Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(7):898-910. doi:10.7150/ijbs.14430 This issue Cite
Research Paper
Long Noncoding RNA: Recent Updates in Atherosclerosis
1. Department of Cardiology, Shanghai Tenth People's Hospital, Tongji University School of Medicine, Shanghai 200072, China.
2. Translational Medical Center for Stem Cell Therapy & Institute for Regenerative Medicine, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China.
3. Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital, Fudan University, Shanghai 200032, China.
Received 2015-11-15; Accepted 2016-4-20; Published 2016-6-7
Abstract
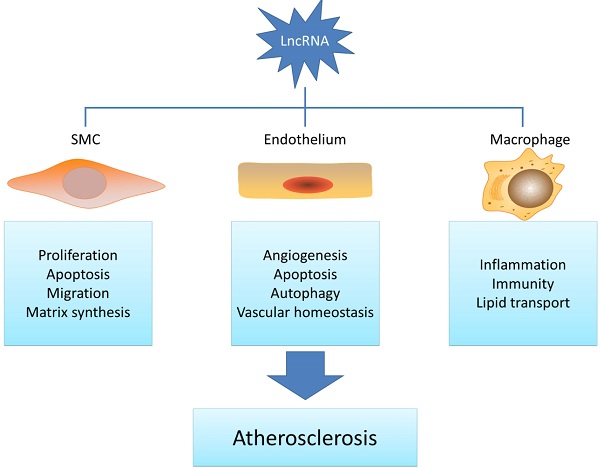
Long noncoding RNAs belong to a class of noncoding RNAs longer than 200 nucleotides with the epigenetic regulation potential. As a novel molecular regulator, lncRNAs are often dysregulated in various pathological conditions and display multiple functions in a wide range of biological processes. Given that recent studies have indicated that lncRNAs are involved in atherosclerosis-related smooth muscle cell, endothelial cell, macrophage and lipid metabolism regulation, it is pertinent to understand the potential function of lncRNAs in atherosclerosis development. This review will highlight the recent updates of lncRNAs in atherogenesis and also discuss their potential roles as novel therapeutic targets.
Keywords: Long noncoding RNA, Atherosclerosis, Gene regulation
Introduction
For a long time, gene regulatory network has been mainly focusing on protein coding genes [1]. However, the fact that about 90% of the human genome is transcribed at specific time points during the development yet only 1% is protein coding, suggests that abundant remaining gene sequences are still unknown [2]. Although they were considered as junk in the past, recent studies have revealed that these noncoding transcripts, which are lowly-expressed, unstable and poorly conserved, play a significant role in the evolution process. Due to the rapid development of high-throughput RNA sequencing technology, a vast number of new noncoding RNAs have been uncovered. Recent studies have also shown that some of these noncoding RNAs play vital regulatory roles in multiple biological processes. Nonetheless, the precise biological function is still far from understood due to their complexity [3]. Noncoding RNAs can be divided by their size into small noncoding RNAs (< 200 nucleotides), which include miRNAs, transfer RNAs and small nucleolar RNAs, and long noncoding RNAs (> 200 nucleotides), which include long intergenic noncoding RNAs, intronic lncRNAs, bidirectional lncRNAs, sense lncRNAs, antisense lncRNAs, and enhancer RNAs according to the adjacent protein-coding genes. LncRNAs can be further distinguished by their cellular location to nucleus or cytoplasm and by their action type to in cis or in trans. Notably, the distinction is not very clear that some lncRNAs may locate in both nucleus and cytoplasm or work both using in cis and in trans dogma [2, 3]. The majority of lncRNAs are located in the nucleus with only 15% in the cytoplasm [4]. For the nuclear lncRNAs, most show transcriptional repression effects by guiding chromatin modifiers like PRC2 to specific genomic targets [3]. Khalil et al., suggest that about 20% of lincRNAs are bound by PRC2 [5]. Xist is the typical example which involves in X-chromosome inactivation. It specifically captures PRC2 through a conserved repeat motif and takes it to the inactive X chromosome with the help of YY1 protein, which binds both Xist and the target gene loci, thus initiating X-chromosome inactivation in cis [6, 7]. It could also act in trans to inactivate the distal X-chromosome by identifying the three-dimensional structure of the X-chromosome [8]. Fendrr specifically binds to PRC2 and guides PRC2 to the promoters of Foxf1 in cis and Pitx2 in trans [9]. ANRIL could work as a scaffold to bind both PRC1 and PRC2 to repress its targets [10]. LncRNAs could also indirectly regulate target genes by acting as a decoy to sequester regulatory factors, e.g. MALAT1 works as a decoy to trap and accumulate specific splicing factors in nuclear speckles. The knocking-down of MALAT1 results in homogeneous distribution of splicing factors in the nucleus thus inhibiting pre-mRNA alternative splicing in trans [11]. For cytoplasmic lncRNAs, most of them regulate on translation level by base pairing with their targets [3]. One of the targets is miRNAs. These lncRNAs are also called ceRNAs which act like sponges to prevent miRNAs from binding to their targets. CARL functions mainly in cytoplasm and works as a sponge to target miR-539 thus blocking its repression on Phb2 [12]. In some cases, lncRNAs serve as the precursor gene of miRNAs with which they are co-transcribed. Some studies show that knocking down the lncRNA could concomitantly influence miRNA transcription [13]. Other studies also indicate that lncRNAs could regulate miRNAs on transcriptional levels. HULC down-regulates miR-9 by inducing methylation of the miR-9 promoter [14]. Another target is mRNA, LincRNA-p21 targets CTNNB1 and JUNB mRNAs and represses them [15]. TINCR can influence the stability of its target mRNA to modulate its expression [16]. LncRNAs could also be co-activators or enhancers by acting as essential elements in target gene activation. Jpx knocking-down inhibits Xist activation and expression of Jpx rescues Xist activation [17]. SRA is a component of a ribonucleoprotein complex which contains the typical co-activator protein SRC-1 and SRA works as a RNA co-activator to selectively enhance transcriptional activation [18]. LncRNAs could specifically bind to other proteins rather than chromatin modifiers. LincRNA-p21 binds with hnRNP-K and guides it to the promoters of p53 target genes thus suppressing p53 target genes [19]. LincRNA-p21 also binds to MDM2 thus enhancing p53 activities [20]. It is therefore obvious that the regulatory roles of lncRNAs are complicated due to abundant unknown areas. Yet, exiting data have already indicated that lncRNAs may selectively bind with proteins, mRNAs or miRNAs to sequester or guide them to target genes to participate in various cellular and molecular regulations. Importantly, as many lncRNAs are poorly conserved between different species even different cells, they may serve as potential cell specific targets.
Although the mortality rate has declined, cardiovascular disease is still the leading cause of death worldwide. In 2011, cardiovascular disease accounts for one out of every three deaths in the United States. Medical cost for cardiovascular diseases is still a heavy economic burden [21]. Atherosclerosis concerns the fundamental vascular change in cardiovascular diseases. Previous studies have shown that atherosclerosis is a combination of lipid disorder and chronic inflammatory disease [22]. Endothelial cell dysfunction caused by oxidative, hemodynamic, or biochemical stimuli initiates the progress of atherosclerosis. Following that, EC's permeability changes and macrophages start to accumulate and release inflammation factors. Smooth muscle cells are then activated and begin proliferation and migration. Therefore, the pathogenesis of atherosclerosis is a result of various changes and interactions in multiple cell types in the artery walls which mainly includes lipid deposition, endothelial cell dysfunction, macrophages activation, and SMC alternation [22]. As a deadly disease, new cellular and molecular mechanisms are urgent to find in order to provide novel therapeutic targets.
Previous reports have shown that miRNAs play a significant role in the development of atherosclerosis [23]. Due to the various functions of lncRNA and its regulatory role as the ceRNA, it may be reasonable to speculate that lncRNAs may directly or indirectly contribute to the development of cardiovascular disease and atherosclerosis. This review will thus summarize the current findings of lncRNAs in atherosclerosis-related cells and provide an overview of lncRNA's regulation in atherosclerosis.
LncRNAs in SMC regulation
The proliferation and migration of SMCs are thought to be the key events in atherosclerotic lesion progression and restenosis. At the initial stage of atherogenesis, SMCs switch their phenotype from contractile to synthetic and move into the intima resulting in intimal hyperplasia [24, 25]. A number of studies have examined the potential function of lncRNAs in SMC's proliferation and migration.
The chromosome 9p21 locus SNPs have been reported to increase the risk of atherosclerosis and myocardial infarction [26, 27]. The nearest protein-coding gene of the 9p21 locus is CDKN2A/B which encodes cell proliferation regulators: p16INK4a, p14ARF and p15INK4b. Motterle et al. find that the 9p21 locus risk SNPs promote VSMC proliferation and reduce p16INK4a and p15INK4b expressions in VSMCs. Furthermore, in atherosclerotic lesions, there are lower levels of p15INK4b expression and higher levels of VSMC proliferation [28]. This and some other studies also suggest that the 9p21 locus risk SNPs influence the long noncoding RNA ANRIL transcription which overlaps the region [29]. The risk SNPs increase the expression of ANRIL transcripts both in plaque and peripheral blood. Meanwhile, ANRIL is closely associated with the severity of atherosclerosis [30]. Further studies demonstrate that ANRIL regulates VSMC growth through CDKN2A/B. The knocking-down of ANRIL in VSMC increases the expression of CDKN2B and inhibits VSMC proliferation [28, 31]. Kotake et al. prove that ANRIL binds to SUZ12, a component of PRC2, and recruits it to p15INK4b thus repressing p15INK4b expression by RNA immunoprecipitation. Using similar technology, Yap and colleagues report another mechanism through which ANRIL interacts with CBX7, a component of the PRC1 thus repressing p16INK4a whereas p15INK4b is not affected [32, 33]. Although there are conflicting binding proteins and functions, ANRIL inhibits CDKN2A/B expression in the end and reduces cell growth. Despite the cis effect, Sato et al. suggest a trans effect by overexpressing ANRIL and find some regulated genes in trans in HeLa and lung tumour cells [34]. Holdt and co-workers, through the gain and loss of function analysis, further prove that the regulated genes in trans are associated with proliferation, cell adhesion, and apoptosis and ANRIL regulates these genes probably by recruiting CBX7 and SUZ12 to the promoters in trans [35]. Finally, it is still uncertain whether ANRIL regulates VSMC proliferation through the mechanisms mentioned above as these studies are conducted in tumour cells. ANRIL's exact mechanism in VSMC needs further investigation. Nonetheless, ANRIL has been proved to regulate CDKN2A/B in VSMC and has revealed a strong regulatory function in VSMC proliferation. As a potential atherosclerosis risk factor and therapeutic target, future studies may concentrate on the circulating ANRIL for biomarkers of atherosclerosis severity and the plaque's ANRIL for pharmacological targets.
The H19 gene is a well-known imprinted gene important in cell differentiation and growth. Its noncoding RNA transcript is H19 lincRNA and H19 lincRNA is the precursor of miR-675. H19 lincRNA has been found to enhance or weaken tumour growth and invasion, respectively, in different types of tumours [36-38]. H19 lincRNA is also highly expressed in neointima after injuries and human atherosclerotic lesions but barely expressed in normal arteries [39, 40]. Gao and co-workers examine the polymorphisms in H19 and find that individuals carrying the risk alleles of H19 display a higher risk of coronary artery disease [41]. It is thus reasonable to assess H19 lincRNA's role in atherosclerosis. MiR-675 is encoded by H19 lincRNA and previous studies indicate that H19 lincRNA controls the expression of miR-675 in regulating the growth of placenta [42]. Recent studies suggest that H19 lincRNA functions as a sponge of the let-7 family miRNA and let-7 miRNAs have been indicated to protect VSMC from oxidative damage [43, 44]. Whether H19 lincRNA regulates SMC through the let-7 pathway or by miR-675, thus promoting atherosclerosis is still ambiguous. More studies are warranted to assess the relationship between H19 lincRNA level and the plaque severity. Furthermore, more precise mechanism under H19 lincRNA in VSMC is needed. Leung et al. conduct an RNA-seq of Ang II-stimulated VSMC and identify a novel lncRNA named Lnc-Ang362 which may promote VSMC proliferation [45]. It is the host gene of miR-221 and miR-222 which have been proven to be regulators in VSMC proliferation and neointimal hyperplasia [46]. Knocking-down of Lnc-Ang362 reduces miR-221 and miR-222 expression and inhibits SMC proliferation. Therefore, Lnc-Ang362 could promote VSMC proliferation through these two miRNAs. [45].
p53 is a tumour repressor gene involved in cell proliferation and apoptosis. Previous studies also indicate that p53 plays an important role in atherosclerosis progression [47]. Recent studies reveal that lincRNA-p21 is controlled by p53. LincRNA-p21 directly binds with hnRNP-K and guides it to the promoters of p53 repressed genes thus involves in p53 downstream repression [19]. Cytoplasmic lincRNA-p21 also modulates translation process by direct binding to its target mRNAs [15]. Wu et al. further identify the role for lincRNA-p21 in SMC proliferation and apoptosis by revealing a novel mechanism. The direct binding between lincRNA-p21 and the protein of a p53 downstream gene MDM2 is firstly observed. Knocking-down of lincRNA-p21 changes the balance between MDM2/p53 complex and p300/p53 complex and as a result, suppressing p53 activity. Above all, p53 regulates SMC proliferation and apoptosis in a lincRNA-p21 dependent manner. In vivo, siRNA-induced lincRNA-p21 inhibition stimulates SMC proliferation and neointimal hyperplasia. Clinical findings also reveal that coronary tissues from CAD patients exhibit a reduced level of lincRNA-p21 compared to aorta tissues from non-CAD patients [20]. Similar to ANRIL and H19 lincRNA which have been tested in lesion area, lincRNA-p21 may also serve as a potential biomarker of atherosclerosis and a therapeutic target.
Hyaluronan is an important component of the arterial walls and excess hyaluronan enhances atherosclerosis progression [48]. Recent findings show both HAS2 and HAS2-AS1, which is an antisense transcript of HAS2 are related to the severity of atherosclerosis. HAS2 and HAS2-AS1 are modulated under metabolic disorders like diabetes. Using nuclease accessibility assays, Vigetti et al. find that hyperglycemic stress induces HAS2-AS1 expression and HAS2-AS1 transcripts could identify specific region on HAS2 and bring chromatin modifiers to the promoters of HAS2 thus up-regulating HAS2 transcription in cis [49]. The exact chromatin modifier is still unknown but targeting artery substrate is an important aspect in preventing atherosclerosis progression.
More other lncRNAs are also found to be associated with SMC although studies on these lncRNAs are still limited. Zhao et al. screen differentially expressed lncRNAs in patients of aorta aneurysm and find HIF1α-AS1. By knocking down HIF1α-AS1 in SMC, apoptosis and apoptosis-related proteins are down-regulated in SMC indicating its role in SMC apoptosis [50]. Li et al. screen lncRNAs in varicose veins and find lncRNA-GAS5. In vitro studies depict that it may inhibit proliferation, migration and cell cycle of SMC. Furthermore, Li et al. identify a direct binding between lncRNA-GAS5 and the protein Annexin A2. Overexpressing Annexin A2 could block lncRNA-GAS5's function on proliferation and migration [51]. Bell et al. find a new lncRNA called SENCR through the RNA-seq of SMC. It is an antisense of FLI1 but there is no cis effect on it. Instead, reduced SENCR could down-regulate contractile related genes (Myocd) and up-regulate migratory genes (Mdk and Ptn) in trans. Therefore, SENCR promotes a synthetic SMC phenotype and cell migration [52].
Overall, a growing number of studies have reported that lncRNA is capable of regulating proliferation, migration and matrix synthesis in SMC, all of which participate in different aspects of lesion formation. To this end, interventions targeting specific lncRNAs may suppress lesion development and luminal stenosis. Some of these lncRNAs have already been identified in lesion area and display an association with the severity of atherosclerosis. However, the research on a wide set of newly identified lncRNAs and their regulating pathways are still in its infancy. Meanwhile, mining the abundant remaining sequence data from RNA sequencing results is a promising approach to identify new gene regulators.
LncRNA in EC function
Endothelial dysfunction stemmed from EC injury represents an early step in the onset and development of atherosclerosis. Injured ECs result in increased permeability and accumulation of adhesion proteins, thereby stimulating migration of leukocytes into the vessel walls [22]. A unique role for LncRNA in ECs has been depicted, many of which indicate that lncRNAs may regulate EC functions especially in angiogenesis. Further evidence reveals that lncRNAs are induced in hypoxic or hyperglycemic conditions which should provide new theory to regulate EC injuries.
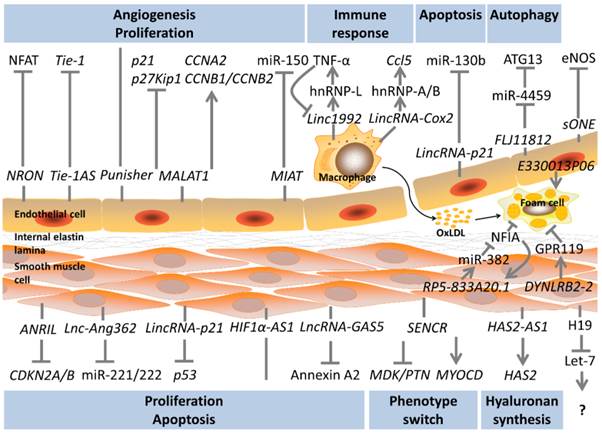
NFAT plays a significant role in cardiac valvular formation. Upon stimulation, NFAT is dephosphorylated and moves into the nucleus to function as a transcriptional factor [53]. The long noncoding RNA NRON is identified as a new cytoplasmic lncRNA which comprises a NRON complex by binding to KPNB1/PPP2R1A/PSMD11/IQGAP1. NRON acts as a decoy to trap NFAT thus inhibiting NFAT's nuclear distribution [54, 55]. Ets-1 could accumulate in the cytoplasm and competitively displace NFAT from the NRON complex thus weakening NRON's function [56]. Tang et al. investigate NRON's role in ECs through the gain and loss of function analysis and find that NRON overexpression inhibits NFAT expression which reduces ECs' proliferation, tube formation and migration. On the other hand, siRNA-induced NRON inhibition results in activation of NFAT which promotes ECs' proliferation, tube formation, and migration [57]. Another lncRNA which may participate in vessel formation is Tie-1 AS. Tie-1 was previously found to be important in vessel formation [58]. Li et al. find that Tie-1 AS and Tie-1 transcripts are expressed in a manner reminiscent of zebrafish embryonic development indicating its role in Tie-1 regulation. By bioinformatics and hybridisation analysis, Li et al. further confirm that Tie-1 AS binds to Tie-1 mRNA to down-regulate Tie-1 translation. In HUVECs, Tie-1 AS overexpression inhibits Tie-1, thus affecting tube formation [59]. Recently, three new lncRNAs are identified in EC's differentiation and angiogenesis. During the process when the pluripotent stem cell differentiates to ECs, three novel lncRNAs are significantly induced. Terminator is mainly expressed in pluripotent stem cell period, while Alien and Punisher are specifically expressed in cardiovascular progenitors and endothelial cells, respectively. In mice and zebrafish, similar trends are found. Loss of function analysis shows that Terminator knockdown mainly influences genes related to pluripotent maintenance. Alien knockdown mainly affects cell adhesion, extracellular matrix remodelling, angiogenesis and blood vessel development. Punisher knockdown mainly impacts mitosis, cell adhesion and extracellular interaction. In vitro analysis of Punisher knockdown shows that EC proliferation and tube formation are significantly reduced [60]. NRON, Tie-1 AS and Punisher are involved in EC tube formation through different mechanisms. Due to their essential roles in angiogenesis, these lncRNAs might be potential regulators in neovascularization.
MALAT1 was firstly identified in lung cancer [61]. Later, increased MALAT1 levels are found in several other types of tumours and overexpression of MALAT1 results in proliferation and metastasis [62, 63]. Recent studies show that MALAT1 are induced in stressful conditions like hypoxia and hyperglycemia in ECs [64-66]. MALAT1 plays a significant role in alternative splicing of pre-mRNAs by maintaining specific splicing factors in the nuclear speckles. Depletion of MALAT1 results in homogeneous distribution of SR proteins and higher ratios of dephosphorylated SR proteins. In a word, MALAT1 regulates alternative splicing by controlling the distribution and activity of SR proteins [11, 67]. In ECs, Michalik et al. find that the expression of MALAT1 is significantly induced in hypoxia-treated HUVEC and that silencing MALAT1 promotes EC's migration, sprouting and inhibits EC proliferation. In vivo studies reveal that both neovascularization in the retina and capillary density after hind limb ischemia are reduced in MALAT1 knockout mice. Microarray analysis following knocking down MALAT1 shows that cell cycle regulators such as CCNA2, CCNB1, and CCNB2 are down-regulated and p21, p27Kip1 are increased. There is little sign of splicing-related genes getting affected [64]. In diabetic mice, MALAT1 expression is up-regulated in retinas and kidneys and silencing MALAT1 is found to inhibit diabetic retinopathy such as pericyte loss, capillary degeneration and microvascular leakage. In ECs stimulated with hyperglycemia, MALAT1 is found not only involved in angiogenesis but also in inflammation. The knocking-down of MALAT1 decreases the proliferation, migration, and tube formation in hyperglycemic ECs. Meanwhile, inflammation factors such as IL-6 and TNF-α are also reduced [65, 66]. These studies show conflicting results with Michalik et al. in in vitro studies especially in migration result which is probably because different cell types (HUVEC and retinal endothelial cell) and stimuli. Nonetheless it is obvious that MALAT1 plays an essential role in macro- and micro-vascular angiogenesis by regulating EC in stressful conditions. As MALAT1 is also induced in stressful stimuli, there is a possible link between MALAT1 and the severity of EC injury. Meanwhile, targeting MALAT1 is a new approach to reduce diabetic retinal neovascularization and inflammation. MIAT has been found to be associated with myocardial infarction previously [68]. Recently, it is also found involved in diabetic microvascular disease. MIAT level is up-regulated in diabetic retinas and hyperglycemic ECs. MIAT knockdown could also inhibit EC proliferation, migration, and tube formation. In vivo analysis shows that MIAT knockdown attenuates diabetes-induced pericyte loss, capillary degeneration and microvascular leakage. But the underlying mechanism is miR-150 dependent. VEGF is the repression target of miR-150. MIAT up-regulates VEGF by working as a decoy to suppress miR-150 [69]. LincRNA-p21, like its role in SMC, also up-regulates EC apoptosis and inhibits cell cycle but the underlying mechanism is a sponge of miR-130b [70]. Another sponge lncRNA is FLJ11812. Ge et al. investigate the role of FLJ11812 following a microarray analysis with 3DBO-treated ECs. Through luciferase assay, FLJ11812 is proved to target miR-4459 as a sponge. ATG13 is the repression target of miR-4459. Via the FLJ11812/miR4459/ATG13 pathway, FLJ11812 could regulate ATG13 levels thus involving in autophagy in the 3DBO downstream pathways [71]. Reduced nitric oxide synthase levels have been found in atherosclerosis. One of the nitric oxide synthase, which is specially expressed in endothelial cells, termed eNOS, is a key regulator in vascular wall homeostasis [72, 73]. Robb et al. thus investigate the regulation of eNOS and report an antisense transcript to eNOS called sONE. It is interesting because sONE is expressed in various types of cells except ECs in normal conditions. Through the gain and loss of function analysis in ECs, sONE is found to reduce eNOS protein but not the mRNA levels indicting its role in posttranscriptional regulation. A lower level and stability of eNOS mRNA is observed in hypoxia ECs, and sONE is reciprocally expressed in hypoxia stimuli [72]. It is reasonable to confer that sONE plays a role in regulating eNOS in this process. Fish et al. find that hypoxia induces sONE by increasing its RNA stability rather than its transcription regulation. In addition, hypoxia induces sONE cytoplasmic accumulation. sONE knockdown reduces the attenuation of eNOS mRNA during hypoxia [74]. Targeting sONE could be a new approach to increase eNOS thus maintaining vascular homeostasis.
Currently, lncRNAs are involved in EC proliferation, tube formation and migration. Some lncRNAs are angiogenesis regulators under normal conditions, while others also function under hypoxic or hyperglycemic stress which may stimulate atherosclerosis process more aggressively. For those with stressful stimuli, there remains the possibility to use them as biomarkers for early EC injury or new molecular targets for EC injury regulation. The two very important lncRNAs MALAT1 and MIAT are induced in diabetic retinal disease and targeting them could reduce the retinal vessel impairment and ameliorate retinal function.
LncRNA in macrophage and immunity
Atherosclerotic lesion is composed of accumulation of lipoproteins, macrophages-derived foam cells within the arterial wall. Macrophages are activated and move into the vessel wall, devour cholesterol to form foam cells, and then release inflammatory factors thus exacerbating macrophage accumulation [22]. This suggests that there is a linkage between lipid metabolism and innate immune response. It is important to find new targets in cellular lipid transport and immunity. Although only a few macrophage-related lncRNAs are identified, researchers have revealed their significant regulatory role in macrophages and inflammation.
Cellular lipid mechanism especially in foam cells is an important issue in atherosclerosis progress. Hu et al. find that lncRNA is a novel regulator in cholesterol metabolism and inflammation. Through microarray analysis, Hu et al. try to identify the expression profile of macrophages and macrophage-derived foam cells. LincRNA-DYNLRB2-2 and lncRNA RP5-833A20.1 are greatly induced during this process, both of which could regulate cellular cholesterol metabolism and inflammation, but in different pathways. LincRNA-DYNLRB2-2 decreases cellular inflammatory cytokines (TNF-α, IL-1β and IL-6) and promotes cholesterol efflux in macrophages under hyperlipidemic stress. Further studies show that lincRNA-DYNLRB2-2 regulates ABCA1, which is an important role in the efflux of cellular lipid, in a GPR119-dependent manner [75]. Another long noncoding RNA RP5-833A20.1 is located in intron 2 of the NFIA gene and an antisense of it. It increases inflammatory cytokines (TNF-α, IL-1β and IL-6) and reduces cholesterol efflux via the miR-382-mediated NFIA pathway. Further studies show that ABCA1 is also involved in this regulation. LncRNA RP5-833A20.1 could attenuate ABCA1 levels thus reduce cholesterol efflux [76]. It is an interesting finding here because unlike most of the lncRNA-miRNA relationships, lncRNA RP5-833A20.1 up-regulates miR-382 thus suppressing NFIA. This is a conflicting result with previous findings which probably in-part because miR-382 may not be the direct downstream target of lncRNA RP5-833A20.1 and they both participate in the regulatory network of cellular lipid. Both of these two lncRNAs show a regulatory function in cellular lipid mechanism and inflammation. Hu et al. have proven the role of GPR119 and NFIA in atherosclerosis. However, it is still not clear whether the two lncRNAs could show the same function in vivo by regulating in vitro proved targets. Mechanism studies on these two important lncRNAs are rather limited. A recent RNA-seq conducted by Reddy et al. [77] compares the expression pattern of macrophages from diabetic and non-diabetic mice and a new pro-inflammatory lncRNA E330013P06 is identified. E330013P06 is the host gene of miR-143 and miR-145. Overexpressing E330013P06 increases inflammatory genes (Nos2, Il6 and Ptgs2) and scavenger receptor CD36, thus promoting foam cell formation. Knocking down E330013P06 could only inhibit CD36 expression indicating its partial function in reversing inflammation. MiR143/145 has been found a key regulator in SMC contractile phenotype acquisition [78]. Loss of miR143/145 in mice results in impaired vasocontraction [79]. As E330013P06 is the host gene of these two miRNAs, it may speculate that E330013P06 plays a role in SMC phenotype regulation.
LncRNAs have also been found to be a novel regulator in the immune response which can inhibit or promote inflammatory signaling [80]. Carpenter et al. conduct a RNA-seq in macrophages stimulated with TLR2 and identify a novel lncRNA called lincRNA-Cox2, which locates proximal to the COX2 gene. In loss of function analysis, lincRNA-Cox2 is proved to regulate various kinds of immune genes before (e.g. Ccl5) and following TLR stimulation (e.g. Il6). Previous studies have indicated that lncRNAs could bind to proteins to regulate transcription process [5]. To further understand the underlying mechanism, mass spectrometry is used to screen the possible specific proteins which lincRNA-Cox2 interacts with. HnRNP-A/B and hnRNP-A2/B1 are finally identified. Recruitment of RNA polymerase II to the promoters of Ccl5 is found when knocking down hnRNP-A/B and hnRNP-A2/B1, no similar trend is found in Il6 promoters [80]. Therefore, it can be concluded that one of the regulatory pathways is: lincRNA-Cox2 binds with hnRNP-A/B and hnRNP-A2/B1 to form an RNA-protein complex thus, influencing the recruitment of RNA polymerase II to regulate the transcription of inflammatory genes. Recently, Li et al. have also identified a similar altered lncRNA through a microarray analysis in TLR2-stimulated macrophages termed linc1992. Linc1992 is widely expressed in various tissues. Meanwhile, TNF-α mRNA and protein are strongly repressed after linc1992 knockdown. Linc1992 expression could also be inhibited by exogenous TNF-α indicating a feedback loop between them. To understand the underlying mechanism, a similar method is used to investigate the interaction between linc1992 and hnRNP. Only hnRNP-L could bind specifically to linc1992. The knocking-down of hnRNP-L could decrease TNF-α expression, and through chromatin immunoprecipitation, hnRNP-L is proved to bind directly to the promoter of TNF-α. Therefore, by binding with hnRNP-L and forming a linc1992/hnRNP-L complex, linc1992 promotes the transcriptional process of TNF-α [81]. In this study, Li et al. also find that the linc1992 knockdown could influence many other immune-associated genes which future studies may evaluate. These two studies promote the role of lncRNAs in immune response in macrophages. In particular, the hnRNP family is a significant mediator between lncRNA and its targets.
Another lncRNA found to be induced in immune response is PTPRJ-AS1, a long noncoding RNA on the antisense strand of the Ptprj. LPS could induce a transient increase in PTPRJ-AS1 expression. The exact targets and the regulatory role of PTPRJ-AS1 are however, still unknown [82].
In the end, lncRNAs may regulate macrophages or foam cells in a number of ways predominantly via inflammatory and cellular cholesterol efflux mechanisms. LincRNA-DYNLRB2-2 and lncRNA RP5-833A20.1 have revealed that lncRNAs could regulate foam cell formation. Studies on lincRNA-Cox2 and linc1992 may further focus on foam cell regulations. Meanwhile, in vivo studies are still limited.
LncRNA with lipid metabolism
It is well known that lipid imbalance is a significant risk factor of atherosclerosis. Lipid disorders cause lipoprotein accumulation within artery walls thus triggering atherosclerosis [22]. Some studies have also indicated that lncRNA is involved in lipid metabolism.
HULC was firstly identified in hepatocellular carcinoma [83]. It has been described to promote proliferation and inhibit the tumour suppressor p18. Meanwhile, it could be a sponge of miR-372 [84, 85]. Cui et al. identify a novel role of HULC which regulates the lipid afflux in hepatoma cells. The knocking-down of HULC affects PPARA which is the transcriptional factor of ACSL1 and ACSL1 is essential in cellular long-chain fatty acid metabolism. Cui et al. further predict a miRNA which targets PPARA and identifies miR-9. HULC could repress miR-9 but not as a ceRNA. Instead, methylation-specific PCR and bisulfitesequencing analysis reveal that the CpG sites of miR-9 are highly methylated when overexpressing HULC. Meanwhile, HULC up-regulates DNMT1 which is a key role in inducing methylation of CpG islands in the miRNA promoter. Therefore, HULC targets miR-9 on transcriptional level by up-regulating DNMT1. Above all, by reducing miR-9 expression, HULC up-regulates PPARA thus increasing ACSL1 which in the end results in triglyceride and cholesterol accumulation in tumour cells [14]. Although HULC plays an important role in cell lipid afflux, no one has ever reported its roles in atherosclerosis especially in foam cells. Hu et al. [75, 76] find two lncRNAs in foam cells: lncRNA-RP5-833A20.1 and lincRNA-DYNLRB2-2 which could both regulate foam cell cholesterol efflux and inflammation. Meanwhile, their down-stream targets have been shown to regulate plasma lipid and atherosclerosis which indicate their important role in regulating atherosclerosis progression. SRA was firstly identified as a RNA coactivator involved in steroid receptor-dependent gene expression [18]. Other studies also indicate that SRA is involved in adipogenesis [86], tumourigenesis [87] and myogenesis [88]. Liu et al. further show its role in SRA knockout mice and reveal that following high-fat diet, SRA knockout mice result in reduced adiposity with decreased body fat. Moreover, decreased lipogenesis-associated genes such as Ppara, Pparg, Fabp4 and Lipe are found [89]. Considering all above functions, it may be speculated that SRA is involved in lipid metabolism. Halley et al. [90] identify an antisense in the ApoA1 locus called APOA1-AS. The knocking-down of APOA1-AS with siRNA could increase ApoA1 in cis. Chromatin immunoprecipitation analysis shows that after knocking down APOA1-AS, H3K4 trimethylation is active around ApoA1 promoter, and H3K27 trimethylation is found to be inactive around ApoA1, ApoA4, and ApoC3. Meanwhile, LSD1 and SUZ12 recruitments are reduced. APOA1-AS could regulate ApoA1 by guiding LSD1 and SUZ12 to the promoter of ApoA1 which causes decreased H3K4 trimethylation and increased H3K27 trimethylation. As ApoA1 is important in lipid efflux and retains atherosclerosis, APOA1-AS might be a risk factor in lipid accumulation.
LncRNAs show their potential function in lipid metabolism. Studies have shown that some important lipid transport and lipogenesis genes are regulated in gain and loss of function analysis. However, until now, studies have only discussed this issue on the cellular level. In vivo function studies of lncRNA in plasma cholesterol, triglycerides or lipoproteins are still lacking.
LncRNAs as cardiovascular biomarkers
Circulating biomarkers have greatly improved the diagnosis accuracy of acute coronary disease. The identified biomarkers today are mostly proteins and peptides. MiRNAs have been discovered to include in the diagnosis and prognosis of patients with myocardial infarction and heart failure [91]. As lncRNAs are involved in various cardiovascular disease progresses, it has also been potential biomarkers in cardiovascular disease [92].
ANRIL has long been investigated as a potential biomarker as it is found increased in plaque and plasma of atherosclerosis patients [30, 93]. In a cohort study, three of the ANRIL transcripts (NR_003529, DQ485454, and EU741058) are investigated. It reveals that NR_003529 and DQ485454 increase in carriers of the risk SNPs individuals, while DQ485454 stays unaffected indicating that different ANRIL transcripts may vary in their functions. Moreover, NR_003529 and DQ485454 correlate with severity of atherosclerosis [30]. Kumarswamy et al. identify a differently expressed lncRNA in patients' plasma with and without cardiac remodelling following myocardial infarction. A lincRNA called LIPCAR is consistently detectable and increases significantly in patients with cardiac remodelling. It is further evaluated in another two studies, one of which is a cohort and the other is a case-control study. LIPCAR is found higher both in ischemic heart failure and non-ischemic heart failure. Higher LIPCAR level is associated with cardiovascular mortality. In the latter study, LIPCAR is further evaluated and after adjusting for other risk factors, LIPCAR is still an independent predictor of 3-year cardiovascular death [94]. Vausort et al. further compare five plasma-detectable lncRNAs between myocardial infraction and healthy volunteers, and find four differently expressed lncRNAs including HIF1αAS2, KCNQ1OT, MALAT1 which are up-regulated and ANRIL which is down-regulated. MIAT does not differ between the two groups. Although they are induced in myocardial infraction, only KCNQ1OT1 and ANRIL show predictive values of LV dysfunction [95].
The clinical use of lncRNA as biomarkers in atherosclerosis and cardiovascular disease seems constraint until now probably due to the relatively low level of lncRNAs in plasma and the weak predictive effect (Summary See Table 2). It is obvious that the study on lncRNAs as biomarkers is just starting. ANRIL has shown to be induced and associated with atherosclerosis and LV dysfunction, LIPCAR, KCNQ1OT and MIAT reveal to predict cardiovascular outcome. Whether the vast amount of unannotated lncRNAs are better predictive biomarkers than the existing ones are to be determined. The stability and detectability of lncRNAs in plasma also need to be taken into account [92].
Summary of atherosclerosis-related lncRNAs
| Cell type/ process | lncRNA | Class | Location | Roles / targets | Function |
|---|---|---|---|---|---|
| SMC | ANRIL[28-35] | Antisense | Nucleus | Guide, scaffold CDKN2A/B | Proliferation ↑/ adhesion ↑/ apoptosis ↓ |
| H19 lincRNA[42-44] | LincRNA | Cytoplasm | Precursor, ceRNA miR-675 Let-7 miRNA | Unknown | |
| Lnc-Ang362[45] | LincRNA | Unknown | Precursor miR-221/222 | Proliferation ↑ | |
| SENCR[52] | Antisense | Cytoplasm | Myocd/ Mdk/ Ptn | Synthetic SMC ↑ | |
| LincRNA-p21[20] | LincRNA | Nucleus | RNA-protein p53 | Proliferation ↓ / apoptosis ↑ | |
| HAS2-AS1[49] | Antisense | Unknown | Guide HAS2 | Hyaluronan ↑ | |
| HIF1α-AS1[50] | Antisense | Unknown | Unknown | Apoptosis ↑ | |
| LncRNA-GAS5[51] | LincRNA | Cytoplasm | RNA-protein Annexin A2 | Proliferation ↓/ migration ↓ | |
| EC | NRON[54-57] | LincRNA | Cytoplasm | Decoy, scaffold NFAT | Proliferation ↓/ tube formation ↓/ migration ↓ |
| Tie-1AS[59] | Antisense | Cytoplasm | lncRNA-mRNA Tie-1 | Tube formation ↓ | |
| Terminator[60] | LincRNA | Nucleus | Unknown | Pluripotent maintenance | |
| Alien[60] | LincRNA | Nucleus+ Cytoplasm | Unknown | Cell adhesion/ extracellular matrix remodelling/ vessel development | |
| Punisher[60] | Antisense | Nucleus+ Cytoplasm | Unknown | Proliferation ↑/ tube formation ↑ | |
| MALAT1[64-66] | LincRNA | Nucleus | Cyclins A2, B1/B2 | Angiogenesis ↑/ diabetic vascular damage ↑ | |
| MIAT[69] | LincRNA | Nucleus | ceRNA miR-150 | Angiogenesis ↑/ diabetic vascular damage ↑ | |
| LincRNA-p21[70] | LincRNA | Nucleus | ceRNA miR-130b | Apoptosis ↑ | |
| FLJ11812[71] | Sense | Unknown | ceRNA miR-4459 | Autophagy ↑ | |
| sONE[72,74] | Antisense | Nucleus+ Cytoplasm | eNOS | eNOS ↓ | |
| Macrophage | LincRNA-DYNLRB2-2[75] | LincRNA | Uknown | GPR119 | Inflammation ↓/ cholesterol efflux ↑ |
| LncRNA RP5-833A20.1[76] | Antisense | Nucleus+ Cytoplasm | miR-382 | Inflammation ↑/ cholesterol efflux ↓ | |
| E330013P06[77] | LincRNA | Unknown | Unknown | Foam cell formation ↑ | |
| Linc1992[81] | LincRNA | Unknown | RNA-Protein hnRNP-L | TNF-α expression ↑ | |
| LincRNA-Cox2[80] | LincRNA | Nucleus+ Cytoplasm | RNA-Protein hnRNP-A/B & hnRNP-A2/B1 | Immune related genes like Ccl5, Il-6 | |
| PTPRJ-AS1[82] | Antisense | Cytoplasm | Unknown | Unknown | |
| Lipid metabolism | HULC[14] | LincRNA | Cytoplasm | Guide miR-9 | Lipid afflux ↑ |
| SRA[89] | Sense | Nucleus | Coactivator Ppara, Pparg, Fabp4, Lipe | Adiposity ↑ | |
| APOA1-AS[90] | Antisense | Unknown | Guide ApoA1 | Lipid afflux ↑ |
Summary of lncRNAs as cardiovascular biomarkers
| LncRNA | Disease | Application | Study type | Study size | Follow-up |
|---|---|---|---|---|---|
| ANRIL (NR_003529 & DQ485454) | CAD[30] | Risk factor | Cross-sectional | 1134 atherosclerosis patients | None |
| ANRIL (all transcripts) | MI[95] | LV dysfunction predictors | Cohort | 414 MI; 86 healthy | 4 month |
| LIPCAR | MI & HF[94] | Cardiac death and cardiac remodelling predictors | Cohort | 246 MI | 12 months |
| Cohort | 344 HF | 3 years | |||
| Case-control | 99 died and 99 alive from HF | 3 years | |||
| KCNQ1OT | MI[95] | LV dysfunction predictors | Cohort | 414 MI; 86 healthy | 4 month |
| HIF1αAS2 | MI[95] | None | Cohort | 414 MI; 86 healthy | 4 month |
| MALAT1 | MI[95] | None | Cohort | 414 MI; 86 healthy | 4 month |
| MIAT | MI[95] | None | Cohort | 414 MI; 86 healthy | 4 month |
| MIAT | MI[68] | Risk factor | Case-control | 3464 MI; 3819 healthy | None |
LncRNAs in the biology of atherosclerosis
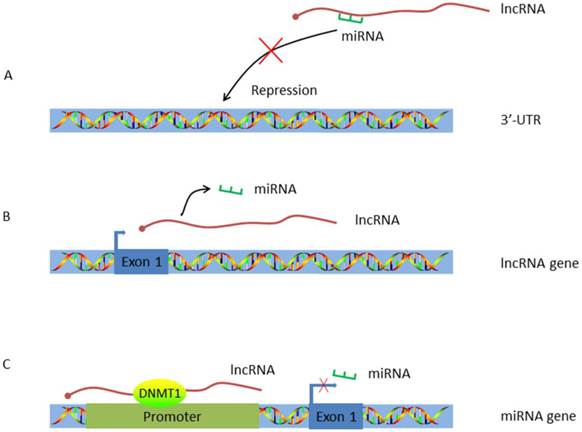
Work pattern of LncRNA-miRNA crosstalk. A. LncRNA acts as a ceRNA. By working as a sponge, lncRNA traps miRNA and inhibits miRNA's repression effect; B. LncRNA acts as the host gene of miRNA. MiRNA is encoded by lncRNA and alternations on lncRNA expression will concomitantly influence miRNA transcription; C. LncRNA targets the promoter of miRNA. HULC guides DNMT1 to the promoter of miRNA thus inducing methylation of CpG islands in the miRNA promoter.
Conclusion and Perspective
In conclusion, long noncoding RNAs are newly identified as vital regulators in multiple biological processes. Recognition of their roles in human disease has unveiled new mechanistic understanding and will lead to novel diagnostic and therapeutic approaches. While recent advances in lncRNA studies of cardiovascular disease indicate progress toward incorporation of lncRNAs into the pre-existing miRNA-mRNA-protein regulatory network, many pressing questions persist.
A challenge for research in this area is that most of the available data are in vitro findings. Poor sequence conservation and challenging gain-and-loss method still hinder in vivo studies. Specifically, for the well-attested functional lncRNA like ANRIL and MALAT1, future studies may focus on further elucidating their signaling pathways in vivo. Another remaining challenge will be to optimize the techniques for lncRNA capture and identification in biofuluids, which may consequently shield the potential clinical values of lncRNAs as biomarkers.
Abbreviations
lncRNA: Long noncoding RNAs; lincRNA: Long intergenic noncoding RNA; microRNA: miRNA; AS: Antisense strand; ANRIL: Antisense noncoding RNA in the INK4 locus; PRC1: Polycomb repressive complex; ceRNAs: competing endogenous RNAs; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; Fendrr: Fetal-lethal non-coding developmental regulatory RNA; HULC: Highly up-regulated in liver cancer; SRC-1: Steroid receptor coactivator 1; MiR‑539: MicroRNA 539; EC: Endothelial cell; SMC: Smooth muscle cells; SNP: Single nucleotide polymorphism; CDKN2A/B: Cyclin-dependent kinase inhibitor 2A/B; IGF1R: Insulin-like growth factor 1 receptor; KSRP: Protein K homology-type splicing regulatory protein; RAS: Renin-angiotensin system; Ang II: Angiotensin II; Mcm7: Minichromosome maintenance complex; SENCR: Smooth muscle and endothelial cell enriched migration /differentiation associated long noncoding RNA; FLI1: Friend leukaemia virus integration 1; ER: Endoplasmic reticulum; CAD: Coronary artery disease; HAS2: Hyaluronan synthases 2; HIF1α-AS1: Hypoxia inducible factor 1α antisense RNA 1; LncRNA-GAS5: Growth arrest-specific transcript 5; NFAT: Nuclear factor of activated T cells; NRON: Noncoding repressor of NFAT; Ets-1: E26 transformation-specific sequence 1; HUVECs: Human umbilical vein endothelial cell; SR proteins: Serine/arginine-rich family of nuclear phosphoprotein; CCN: Cyclin; MIAT: Myocardial infarction-associated transcripts; 3DBO: [3-benzyl-5-((2-nitrophenoxy)methyl)-dihydrofuran-2(3H)-one]; eNOS: Endothelial cell nitric oxide synthase; ABCA1: ATP binding cassette transporter A1; GPR119: G protein-coupled receptor 119; Apo: Apolipoprotein; NFIA: Nuclear factor I A; LDL: Low-density lipoprotein cholesterol; HDL: High-density lipoprotein cholesterol; TLR2: Toll-like receptors; HnRNP: Heterogeneous nuclear ribonucleoprotein; Ptprj: CD148, DEP-1, PTPη, Byp or PTPβ-like tyrosine phosphatase locus; IGF2BP1: IGF2 mRNA-binding proteins; ACSL1: Acyl-CoA synthetase long-chain family; PPARα: peroxisome proliferator-activated receptor alpha; DNMT1: DNA (cytosine-5)-methyltransferase 1; SRA: Steroid receptor RNA activator; LSD1: LSD1 histone demethylase; SUZ12: SUZ12 polycomb repressive complex 2 subunit; LIPCAR: Long intergenic noncoding RNA predicting cardiac remodelling; KCNQ1OT: Potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1; MI: Myocardial infarction; HF: Heart failure; LV: Left ventricle.
Acknowledgements
This work was sponsored by Shanghai Sailing Program (16YF1409500) and the Fundamental Research Funds for the Central Universities (No. 2014KJ086).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mattick JS. The genetic signatures of noncoding RNAs. PLOS GENET. 2009;5:e1000459
2. Lee JT. Epigenetic regulation by long noncoding RNAs. SCIENCE. 2012;338:1435-9
3. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. NAT REV GENET. 2014;15:7-21
4. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. SCIENCE. 2007;316:1484-8
5. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea MD. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667-72
6. Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. CELL. 2011;146:119-33
7. Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK. et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. NATURE. 2013;504:465-9
8. Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. SCIENCE. 2013;341:1237973
9. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. DEV CELL. 2013;24:206-14
10. Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. CANCER RES. 2011;71:5365-9
11. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. MOL CELL. 2010;39:925-38
12. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY. et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. NAT COMMUN. 2014;5:3596
13. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491-501
14. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X. et al. Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells through an miR-9-Mediated RXRA Signaling Pathway. CANCER RES. 2015;75:846-57
15. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S. et al. LincRNA-p21 suppresses target mRNA translation. MOL CELL. 2012;47:648-55
16. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. NATURE. 2013;493:231-5
17. Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. CELL. 2010;143:390-403
18. Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY. et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. CELL. 1999;97:17-27
19. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. CELL. 2010;142:409-19
20. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C. et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. CIRCULATION. 2014;130:1452-65
21. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M. et al. Heart disease and stroke statistics-2015 update: a report from the American Heart Association. CIRCULATION. 2015;131:e29-322
22. Nabel EG, Braunwald E. A tale of coronary artery disease and myocardial infarction. N Engl J Med. 2012;366:54-63
23. Andreou I, Sun X, Stone PH, Edelman ER, Feinberg MW. miRNAs in atherosclerotic plaque initiation, progression, and rupture. TRENDS MOL MED. 2015;21:307-18
24. Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. CARDIOVASC RES. 2012;95:156-64
25. Schwartz SM. Smooth muscle migration in atherosclerosis and restenosis. J CLIN INVEST. 1997;100:S87-9
26. Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196-206
27. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B. et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443-53
28. Motterle A, Pu X, Wood H, Xiao Q, Gor S, Ng FL. et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. HUM MOL GENET. 2012;21:4021-9
29. Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T. et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671-7
30. Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H. et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620-7
31. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E. et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. ATHEROSCLEROSIS. 2012;220:449-55
32. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. ONCOGENE. 2011;30:1956-62
33. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. MOL CELL. 2010;38:662-74
34. Sato K, Nakagawa H, Tajima A, Yoshida K, Inoue I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. ONCOL REP. 2010;24:701-7
35. Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K. et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLOS GENET. 2013;9:e1003588
36. Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS. et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. CARCINOGENESIS. 2013;34:577-86
37. Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S. et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. ONCOGENE. 2015;34:3076-84
38. Li H, Yu B, Li J, Su L, Yan M, Zhu Z. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. ONCOTARGET. 2014;5:2318-29
39. Han DK, Khaing ZZ, Pollock RA, Haudenschild CC, Liau G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J CLIN INVEST. 1996;97:1276-85
40. Kim DK, Zhang L, Dzau VJ, Pratt RE. H19, a developmentally regulated gene, is reexpressed in rat vascular smooth muscle cells after injury. J CLIN INVEST. 1994;93:355-60
41. Gao W, Zhu M, Wang H, Zhao S, Zhao D, Yang Y. et al. Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat Res. 2015;772:15-22
42. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G. et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. NAT CELL BIOL. 2012;14:659-65
43. Ding Z, Wang X, Schnackenberg L, Khaidakov M, Liu S, Singla S. et al. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. INT J CARDIOL. 2013;168:1378-85
44. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. MOL CELL. 2013;52:101-12
45. Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P. et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. CIRC RES. 2013;113:266-78
46. Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. CIRC RES. 2009;104:476-87
47. Guevara NV, Kim HS, Antonova EI, Chan L. The absence of p53 accelerates atherosclerosis by increasing cell proliferation in vivo. NAT MED. 1999;5:335-9
48. Chai S, Chai Q, Danielsen CC, Hjorth P, Nyengaard JR, Ledet T. et al. Overexpression of hyaluronan in the tunica media promotes the development of atherosclerosis. CIRC RES. 2005;96:583-91
49. Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M. et al. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J BIOL CHEM. 2014;289:28816-26
50. Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643-52
51. Li L, Li X, The E, Wang LJ, Yuan TY, Wang SY. et al. Low expression of lncRNA-GAS5 is implicated in human primary varicose great saphenous veins. PLOS ONE. 2015;10:e120550
52. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL. et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249-59
53. Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T. et al. The transcription factor NF-ATc is essential for cardiac valve formation. NATURE. 1998;392:186-90
54. Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z. et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. 2011;108:11381-6
55. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P. et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. SCIENCE. 2005;309:1570-3
56. Tsao HW, Tai TS, Tseng W, Chang HH, Grenningloh R, Miaw SC. et al. Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proc Natl Acad Sci U S A. 2013;110:15776-81
57. Tang YY, Wo LK, Chai H. Effects of noncoding RNA NRON gene regulation on human umbilical vein endothelial cells functions. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:245-50
58. Tsiamis AC, Morris PN, Marron MB, Brindle NP. Vascular endothelial growth factor modulates the Tie-2: Tie-1 receptor complex. MICROVASC RES. 2002;63:149-58
59. Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR. et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. BLOOD. 2010;115:133-9
60. Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB. et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. CIRCULATION. 2015;131:1278-90
61. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. ONCOGENE. 2003;22:8031-41
62. Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K. et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. CARCINOGENESIS. 2014;35:2731-9
63. Huang Z, Huang L, Shen S, Li J, Lu H, Mo W. et al. Sp1 cooperates with Sp3 to upregulate MALAT1 expression in human hepatocellular carcinoma. ONCOL REP. 2015;34:2403-12
64. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T. et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. CIRC RES. 2014;114:1389-97
65. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ. et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. CELL DEATH DIS. 2014;5:e1506
66. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J CELL MOL MED. 2015;19:1418-25
67. Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLOS GENET. 2013;9:e1003368
68. Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A. et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J HUM GENET. 2006;51:1087-99
69. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ. et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. CIRC RES. 2015;116:1143-56
70. He C, Ding JW, Li S, Wu H, Jiang YR, Yang W. et al. The Role of Long Intergenic Noncoding RNA p21 in Vascular Endothelial Cells. DNA CELL BIOL. 2015;34:677-83
71. Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z. et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. AUTOPHAGY. 2014;10:957-71
72. Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T. et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J BIOL CHEM. 2004;279:37982-96
73. Wilcox JN, Subramanian RR, Sundell CL, Tracey WR, Pollock JS, Harrison DG. et al. Expression of multiple isoforms of nitric oxide synthase in normal and atherosclerotic vessels. Arterioscler Thromb Vasc Biol. 1997;17:2479-88
74. Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K. et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J BIOL CHEM. 2007;282:15652-66
75. Hu YW, Yang JY, Ma X, Chen ZP, Hu YR, Zhao JY. et al. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J LIPID RES. 2014;55:681-97
76. Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR, Ma X. et al. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2015;35:87-101
77. Reddy MA, Chen Z, Park JT, Wang M, Lanting L, Zhang Q. et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. DIABETES. 2014;63:4249-61
78. Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L. et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J CLIN INVEST. 2009;119:2634-47
79. Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G. et al. MicroRNA 143-145 deficiency impairs vascular function. Int J Immunopathol Pharmacol. 2012;25:467-74
80. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL. et al. A long noncoding RNA mediates both activation and repression of immune response genes. SCIENCE. 2013;341:789-92
81. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C. et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002-7
82. Dave RK, Dinger ME, Andrew M, Askarian-Amiri M, Hume DA, Kellie S. Regulated expression of PTPRJ/CD148 and an antisense long noncoding RNA in macrophages by proinflammatory stimuli. PLOS ONE. 2013;8:e68306
83. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. GASTROENTEROLOGY. 2007;132:330-42
84. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y. et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J BIOL CHEM. 2012;287:26302-11
85. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y. et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. NUCLEIC ACIDS RES. 2010;38:5366-83
86. Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R. et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLOS ONE. 2010;5:e14199
87. Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L. et al. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. CANCER RES. 2000;60:6266-71
88. Hube F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. NUCLEIC ACIDS RES. 2011;39:513-25
89. Liu S, Sheng L, Miao H, Saunders TL, MacDougald OA, Koenig RJ. et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J BIOL CHEM. 2014;289:13000-9
90. Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O. et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. CELL REP. 2014;6:222-30
91. Goretti E, Wagner DR, Devaux Y. miRNAs as biomarkers of myocardial infarction: a step forward towards personalized medicine? TRENDS MOL MED. 2014;20:716-25
92. Skroblin P, Mayr M. "Going long": long non-coding RNAs as biomarkers. CIRC RES. 2014;115:607-9
93. Folkersen L, Kyriakou T, Goel A, Peden J, Malarstig A, Paulsson-Berne G. et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLOS ONE. 2009;4:e7677
94. Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A. et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. CIRC RES. 2014;114:1569-75
95. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. CIRC RES. 2014;115:668-77
Author contact
![]() Corresponding authors: Hongming Zhu, PhD and Junbo Ge, MD, PhD. E-mail: Hongming Zhu: zhmedu.cn; Junbo Ge: junbogecom
Corresponding authors: Hongming Zhu, PhD and Junbo Ge, MD, PhD. E-mail: Hongming Zhu: zhmedu.cn; Junbo Ge: junbogecom

 Global reach, higher impact
Global reach, higher impact