstream of our tested genes, had by far the strongest effect on wing phenotypes - resulting in almost every instance in complete production of long-winged adults, regardless of what this treatment was paired with. This result makes sense if changes to FOXO effectively erase any alteration to pathway activity occurring upstream. Perturbation of genes predicted to act further upstream, on the other hand, had less dramatic phenotypic effects. Knockdown of NlPI3K and NlPDK1, both relatively upstream, had a smaller combined effect on female wing growth than did combinations of either of these genes with NlAkt, which is farther downstream (Fig. 3B).
In natural populations, the ratio of wing morphs is different in females and males [18, 20]. Variation in sensitivity of female and male brown planthoppers to condition is also seen in the disruption of the PI3K/Akt/FOXO signaling. This sex specific response to condition has been shown in other insect polyphenisms such as in the dung beetle genus Onthophagus [48-50], the pea aphid Acyrthosiphon pisum, [5, 12], and the wing polymorphic field cricket genus Gryllus [2, 5]. Taken together, our results suggest that insulin signaling is required for the wing-morph change of the brown planthopper, through the consecutive action of NlPI3K/NlPDK1/NlAkt and their negative regulation of NlFOXO.
When environmental conditions or the nutritional of the rice deteriorate, favoring development of full-length, dispersal-capable wings, then activation of NlPI3K and NlPDK1 within the wing primordia would activate NlAkt. Activated NlAkt would, in turn, phosphorylate NlFOXO, and subsequent translocation of phosphorylated NlFOXO into the cytoplasm would inhibit its transcriptional activity, promoting cell proliferation and leading to the formation of the long-winged, dispersal form (left, Fig. 5). Alternatively, when environmental conditions or the nutrition of rice favor reproduction rather than dispersal, then inactivation of NlPI3K, NlPDK1 and NlAkt in wing primordia would lead to the nuclear localization of NlFOXO, activating transcription of NlFOXO target genes, inhibiting cellular proliferation, and leading to the formation of the short-winged, reproductive form (right, Fig. 5).
Brown planthopper wing-morph is determined not only genetically, but also by many other factors including hormones, population density, temperature, nutrition and even sub-lethal doses of commonly used insecticides such as imidacloprid and dinotefuran [51]. Therefore, stimulated by the action of both intrinsic and extrinsic factors, these unknown upstream response factors or receptors regulate signaling activity of the PI3K/Akt/FOXO signaling pathway and thus wing polyphenism of the brown planthopper. It will be important to link juvenile hormone signaling to insulin signaling in the regulation of this important wing polyphenism in animals.
Materials and Methods
Insect rearing
The N. lugens brown planthopper population was maintained in the laboratory at China Jiliang University, Hangzhou, China. The criteria for categorizing the long- and short wing brown planthopper was based on the length of forewings and hind wings: long-winged individuals possessed wings that extended longer than posterior end of abdomen, while short-winged individuals posessed forewings shorter than the sixth abdominal segment and hind wings shorter than the first abdominal segment. The brown planthoppers were fed with rice seedlings of IIyou-023 (Oryza sativa L. cv.) in the lab at 25ºC, under 14 hrs:10 hrs light:dark cycle at 70%-80% humidity. Animals were reared at densities of 8-12 animals per 110 cm3 space, the ratios of planthopper wing forms under this condition without treatment were shown in Fig. 2 and Fig. 3(NC).
RNA preparation, cloning and sequence analysis
Total RNA was extracted from equally mixed brown planthopper nymphs and adults using Trizol-based RNAiso Plus total RNA extraction kit (Takara, Dalian). First strand cDNA was synthesized using the Transcriptor First strand cDNA synthesis kit (Roche, Shanghai) and used as a template. The brown planthopper homologues of PDK1, PI3K, Akt, and FOXO were cloned(NlFOXO, KM250122; NlAkt, KM250121; NlPDK1, KM373312; NlPI3K, KM373311). Cloning was performed as previously described [52, 53].The NlFOXO, NlAkt and the NlPDK1, NlPI3K fragment used for dsRNA synthesis were amplified by PCR using Ex-Taq polymerase (Takara, Dalian). The primers used were:
NlFOXOF: 5'TGCTGTGCTTGTTATCATCA3', NlFOXOR: 5'ATTGACGTACCGCTAATGAA3'; NlAktF: 5'TGCTCAGGACTACCAACCATC3', 5'GTTGTGTATGAGCGAATGCG3',
NlPDK1F: 5'CAGTGATGTCGCCGGTGACA3', NlPDK1R: 5'AGCCGCGTCATCTGCTTGTC3'; NlPI3KF: 5'TGAGAGGTTTACATTTCTCC3', NlPI3KR: 5'ACATCAACCACATTCAGAGT3'.
The fragments were then purified with a gel purification kit (Omega, USA) and cloned into the PMD18-T vector (Takara, Dalian) and the sequences were analyzed. Homologous sequences were identified with the Blast program at the NCBI (http://blast.ncbi.nlm.nih.gov). The sequences were analyzed and phylogenic trees were constructed using ClustalW, MEGA5.1.
RNA interference
Double stranded RNA was synthesized using the synthesis kit RiboMAX™ Large Scale RNA Production System-T7 (Promega, Beijing). The template of dsRNA synthesis was amplified by PCR using the PMD18-T plasmid (Takara, Dalian) inserted with a DNA fragment of the gene of interest, and then the PCR fragment was purified with a DNA purification kit (Omega bio-tek, USA). dsRNA synthesis was carried out as described in the Promega technical bulletin TB166 and in our previous paper [52, 53]. Primers used for dsRNA synthesis, including primers for the control gene Green Fluorescent Protein (GFP), are listed in Table 1.
Primers used for dsRNA synthesis.
| Name | Sequence(5′-3′) |
|---|---|
| dsNlPI3KF | TAATACGACTCACTATAGGGAGACCACTGTCGCTCGACTCGGTCGTT |
| dsNlPI3KR | TAATACGACTCACTATAGGGAGACCACGCGCCACCTTTTGTAGCAGG |
| dsNlPDK1F | TAATACGACTCACTATAGGGAGACCACCGCCTCTACTTTGTGCTGAC |
| dsNlPDK1R | TAATACGACTCACTATAGGGAGACCACCTTGGCTCCAGAACCAACAG |
| dsNlAktF | TAATACGACTCACTATAGGGAGACCACTGAACGGAGGGGAGCTGTTCTT |
| dsNlAktR | TAATACGACTCACTATAGGGAGACCACGCAAAGAAGGGATGCGCCAT |
| dsNlFOXOF | TAATACGACTCACTATAGGGAGACCACCTGTTCCCTGAATCGCCGCT |
| dsNlFOXOR | TAATACGACTCACTATAGGGAGACCACCGTTGCAGTCGAATCCGTCG |
| dsGFPF | TAATACGACTCACTATAGGGAGATTTGTATAGTTCATCCATGCCATGT |
| dsGFPR | TAATACGACTCACTATAGGGAGAATGAGTAAAGGAGAAGAACTTTTCA |
Primers used for quantitative real-time PCR.
| Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| NlRPS15 | TAAAAATGGCAGACGAAGAGCCCAA | TTCCACGGTTGAAACGTCTGCG |
| NlPI3K | TACAAAAGGTGGCGCTCTAC | GCGAGCAAGAGAACTGATGA |
| NlPDK1 | ACATTCTGCGCGAACACAAGG | TGTCGCGAACAGCTTCACGA |
| NlAkt | CCTGCATCAGGAGGGCATCA | CTTGCACAGGCCAAAGTCGG |
| NlFOXO | ACCGGTTCATGCGCGTACAG | CTCGACGGCGAGCTGATTTG |
dsRNA was injected into the thorax of CO2-anesthesized 4th and 5th instar nymphs using a Nikon microscope and Narishige injection system (MN-151, Narishige) as previously described [54]. 0.1 μg dsRNA was injected for each insect. The concentration and volume of the dsRNA injections were chosen based on previous studies [53, 54]. Nymphs were reared on rice seedlings after injection and cultured under conditions described above. The percentage of individuals developing with long and short wings was then compared between treatment and control populations using Chi Square tests in SPSS 20.0.
Pharmacological Inhibitor Experiments
Both Phosphatidylinositol 3-kinase (PI-3K) inhibitor LY294002(MW=307, MedChem Express, USA) and AKT Inhibitor Perifosine(MW=462, MedChem Express, USA) were dissolved in ultrapure water at a final concentration of 10 μM. Injection of ultrapure water was used as a control. 1 μl inhibitor was used for each brown planthopper nymph. The mortality, wing-morph and sex were recorded.
Trait measurement
Wing length was measured using a C7 microscope eyepiece ocular micrometer (Shanghai Optics, Shanghai). Wings were examined under a dissection microscope (Nikon SMZ745T) at 3x magnification. Wing length was measured from midway along the vein to the distal end of the wing.
Whole brown planthoppers were imaged using a Nikon microscope (Nikon SMZ745T) with NIS-elements. The wing was removed from the thorax and mounted in euparal on a microscope slide and covered with a cover slip. Then the long wing was imaged with a Nikon microscope (AZ100, 2X) with NIS-elements; short wings were imaged with a Nikon microscope (Eclipse 80i, 4X) with NIS-elements. Images were processed with Adobe Photoshop CS5. Wing area was measured using NIH ImageJ software (NIH, Bethesda, MD, USA). Bristles were counted and recorded for each of the wing types.
Hierarchical Cluster analyses using SPSS 20.0 were used to contrast wing morphologies for natural and experimentally-induced long and short-winged forms. For cluster analysis, a between-group linkage method with Euclidean distance measures to discriminate clusters was used.
Quantitative Real-time PCR
Quantitative real-time PCR was carried out using Roche SYBR® Green PCR Master Mix and SYBR® Green RT-PCR Reagents Kit (Roche Applied Science, Shanghai) and the procedures were similar to that described in our previous paper [52, 53]. Reverse transcription was carried out as described by the supplier. A 25 μl reaction was used as described (Roche Applied Science, Shanghai) and 2 μl of diluted cDNA (20× dilution of the first strand cDNA synthesis reaction) was used for each reaction. We used the 2-ΔΔCt method[55] to compare the relative expression levels of our genes of interest at different developmental stages, in different tissues or before and after gene silencing. NlRPS15 was used as reference genes for quantitative real-time PCR, which were selected according to those previously reported for brown planthoppers [56].
Supplementary Material
Supplementary tables and figures.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31471771), Natural Science Foundation of Zhejiang Province (No. Y14C140007) and National Basic Research Program (973) of China (No. 2010CB126200).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Price TD, Qvarnstrom A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc Biol Sci. 2003;270:1433-40
2. Zera AJ, Denno RF. Physiology and ecology of dispersal polymorphism in insects. Annu Rev Entomol. 1997;42:207-30
3. Zera AJ. The endocrine regulation of wing polymorphism in insects: state of the art, recent surprises, and future directions. Integrative and comparative biology. 2003;43:607-16
4. Zera AJ. Physiological, biochemical, and molecular bases of a nutrient allocation trade-off that underlies a life history trade-off in a wing-polymorphic cricket. Integrative and comparative biology. 2014;54:E233-E
5. Brisson JA. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos Trans R Soc Lond B Biol Sci. 2010;365:605-16
6. Ishikawa A, Gotoh H, Abe T, Miura T. Juvenile hormone titer and wing-morph differentiation in the vetch aphid Megoura crassicauda. Journal of insect physiology. 2013;59:444-9
7. Zera AJ. Differences in survivorship, development rate and fertility between the longwinged and wingless moprhos of the waterstrider, Limnoporus canaliculatus. Evolution. 1984;38:10
8. Zera AJ. B. Induction and function of polyphenic morphs: proximate regulatory mechanisms and evolutionary implications. Integr Organismal Biology. 2014:10
9. Ishikawa A, Miura T. Differential regulations of wing and ovarian development and heterochronic changes of embryogenesis between morphs in wing polyphenism of the vetch aphid. Evolution & development. 2009;11:680-8
10. Ishikawa A, Miura T. Transduction of high-density signals across generations in aphid wing polyphenism. Physiol Entom. 2013;38:7
11. Ishikawa A, Ishikawa Y, Okada Y, Miyazaki S, Miyakawa H, Koshikawa S. et al. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda. J Exp Zool A Ecol Genet Physiol. 2012;317:194-203
12. Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity (Edinb). 2006;97:192-9
13. Cisper G, Zera AJ, Borst DW. Juvenile hormone titer and morph-specific reproduction in the wing-polymorphic cricket, Gryllus firmus. Journal of insect physiology. 2000;46:585-96
14. Bertuso AG, Morooka S, Tojo S. Sensitive periods for wing development and precocious metamorphosis after precocene treatment of the brown planthopper, Nilaparvata lugens. Journal of insect physiology. 2002;48:221-9
15. Morooka S, Tojo S. Maintenance and selection of strains exhibiting specific wing form and body colour under high density conditions in the brown planthopper, Nilaparvata lugens (Homoptera:Delphacide). Appl Entomol and Zool. 1992;27:445-54
16. Morooka S, Ishibashi N, Tojo S. Relationship between wingform response to nymphal density and black colouration of adult body in the brown planthopper, Nilaparvata lugens (Homoptera:Delphacidae). Appl Entomol and Zool. 1988;23:449-58
17. Kisimoto R. Effect of crowding during the larval period on the determination of the wing-form of an adult plant-hopper. Nature. 1956;178:641-2
18. Kisimoto R. Studies on polymorphism and its role playing in the population growth of brown planthopper, Nilaparvata lugens (Stal). Bull Shikoku Agric Exp Stn. 1965;13:101-6
19. Iwanaga K, Tojo S. Effects of juvenile hormone and rearing density on wing dimorphism and oocyte development in the brown planthopper, Nilaparvata lugens. Journal of insect physiology. 1986;32:585-90
20. Iwanaga K, Tojo S, Nagata T. Immigration of the brown planthopper, Nilaparvata lugens, exhibiting various responses to density in relation to Wing morphism. Entomologia Experimentalis et Applicata. 1985;38:101-8
21. Ayoade O, Morooka S, Tojo S. Enhancement of short wing formation and ovarian growth in the genetically defined macropterous strain of the brown planthopper, Nilaparvata lugens. Journal of insect physiology. 1999;45:93-100
22. Ayoade O, Morooka S, Tojo S. Metamorphosis and wing formation in the brown planthopper, Nilaparvata lugens, after topical application of Precocene II. Archives of Insect Biochemistry and Physiology. 1996;32:485-91
23. Ayoade O, Morooka S, Tojo S. Induction of macroptery, precocious metamorphosis, and retarded ovarian growth by topical application of Precocene II with evidence of its non-systemic allaticidal effects in the brown planthopper, Nilaparvata lugens. Journal of insect physiology. 1996;42:529-40
24. Ma Y, Wang H. PI3K/Akt/FoxO: a novel participant in signal transduction in bone cells under mechanical stimulation. Cell biology international. 2012;36:923-6
25. Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL. et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590-603
26. Duronio V, Scheid MP, Ettinger S. Downstream signalling events regulated by phosphatidylinositol 3-kinase activity. Cell Signal. 1998;10:233-9
27. Lee HH, Chang CC, Shieh MJ, Wang JP, Chen YT, Young TH. et al. Hypoxia enhances chondrogenesis and prevents terminal differentiation through PI3K/Akt/FoxO dependent anti-apoptotic effect. Sci Rep. 2013;3:2683
28. Cabodi S, Morello V, Masi A, Cicchi R, Broggio C, Distefano P. et al. Convergence of integrins and EGF receptor signaling via PI3K/Akt/FoxO pathway in early gene Egr-1 expression. J Cell Physiol. 2009;218:294-303
29. Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & development. 2003;17:2006-20
30. Puig O, Tjian R. Nutrient availability and growth: regulation of insulin signaling by dFOXO/FOXO1. Cell Cycle. 2006;5:503-5
31. Mattila J, Bremer A, Ahonen L, Kostiainen R, Puig O. Drosophila FoxO regulates organism size and stress resistance through an adenylate cyclase. Mol Cell Biol. 2009;29:5357-65
32. Straus DS. Nutritional regulation of hormones and growth factors that control mammalian growth. FASEB J. 1994;8:6-12
33. Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293-300
34. Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1-24
35. Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562-6
36. Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF. et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865-75
37. Tang HY, Smith-Caldas MS, Driscoll MV, Salhadar S, Shingleton AW. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS genetics. 2011;7:e1002373
38. Snell-Rood EC, Moczek AP. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS One. 2012;7:e34857
39. Xu HJ, Xue J, Lu B, Zhang XC, Zhuo JC, He SF. et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519:464-7
40. Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol. 2003;3:5
41. Cho KS, Lee JH, Kim S, Kim D, Koh H, Lee J. et al. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc Natl Acad Sci U S A. 2001;98:6144-9
42. Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584-94
43. Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019-29
44. Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500-6
45. Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E. et al. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365-72
46. Nijhout HF, Riddiford LM, Mirth C, Shingleton AW, Suzuki Y, Callier V. The developmental control of size in insects. Wiley interdisciplinary reviews Developmental biology. 2014;3:113-34
47. Lin X, Yao Y, Wang B, Lavine MD, Lavine LC. FOXO links wing form polyphenism and wound healing in the brown planthopper, Nilaparvata lugens. Insect biochemistry and molecular biology. 2015
48. Emlen DJ, Marangelo J, Ball B, Cunningham CW. Diversity in the weapons of sexual selection: horn evolution in the beetle genus Onthophagus (Coleoptera: Scarabaeidae). Evolution. 2005;59:1060-84
49. Emlen DJ, Corley Lavine L, Ewen-Campen B. On the origin and evolutionary diversification of beetle horns. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(Suppl 1):8661-8
50. Emlen DJ, Hunt J, Simmons LW. Evolution of sexual dimorphism and male dimorphism in the expression of beetle horns: phylogenetic evidence for modularity, evolutionary lability, and constraint. Am Nat. 2005;166(Suppl 4):S42-68
51. Bao YY, Li BL, Liu ZB, Xue J, Zhu ZR, Cheng JA. et al. Triazophos up-regulated gene expression in the female brown planthopper, Nilaparvata lugens. Journal of insect physiology. 2010;56:1087-94
52. Lin X, Yao Y, Jin M, Li Q. Characterization of the Distal-less gene homologue, NlDll, in the brown planthopper, Nilaparvata lugens (Stal). Gene. 2014;535:112-8
53. Jin MN, Xue J, Yao Y, Lin XD. Molecular Characterization and Functional Analysis of Kruppel-homolog 1 (Kr-h1) in the Brown Planthopper, Nilaparvata lugens (Stal). J Integr Agr. 2014;13:1972-81
54. Liu S, Ding Z, Zhang C, Yang B, Liu Z. Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect biochemistry and molecular biology. 2010;40:666-71
55. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
56. Yuan M, Lu Y, Zhu X, Wan H, Shakeel M, Zhan S. et al. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One. 2014;9:e86503
Author contact
![]() Corresponding author: Xinda Lin, College of Life Sciences, China Jiliang University, Hangzhou, China, 310018. Telephone: +86-139-5802-8822 Email: linxindaedu.cn.
Corresponding author: Xinda Lin, College of Life Sciences, China Jiliang University, Hangzhou, China, 310018. Telephone: +86-139-5802-8822 Email: linxindaedu.cn.

 Global reach, higher impact
Global reach, higher impact