10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(8):931-943. doi:10.7150/ijbs.15447 This issue Cite
Research Paper
The Protein Elicitor PevD1 Enhances Resistance to Pathogens and Promotes Growth in Arabidopsis
State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Received 2016-3-4; Accepted 2016-5-12; Published 2016-6-7
Abstract
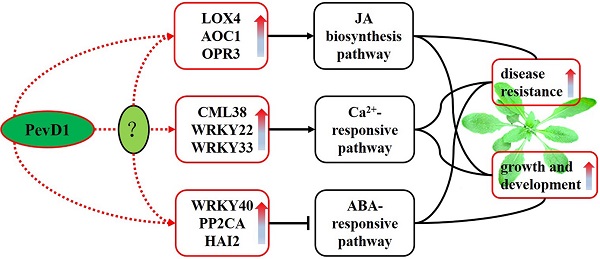
The protein elicitor PevD1, isolated from Verticillium dahlia, could enhance resistance to TMV in tobacco and Verticillium wilt in cotton. Here, the pevd1 gene was over-expressed in wild type (WT) Arabidopsis, and its biological functions were investigated. Our results showed that the transgenic lines were more resistant to Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 than the WT line was. In transgenic plants, both the germination time and bolting time required were significantly shorter and fresh weights and plant heights were significantly higher than those in the WT line. A transcriptomics study using digital gene expression profiling (DGE) was performed in transgenic and WT Arabidopsis. One hundred and thirty-six differentially expressed genes were identified. In transgenic Arabidopsis, three critical regulators of JA biosynthesis were up-regulated and JA levels were slightly increased. Three important repressors of the ABA-responsive pathway were up-regulated, indicating that ABA signal transduction may be suppressed. One CML and two WRKY TFs involved in Ca2+-responsive pathways were up-regulated, indicating that this pathway may have been triggered. In conclusion, we show that PevD1 is involved in regulating several plant endogenous signal transduction pathways and regulatory networks to enhance resistance and promote growth and development in Arabidopsis.
Keywords: PevD1, transgene, disease resistance, growth phenotype, transcriptomics, signaling pathway.
Introduction
Plants use several regulatory mechanisms to protect themselves from pathogens and adverse environmental stress. When subjected to prolonged biotic or abiotic stress, plant innate immune systems detect the exogenous stimulus and initiate signal transduction pathways that play key roles in regulating plant growth and defense [1]. Plant defense is most often coupled with concomitant changes in plant growth, such as senescence [2]. Plant growth and defense processes are regulated by a complicated and finely tuned regulatory networks. Signaling pathways mediated by plant phytohormones, calcium ions (Ca2+) and WRKY transcription factors (TFs) play critical roles in these regulatory networks.
Phytohormones are a diverse group of growth and defense regulators. Jasmonates (JAs), which comprise jasmonic acid (JA) and its derivatives, are involved in many developmental processes, such as seed germination, growth, leaf senescence, and reproduction, and they are also involved in defending the plant against biotic stressors, such as pathogens, herbivores, and nematodes, and regulating plant responses to stimuli and signals that originate in the environment [3,4]. Several key components that catalyze JA biosynthesis are essential for regulating JA-mediated processes by controlling the balance of JA levels [3,4]. Abscisic acid (ABA) is another essential phytohormone that regulates aspects of plant development, including seedling growth, seed maturation, seed dormancy, and stomatal behavior. ABA pathways respond to biotic and abiotic stressors, such as pathogen attack and drought, and have been reported to play both positive and negative roles in plant defenses [5,6]. Several repressors have been identified as key negative regulators of the ABA-responsive pathway by repressing core components of ABA-responsive elements [6]. Some Arabidopsis WRKY TFs are reported to be key components in the ABA-responsive pathway, and WRKY40 has been identified as a central transcriptional repressor of ABA-responsive pathway that acts by directly suppressing the expression of ABA-sensitive genes, such as ABI5 [7,8]. An increasing amount of evidence indicates that phytohormone-mediated signaling pathways are interconnected by synergistic or antagonistic activities [9]. The crosstalk between JA and ABA is complicated and far from fully understood. Briefly, on the one hand, JA and ABA cooperate to induce plant defenses, and ABA plays a role in the synthesis of JA. On the other hand, ABA can also play an antagonistic role in its interactions with JA during plant defense processes [9,10].
Ca2+ is a secondary messenger that acts as a versatile signaling molecule in eukaryotic cells. Ca2+-dependent signaling pathways are involved in almost every biological process in plants, including development and responses to environmental stimuli [11]. Ca2+-mediated stimulation is transmitted by a suite of Ca2+ binding proteins that are called Ca2+ sensors, which include calmodulin (CaM) and members of the calmodulin-like protein (CML) family, the family of Ca2+-dependent protein kinases (CDPK), and the calcineurin B-like protein (CBL) family, which regulate a range of biological processes by targeting different downstream components via Ca2+-dependent protein-protein interactions [12]. Some CaM/CMLs can activate the mitogen-activated protein kinase (MAPK) pathway in response to stress [13,14] and interact with several WRKY TFs in plants [15]. WRKY TFs are one of the largest families of transcriptional regulators in plants. TFs in this family contain a highly conserved amino acid sequence, WRKYGQK, and bind to the W-box (TTGACC/T) in the promoters of their target genes [16,17]. WRKY TFs are important components of plant signaling networks that play diverse biological roles as activators or repressors in various processes during the regulation of plant growth, including seed and trichome development, seed dormancy and germination, embryogenesis, senescence and other developmental processes. They also play roles in responses to biotic stimuli, such as microbial pathogens and herbivores, and to abiotic stimuli, such as drought, cold, salinity, and heat [18,19]. In addition, WRKYs also participate in complex phytohormone signaling pathways and can function as up- or down-stream regulators of hormones [16,18].
Some elements of plant defense responses can be triggered by protein elicitors. These responses are called induced immunity. Protein elicitors have been isolated from a variety of sources, including bacteria, viruses, oomycetes and fungi. For instance, a protein elicitor called Harpin, which was first isolated from Erwinia amylovora, has been shown to induce disease resistance in various plants [20]. Overexpressing the Harpin gene in tobacco and rice has been shown to improve pathogen resistance [21,22]. Harpin has also been reported to activate the expression of genes that are involved in generating nitric oxide (NO) and the JA signaling pathway [23]. Eubacterial flagellin was shown to increase intracellular Ca2+ concentrations and activate WRKY TFs that regulate MAPK signaling and pathogen-related gene expression [24]. Flagellin expression also confers disease resistance in transgenic rice [25]. Hrip1, a HR-inducing protein that was isolated from the necrotrophic fungus Alternaria tenuissima, has been shown to induce calcium influx, the activation of salicylic acid-induced protein kinase, and the expression of several defense-related genes after infiltrating into tobacco leaves [26]. Furthermore, overexpressing Hrip1 may enhance stress tolerance in Arabidopsis [27]. In addition to triggering plant defense responses, some elicitors have been reported to promote growth in treated plants. For example, chitosan has a profound effect on the growth and development of orchid plant tissue [28], and Harpin can stimulate Arabidopsis growth via the ethylene (ET) signaling pathway [29] and act as a growth promoter in Phalaenopsis orchids [23].
PevD1, a novel protein elicitor that was isolated from the fungal pathogen Verticillium dahliae, is an exocrine protein. Its 468 bp open reading frame (GenBank accession no. ABJE 0100044 5.1) encodes a 155-residue polypeptide with an 18-residue signal peptide. In our previous work, we showed that PevD1 induced hypersensitive response (HR), caused necrotic lesions and induced systemic acquired resistance (SAR) in tobacco plants that were exposed to TMV [30]. PevD1 was also shown to trigger innate immunity and to result in the up-regulation of pathogen-related genes, metabolite deposition and cell wall modifications [31]. In this study, the pevd1 gene was inserted into the Arabidopsis genome. We investigated the growth and pathogen response phenotypes in transgenic Arabidopsis and compared them to those in WT plants. We also performed high-throughput RNA-seq digital gene expression profiling (DGE) to analyze the differential expression of genes between transgenic and WT Arabidopsis. Our data provide valuable information that increases our understanding of the functions and mechanisms involving the protein elicitor PevD1 during the regulation of plant defenses and growth.
Materials and methods
Plant growth conditions
Seeds from wild type Arabidopsis thaliana (Columbia Col-0) and transgenic lines of plants were surface-sterilized using 75% ethanol for 10 min, washed with sterile water five times, and then placed on growth medium containing ½-strength Murashige and Skoog, 1% sucrose and 0.8% agar. After stratification at 4°C in the dark for three days, the plates were transferred to a growth chamber. All seedlings grown in growth medium and plantlets grown in nutrient soil were grown in a growth chamber (Percival AR800, USA) with a day/light cycle (16-h light/8-h dark cycle) at 22°C with 75% relative humidity.
Vector construction, plant transformation and transgenic plant generation
A pevd1 gene was truncated to remove its signal peptide sequence and then cloned into the Nco I and BstE II site of the pCAMBIA1301 vector, in which it replaced the GUS gene. This vector was then introduced into Agrobacterium tumefaciens strain GV3101. The Agrobacterium-mediated transformation of Arabidopsis Col-0 plants was performed using the floral dip method [32]. The T1 seeds were harvested from treated Col-0 plants (the T0 generation) and then screened in growth medium supplemented with 25 μg/mL hygromycin. Surviving T1 seedlings were transplanted into soil to obtain T2 seeds. The T1 and T2 seeds were screened using hygromycin and selected according to Mendel's law. The T3 generation was homozygotic, and three independent transgenic lines were used in subsequent experiments.
Genomic DNA PCR and qRT-PCR
Genomic DNA was extracted from Arabidopsis leaves using an EasyPure Plant Genomic DNA Kit (Transgen, Beijing, China) according to the manufacturer`s instructions. The genomic DNA was used as the template for PCR with the forward primer 5`-GCCCCCGCGTCTCCCGGC-3` and the reverse primer 5`-AGCCTCGGCGGGAGCGTCG-3`. The following PCR program was used: pre-denaturation at 94°C for 5 min, 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, and a final extension step at 72°C for 10 min. The reaction product was separated on a 1.0% agarose gel and analyzed under a UV transilluminator (Bio-Rad Laboratories, Hercules, CA, USA). The Arabidopsis Actin2 gene (AT3G18780) was used as an internal control using the following primers: forward 5`-TCAGCCGTTTTGAATCTCCGGC-3` and reverse 5`-TGCATCCTTCTGGTTCATCCCAACC-3`.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, California, USA) and then reverse-transcribed into cDNA using TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Transgen, Beijing, China) according to the manufacturer's instructions. Quantitative RT-PCR (qRT-PCR) was performed using TransStart Green qPCR SuperMix (Transgen, Beijing, China) and an iQ5 real-time PCR instrument (Bio-Rad Laboratories, Hercules, California, USA). The following conditions were used for qRT-PCR: initial denaturation at 95°C for 30 sec followed by 42 cycles of 94°C for 5 sec and 60°C for 30 sec. The following primers were used: PevD1, forward 5`-ACGGCACCATCAAGTACGTC-3` and reverse 5`- TGATGACGTTGGAAGGCAGG-3`; and the internal control, Actin2, forward 5`- CACCACCTGAAAGGAAGTAC-3` and reverse 5`-GATCCACATCTGCTGGAATG-3`.
Protein extraction and western blot analysis
Total protein extractions were performed using Arabidopsis leaves as previously described [33]. Leaves were ground into a powder in liquid nitrogen, and the extraction buffer [20 mM Tris-HCL pH 8.0, 20 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protein inhibitor cocktail] was then added. The solution was then micro-centrifuged at 18,000 g for 15 min at 4°C to extract the crude protein. The crude protein was electrophoresed using SDS-PAGE and then transferred onto a PVDF membrane using semi-dry electrophoresis. Immunoblotting was performed using Anti-PevD1 rabbit polyclonal antibodies (maintained by our lab) or Anti-Plant Actin Mouse Monoclonal Antibodies (Abbkine, Redlands, California, USA) and HRP AffiniPure Goat Anti-Rabbit IgG (H+L) or Goat Anti-Mouse IgG (H+L) (Abbkine, Redlands, California, USA). Proteins were detected using an enhanced chemiluminescence western-blot detection reagent kit.
B. cinerea culture and disease assay
The fungus Botrytis cinerea was cultured on a potato dextrose agar (PDA) (BD Difco, Sparks, Nevada, USA) plate for seven to ten days at 25°C with a 12-h photoperiod before spores were collected. Plant inoculation was performed as previously described [34]. Rosette leaves from four-week-old WT or transgenic Arabidopsis plants grown under the conditions described above were placed in 1% agar-medium plates and inoculated with 5 μL (1×105 cfu·mL-1) of B. cinerea spores. The plates were placed in a growth chamber under the same conditions that were used for the plants. Lesion sizes were determined by measuring the diameters or the major axes of oval lesions in the necrotic area at three days post-inoculation (dpi) [34]. To analyze gene expression using qRT-PCR, plants were sprayed with a spore suspension until all of the leaves were covered with fine droplets, and the plants were then maintained in a growth chamber with a higher relative humidity. At the indicated time points, plant leaves were harvested from three individual plants for each biological replicate.
Pst DC3000 culture and disease assay
Cultures of the bacterial strain Pseudomonas syringae pv. tomato (Pst) DC3000 were grown as previously described [35] using KB medium [36]. Final bacterial suspensions in 10 mM MgCl2 with OD600 = 0.1 were used in the infection assays. The surfactant Silwet L-77 was added at a final concentration of 200 μL·L-1. Four-week-old WT and transgenic Arabidopsis were used, and the bacterial suspension was sprayed until all of the leaves were covered with fine droplets. Incubated plants were maintained in a growth chamber under the same conditions described above except with higher relative humidity [37]. The plant bacterial population was determined according to methods that have been previously described [38]. Leaves were collected at 0 and 5 dpi and sterilized using 75% ethanol followed by three washes with distilled water. Leaf discs of the same size (with a diameter of approximately 0.5 cm) were made using a hole puncher and homogenized in 10 mM MgCl2 using a plastic pestle. Pst DC3000 growth numbers were determined by plating appropriate dilutions in KB medium containing rifampicin (30 mg·L-1) [35]. Each infection assay experiment was performed in triplicate. At each time point, three leaf discs were collected from one plant for each replicate. To analyze gene expression using qRT-PCR, plant leaves were harvested from three individual plants for each biological replicate at the indicated time points.
Growth phenotypic assay of transgenic plants
The phenotypic analysis of growth was carried out as previously described [29,39]. To assay the germination of seeds, Arabidopsis seeds were sown on growth medium, and the germination rate (defined as the emergence of hypocotyl) was scored at the indicated times. To assay seedling growth, the fresh weights (without root) of 10-day-old seedlings grown on growth medium were measured. After the seedlings were transferred into nutrient soil, the fresh weights (without roots) of three-week-old and four-week-old (including the seven days grown on growth medium) Arabidopsis plantlets were measured. The heights of the Arabidopsis plants were measured every three days starting on the day of bolting to assay plant height. All of the Arabidopsis plants were grown in the same growth chamber under the same conditions described above for the growth phenotype analysis.
DGE libraries preparation and sequencing
Total RNA was extracted from Arabidopsis plants (without roots) using TRIzol reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions. The quality and quantity of RNA were determined using a NanoPhotometer® Spectrophotometer (IMPLEN, Carlsbad, California, USA) and a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, California, USA). RNA integrity was assessed using a RNA Nano 6000 Assay Kit in a Bioanalyzer 2100 system (Agilent Technologies, Carlsbad, California, USA). DGE libraries were generated using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB) according to the manufacturer's recommendations. The library fragments were purified using an AMPure XP system (Beckman Coulter, Beverly, California, USA) to preferentially select cDNA fragments that were 150-200 bps in length. The libraries were then enriched using PCR with Phusion High-Fidelity DNA polymerase, universal PCR primers and an Index (X) primer. The PCR products were purified (AMPure XP system), and library quality was appraised using an Agilent Bioanalyzer 2100 system. The index-coded samples were clustered on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. The prepared libraries were sequenced on an Illumina HiSeq 2000 platform.
Analysis of DGE sequence results
High-quality clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from raw data (raw reads), and these were used for all downstream analyses. An index of the Arabidopsis genome was built using Bowtie v2.0.6, and single-end clean reads were aligned to the Arabidopsis genome using TopHat v2.0.9. The RPKM measure (reads per kilobase of predicted exons per million mapped reads) was used to correct the read counts for both sequencing depth and gene length [40], and the results were computed using HTSeq v0.6.1 to quantify gene expression levels. A differential expression analysis of transgenic and WT Arabidopsis (two biological replicates per condition) was performed using the DESeq R package (1.10.1) [41]. The resulting P-values were adjusted using the Benjamini-Hochberg Method [42] for false discovery. Genes with an adjusted P-value of <0.05 according to DESeq were defined as differentially expressed.
A Gene Ontology (GO, http://www.geneontology.org/) enrichment analysis of the differentially expressed genes was implemented using the GOseq R package [43]. GO terms with corrected P-values of <0.05 were considered significantly enriched among the differentially expressed genes. KEGG (http://www.genome.jp/kegg/) enrichment and pathway analysis was performed using KOBAS software. Sequence data were deposited in the Sequence Read Archive (SRA) database at the National Center for Biotechnology Information (NCBI) under access number SRA275647.
To confirm the results of DGE, RNA extraction and qRT-PCR were performed according to the methods described above. Primers were designed using Beacon Designer 8.
Extraction and determination of JA
Exactly 1.0 g of each fresh sample was ground into a fine powder in liquid nitrogen. Two milliliters of 80% pre-chilled methanol (methanol: water = 80:20, V/V) were then added, and the solution was mixed for 15 min. The samples were then stored at 4°C overnight. Subsequently, the samples were mixed for 10 min at 4°C and centrifuged for 10 min at 3000 rpm. The supernatants were transferred to new tubes, washed twice with 2 mL of 80% pre-chilled methanol and then centrifuged again. All of the supernatants were combined together and mixed again for 10 min at 4°C. Two drops of ammonia (approximately 40 µL) were added, and the samples were concentrated using evacuation into the aqueous phase at 35-40°C, but not to complete dryness. The aqueous phase was repeatedly frozen and thawed three times, and then 50 mg of PVPP was added, followed by mixing for 10 min. The mixture was then centrifuged for 25 min at 3000 rpm. The supernatant was transferred to a new tube, its pH was adjusted to 2.5-3.0 using 2 M HCl, and it was then extracted three times using the same volume of ethyl acetate. The upper layer of every extraction was merged, and two drops of ammonia were added. The samples were then concentrated to dryness at 35-40°C using evacuation. The dry sample was dissolved using 5 mL of 0.1 M HAc and filtered through a Sep-Pak C18 solid phase column as follows: a) The column was successively cleaned with 5 mL of methanol and 5 mL of 0.1 M HAc. b) A 5 mL volume of the sample traversed the column, which was then washed with 4 mL of 17% methanol (methanol: 0.1 M HAc = 17:83, V/V). These outflows were discarded. c) The column was successively eluted with 6 mL of 40% methanol (methanol: 0.1 M HAC = 40:60, V/V) and 3 mL of 60% methanol (methanol: 0.1 M HAC = 60:40, V/V) elution. These outflows were pooled. d) To the 9 mL of collected outflow, 2 drops of ammonia were added, and the mixture was concentrated to dryness at 40°C using evacuation. Methyl esterification of the dry sample was performed by incubating it with 100 µL of esterification reagent (HCl (37%): methanol = 1:2, V/V, freshly prepared) for 12 hours at 30°C. The esterification reagent was then completely removed under a stream of N2 gas, and each sample was then dissolved in 1 mL of methanol and filtered through a 0.22-µm Millipore filter for subsequent analysis.
The prepared samples were tested according to standard procedures for gas chromatography-mass spectrometry (GC-MS) using an AB-5MS Quartz capillary chromatographic column (30 m × 0.25 mm × 0.25 µm). JA was determined at m/z = 83, m/z = 151 and m/z = 210, and the mass spectrum of JA was retrieved from the NIST98 spectral library using Xcalibur (version 1.2) utility software. The JA in the sample was identified by comparing its retention time with pure, commercially available standards, and the concentration of JA was determined by computing its peak area using standard curves that were derived from the correlation between the peak area and known standard concentrations.
Results
PevD1 was over-expressed in Arabidopsis
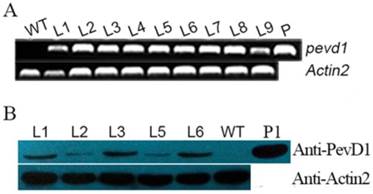
The gene pevd1, with its signal peptide sequence truncated, was subcloned into the pCAMBIA1301 vector and introduced into Arabidopsis Col-0 under the CaMV35s promoter (Figure S1). More than 20 transformants (T0) were obtained by selection for hygromycin resistance. Hygromycin-resistant segregation in T1 transgenic plants showed that nine transgenic lines were produced and that their phenotypic ratios corresponded to the Mendelian ratio of 3:1. T2 homozygotes were then selected, and T3 homozygotes were confirmed using hygromycin resistance. To screen the transgenic lines for consistent expression of PevD1, different transgenic lines (L1-L9) from the T3 generation were assayed using genomic PCR and qRT-PCR. The results showed that the gene of PevD1 was successfully inserted into the genome of Arabidopsis (Figure 1A). A western blot assay, shown in Figure 1B, indicated that PevD1 was constitutively expressed in the transgenic Arabidopsis. To eliminate the phenotypic difference that was caused by different expression level, three lines, L1, L3 and L6, with PevD1 expression in the similar level were therefore used in the follow-up experiments.
Molecular-level analysis of transgenic plants. Genomic PCR results (A), and Western blot analysis (B) of WT Arabidopsis and the indicated transgenic Arabidopsis lines that analyzed PevD1 gene or protein levels. L1-L9: transgenic Arabidopsis lines 1-9. WT: wild type Arabidopsis. P: positive control, the pCAMBIA1301-PevD1 vector. P1: positive control of purified protein. Actin2 was used as the internal control. Each experiment was repeated three times with similar results.
Arabidopsis plants overproducing PevD1 showed enhanced resistance to B. cinerea and Pst DC3000
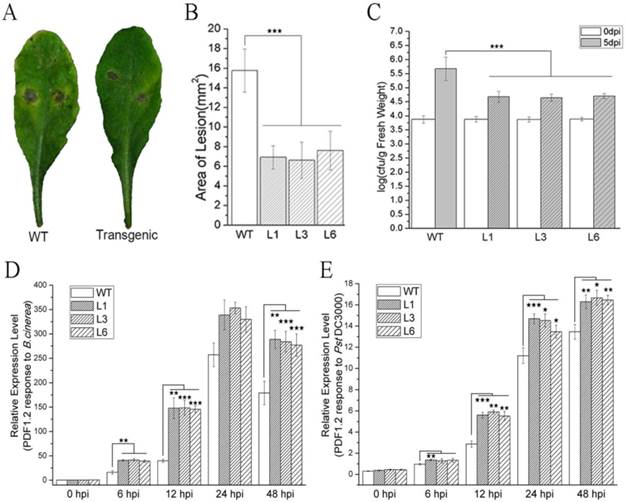
Leaves obtained from WT and transgenic Arabidopsis plants were infected with B. cinerea and subsequently formed necrotic lesions after 3 dpi. The necrotic lesions in the WT plants were bigger than those in the transgenic plants (Figure 2A). The areas of the lesions were computed by measuring the lesion diameter or their major axis. The average lesion area in the WT leaves was 15.8 mm2, whereas the average lesion areas in the L1, L3 and L6 leaves were 6.9, 6.6, and 7.6 mm2, respectively (Figure 2B). The reductions in lesion area therefore ranged from 51.8% to 58.0%. These results indicated that PevD1 significantly enhanced Arabidopsis resistance against the fungal pathogen B. cinerea. This experiment was independently repeated three times with similar results.
WT and transgenic Arabidopsis plants were incubated with the bacterial pathogen Pst DC3000. The bacterial counts on WT and transgenic plants were approximately equal just after incubation (0 dpi) with Pst. DC3000, but at 5 dpi, the bacterial counts in the transgenic L1, L3 and L6 plants were reduced by approximately 54.6% to 57.1% in comparison to the counts on the WT plants (Figure 2C). These results indicated that PevD1 enhanced Arabidopsis resistance against the bacterial pathogen Pst DC3000. This experiment was independently repeated three times with similar results.
The PDF1.2 gene of Arabidopsis encodes a plant defensin that has always been regarded as a marker for JA-dependent defense responses. Its expression is induced by B. cinerea and Pst DC3000 infection [44,45]. To investigate the response to pathogenic infection in WT and transgenic Arabidopsis plants, the expression of PDF1.2 was tested after the plants were inoculated with B. cinerea and Pst DC3000. qRT-PCR was performed using the primers listed in Table S1. Under our experimental conditions, PDF1.2 was strongly induced and maintained at a high expression level within 48 h (Figure 2D and 2E). Moreover, the transgenic plants showed a stronger induction of the expression of PDF1.2 after inoculation with B. cinerea and Pst DC3000 than was observed in the WT plants (Figure 2D and 2E).
Effects of PevD1 overexpression on B. cinerea and Pst. DC3000 infection in transgenic Arabidopsis. (A) B. cinerea infection at 3 dpi. (B) The area of the necrotic lesions caused by B. cinerea infection at 3 dpi. (C) Altered bacterial growth of Pst. DC3000 at the indicated times. All experiments were repeated three times, and the data presented are the means ± SD of three independent experiments. The relative expression level of PDF1.2 that was induced by B. cinerea (D) and Pst. DC3000 (E) infection in WT and transgenic Arabidopsis. The Actin2 gene was used as an internal control to normalize the expression data. The bars represent the standard deviation (n = 3). The asterisks indicate a significant difference between transgenic and WT Arabidopsis (Student's t-test: «, P < 0.1; ««, P < 0.05; «««, P < 0.01).
PevD1 overexpression promoted Arabidopsis growth and development
The germination rates of the WT and transgenic Arabidopsis seeds were scored after they were sown on growth medium. Our results showed that over 90% of the L1, L3, and L6 seeds sprouted at two days after sowing (das), with germination rate values of 93.5%, 90.2% and 90.8%, respectively, whereas the germination value for the WT seeds was only 10.3% (Figure 3A). At 3 das, the germination rate of the WT seeds reached 94.8%, which was not significantly different from the values of the L1, L3, or L6 seeds (97.8%, 98.0%, and 96.9%, respectively) (Figure 3A). These data indicated that PevD1 overexpression promoted Arabidopsis seed germination and significantly shortened the required generation time by approximately one day. The germination rate was calculated using fifty seeds from each Arabidopsis strain sown per plate.
To study the effects of PevD1 overexpression on different growth stages in Arabidopsis, the growth status of the plants was monitored by observing the plants` phenotypes and measuring their fresh weight. The phenotypic analysis of the three growth periods that occur before bolting, specifically including 10-day-old, 21-day-old and 28-day-old plants, showed that the transgenic Arabidopsis plants were larger than the WT Arabidopsis in different periods (Figure 3B).
Effects of PevD1 overexpression on growth and development in transgenic Arabidopsis. (A) Germination rate of Arabidopsis at the indicated times. Phenotype (B) and fresh weight (C) of Arabidopsis at the indicated three growth periods before bolting. Bolting phenotype (D) and bolting time (E). Plant height in WT and transgenic Arabidopsis L3 line at the indicated times, shown in days after sowing (das) (F) and days after bolting (dab) (G). (H) Plant height at the end of growth. All experiments were repeated three times, and the data presented are shown as the means ± SD of three independent experiments. The bars represent the standard deviation (n = 3). The asterisks indicate a significant difference between transgenic and WT Arabidopsis (Student's t-test: «««, P < 0.01).
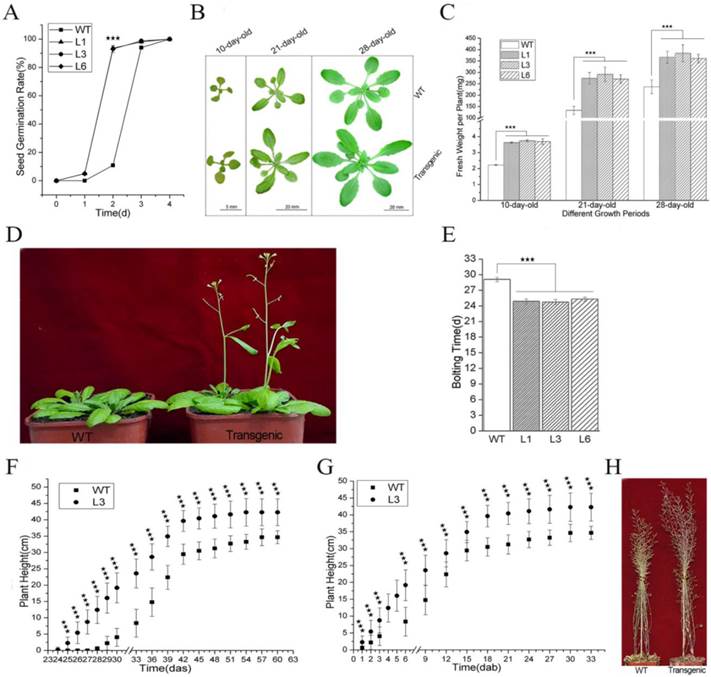
Fifty 10-day-old plantlets that were grown on growth medium were harvested and weighed together. The weight of one plantlet was computed as the average of the fifty plantlets. The average plantlet weights of the transgenic lines L1, L3, and L6 were 3.6 mg, 3.7 mg and 3.7 mg, respectively. These plants were approximately 1.6-1.7 times as heavy as the WT Arabidopsis, which weighed 2.2 mg (Figure 3C). Ten 21-day-old plants that were grown in soil were randomly selected and harvested without their roots and weighed together. The weight of one plant was computed as the average of the ten plants. The average plant weights of the transgenic lines L1, L3, and L6 were 274 mg, 292 mg and 271 mg, respectively. These plants were approximately 2.0-2.2 times as heavy as the WT Arabidopsis, which weighed 133 mg (Figure 3C). The weight of one 28-day-old plant was scored according to the method described above. The values for L1, L3, and L6 were 366 mg, 384 mg and 361 mg, respectively. These plants were approximately 1.5-1.6 times as heavy as the WT Arabidopsis, which weighed 237 mg (Figure 3C).
The transgenic plants started to bolt approximately 25 days after they were sown, which was earlier than the WT plants (approximately 29 days after sowing) (Figure 3D and 3E). Plant height was measured every three days from bolting. On the day after sowing (das), the transgenic plants were taller than the WT plants because the bolted earlier (Figure 3F, S2). On the day after bolting (daf), the transgenic plants were still taller than the WT plants (Figure 3G, S2). At the end of growth, the transgenic plants were approximately 7.6 cm taller than the WT plants (Figure 3F, S2 and 3H). These data indicated that PevD1 not only shorted the required Arabidopsis bolting time but also increased plant height.
Together, these results suggest that PevD1 affects Arabidopsis growth by affecting seed germination time, fresh weight during different growth periods, bolting time and plant height.
Analysis of DGE sequencing results
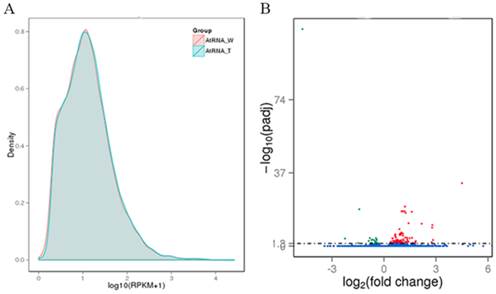
The WT and transgenic L3 strains of Arabidopsis were submitted to DGE sequencing, which was performed on an Illumina HiSeq 2000 platform by Novogene Bioinformatics Technology Co., Ltd (Beijing, China). Two biological replicates of both WT and the L3 line (AtW1, AtW2. AtT1 and AtT2) were simultaneously sequenced. The quality of the sequencing data was evaluated according to the error rate and the base contents (Table S2). Clean reads (Table S2) of the samples were mapped to the genome of Arabidopsis using TopHat v2.0.9 software, and the specific results from the reads that were mapped are shown in Table S3. The expression level of each gene was estimated using RPKM. We defined RPKM > 1 as the threshold for significant gene expression, and more than 50% of the analyzed genes were significantly expressed in each sample (Table S4). Based on the average RPKM value of two biological replicates of each gene, the gene expression levels of WT and L3 was compared (Figure 4A). The average read count data for the WT and L3 (AtRNA_W and AtRNA_T) plants were used to analyze differences in gene expression. The fold change of one gene was calculated as the ratio of its read count value from AtRNA_T and AtRNA_W. A statistical analysis was performed, and a padj value was obtained to estimate the significance of the observed differential expression of each gene. For the biological replicates, the analysis condition padj < 0.05 was used to screen the differentially expressed genes. One hundred and thirty-six differentially expressed genes were identified between AtRNA_T and AtRNA_W, with 106 genes up-regulated and 30 genes down-regulated (Figure 4B).
Gene expression analysis of transgenic and WT Arabidopsis. (A) Capsule RPKM density distribution in transgenic and WT Arabidopsis. The y-axis indicates the density values. The x-axis indicates the log10(RPKM+1) values of genes. Red and blue represent AtRNA_W and AtRNA_T, respectively. (B) Volcano plot of the differentially expressed genes. The y-axis indicates the -log10(padj) values with the indicated significant differences. The x-axis indicates the log2(fold change) values. Blue points represent genes that were not significantly differentially expressed, red points represent significantly up-regulated genes, and green points represent significantly down-regulated genes.
Among these differentially expressed genes, nine were further analyzed. The genes 13-lipoxygenase 4 (LOX4), allene oxide cyclases1 (AOC1) and oxophytodienoic acid (OPDA) reductase 3 (OPR3) are involved in JA biosynthesis and showed 6.9-, 2- and 1.9-fold changes, respectively, in the DGE results. WRKY40, clade A of protein phosphatase 2C (PP2CA) and highly abscisic-acid induced 2 (HAI2) are involved in the abscisic acid (ABA)-responsive pathway and showed 4.5-, 2.1- and 2.3-fold changes, respectively, in the DGE results. One Ca2+ sensor (CML38, Calmodulin (CaM)-like Ca2+-binding protein 38) and two WRKY TFs (WRKY22 and WRKY33) known to be involved in a Ca2+-responsive pathway showed 3.6-, 2.4- and 2.6-fold changes, respectively, in the DGE results.
The differentially expressed genes were further analyzed using GO enrichment to determine their potential functions. The twenty-three GO accessions shown in Figure S3 were the most enriched (pcorrected < 0.05). These twenty accessions were classified according to biological processes, of which the top three significant GO terms were “regulation of cellular process” (GO:0050794), “regulation of biological process” (GO:0050789) and “biological regulation” (GO:0065007). The other 3 accessions, including “histone acetyltransferase activity” (GO:0004402), “RNA helicase activity” (GO:0003724) and “ADP binding” (GO:0043531) were classified as molecular functions. In the analysis of the 23 GO terms, most of the differentially expressed genes in the DGE were involved in biological processes. Among the nine genes mentioned above, the function of both WRKY40 and CML38 were involved in the top three significant GO terms of biological processes mentioned above, while WRKY33 function was involved in all of the remaining GO terms of the most enriched classified into biological processes.
The differentially expressed genes were further analyzed using KEGG enrichment to determine the principal biochemical metabolism pathways or signal transduction pathways that the genes participated in. Among the 20 pathways that were identified, two pathways, named “photosynthesis-antenna proteins” (ath00196) and “α-Linolenic acid metabolism” (ath00592), were significantly enriched at pcorrected< 0.05, followed by “Plant-pathogen interaction” (ath04626) pathway, at pcorrected= 0.0659 (Figure S4). The three genes mentioned above, LOX4, AOC1 and OPR3, are involved in the “α-Linolenic acid metabolism” pathway, which is also known to include the JA biosynthesis pathway. Four identified genes are involved in the “Plant-pathogen interaction” pathway, including CML38, WRKY22 and WRKY33, which were mentioned above. These three genes act downstream of Ca2+ in this pathway. PP2CA and HAI2 are involved in the pathway named “Plant hormone signal transduction”, identified at pcorrected= 0.3016 (Figure S4), in which the two genes act in the ABA-responsive pathway. The most enriched pathway was “Photosynthesis-antenna proteins”, and six light-harvesting chlorophyll a/b-binding (LHCB) proteins in the DGE results were down-regulated and involved in this pathway. However, the fold changes of these six genes were not significant (fold change <1.5). What is interesting is that the expression of the LHCBs is suppressed by WRKY40 to balance the positive function of the LHCBs during ABA signaling [46].
Quantitative PCR analysis of DGE results
To confirm the DGE results, four-week-old transgenic Arabidopsis (L1, L3 and L6) and WT Arabidopsis were harvested. Quantitative PCR was performed according to the MIQE standard to analyze the nine focused genes of interest using the primers shown in Table S1. The relative changes in the expression of the transgenic plants compared to their expression in WT plants are shown in Table 1. Generally, the results of quantitative PCR were in agreement with the results of DGE. The experiment was repeated independently three times with similar results.
Verification of differentially expressed genes by qRT-PCR.
| Samples | Relative expression level# of genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| LOX4 | AOC1 | OPR3 | CML38 | WRKY22 | WRKY33 | WRKY40 | PP2CA | HAI2 | |
| L1 | 9.15 (0.255)※ | 2.67 (0.259) | 2.50 (0.290) | 4.47 (0.372) | 2.89 (0.227) | 2.91 (0.233) | 5.20 (0.293) | 2.32 (0.365) | 2.81 (0.396) |
| L3 | 8.78 (0.292) | 2.58 (0.325) | 2.62 (0.336) | 4.70 (0.352) | 2.96 (0.302) | 2.83 (0.369) | 5.06 (0.247) | 2.77 (0.315) | 2.80 (0.337) |
| L6 | 8.69 (0.234) | 2.94 (0.452) | 2.63 (0.249) | 4.67 (0.327) | 2.82 (0.253) | 2.86 (0.421) | 5.05 (0.327) | 2.64 (0.352) | 2.49 (0.283) |
# The relative expression level of each gene was the fold change in the transgenic Arabidopsis over WT Arabidopsis. The Actin2 gene was used as an internal control to normalize the expression data.
※ The values shown in the brackets denote the standard deviation (SD) of three times independent experiments.
JA levels were slightly enhanced in transgenic Arabidopsis
To determine JA levels in the transgenic and WT Arabidopsis plants, JA was extracted from four-week-old Arabidopsis and quantified using GC-MS. The samples of transgenic Arabidopsis that were used in this experiment were mixed samples from the L1, L3 and L6 lines. The levels of endogenous JA in the transgenic Arabidopsis plants were slightly higher than those in WT Arabidopsis, with mean values of 221.7 ng g-1 FW (fresh weight) and 175.3 ng g-1 FW, respectively (Figure 5). In combination with previous results that showed that JA biosynthesis enzymes were activated and JA levels were increased, our results indicate that JA biosynthesis is triggered by PevD1.
Endogenous JA concentration in leaves of four-week-old transgenic and WT Arabidopsis. Values are shown as the means ± SD of three independent samples. The asterisks indicate significant differences between transgenic and WT Arabidopsis (Student's t test: «««, P < 0.01).

Discussion
The protein elicitor PevD1 was purified in our lab as a hypothetical protein from the cotton Verticillium wilt fungus V. dahliae. Our previous studies showed that PevD1 triggered an immune response in different plants, including tobacco and cotton. Here, we show that the function of PevD1 in Arabidopsis is to enhance resistance and promote growth and development.
As an elicitor, the functions of PevD1 that enhance resistance in plants in vitro have been previously reported [30,31,47]. Other studies are currently in progress to screen for binding proteins or receptors of PevD1 in Arabidopsis. We found that an Arabidopsis Asparagine-Rich Protein (NPR) was a binding protein. An NPR containing a highly conserved DCD (development and cell death) domain was originally found to be activated by pathogen infection or endoplasmic reticulum stress in soybean [48,49]. This indicated that PevD1 has a target in Arabidopsis cells and that it may be involved in plant defenses and growth. To investigate the function of PevD1 in vivo, the pevd1 gene was over-expressed in Arabidopsis, and biological features, including disease resistance and the growth of transgenic Arabidopsis lines, were assayed. Our results showed that PevD1 overexpression in Arabidopsis not only enhanced resistance to B. cinerea and Pst DC3000 (Figure 2) but also promoted plant growth and development (Figure 3). These dual features can be observed in other elicitors identified from different pathogens. For example, Harpin, a famous protein elicitor that has been identified in many pathogens, enhanced resistance to different pathogens in tobacco and rice [21,22]; and could enhance Arabidopsis growth and increase plant fresh weight in Phalaenopsis orchids by modulating plant genes expression, including components of the ethylene (ET) signaling pathway and the JA signaling pathway [23,29,39]. Hrip1, a HR-inducing protein that was isolated from the fungus A. tenuissima, was reported to enhance disease resistance and promote plant growth in Arabidopsis [26,27]. Fungal chitosan could stimulate the immunity of plant and had the ability to induce differentiation in orchid plant tissue and enhance orchid production [28,50]. These elicitors could affect the plant signaling pathways and regulatory network, such as plant phytohormones, calcium ions (Ca2+) and WRKY TF-mediated signaling pathways. Following stimulation by PevD1, the JA biosynthesis pathway, the Ca2+-responsive pathway and some WRKY TFs signaling pathways may be activated, while the ABA-responsive pathway may be suppressed, in PevD1-overexpressing Arabidopsis.
The JA biosynthesis pathway may be activated by PevD1
The JA-mediated signaling pathway is regarded as an important component of the regulatory network that is involved in responses to pathogen attack [3,4]. JA-mediated regulation is embodied in the expression levels of several key components that catalyze JA biosynthesis and maintain the balance of JA levels [3]. For example, 13-lipoxygenases (13-LOXs), AOC and OPR catalyze the three critical steps in JA biosynthesis and are involved in JA-dependent responses [4]. In our study, both the DGE results and the qRT-PCR results showed that the transcription levels of LOX4, AOC1 and OPR3 were significantly higher in transgenic Arabidopsis than in WT Arabidopsis (Table 1). Furthermore, our results showed that the endogenous JA levels were higher in transgenic Arabidopsis than in WT Arabidopsis (Figure 5). All of these data indicate that JA biosynthesis is activated when pevd1 is overexpressed, which may partly account for transgenic Arabidopsis resistance to B. cinerea and Pst DC3000 (Figure 2). Under transitory and strong stimulus with exogenous PevD1 protein, plants, including tobacco and cotton, exhibit resistance to pathogens that is always coupled with a hypersensitive response, ROS burst, H2O2 accumulation and the significant up-regulation of pathogen-related genes and PTI-response genes [30,31,47]. However, these strong responses have not been observed in transgenic Arabidopsis. This may be because PevD1 is endogenously expressed in plants and produces a modest level of stimulus that is much weaker than the level induced by exogenous stimulus. In the list of differently expressed genes we obtained from the DGE results, no genes related to SA biosynthesis or SA signaling pathways were detected. Moreover, marker genes for the SA signaling pathway, such as PR1, displayed no significant difference in transcription level between transgenic Arabidopsis and WT Arabidopsis after infection with B. cinerea and Pst DC3000 (Figure S5). This result indicates that the SA signaling pathway may be not involved in the effects of endogenously expressed PevD1 in Arabidopsis.
The Ca2+-responsive pathway is potentially activated by PevD1
Ca2+ signal cascades are propagated by “Ca2+ sensors”, which are Ca2+ binding proteins that subsequently regulate downstream targets. CML38 is thought to be involved in Ca2+-responsive pathways in which it acts as a “Ca2+ sensor” [51]. The transcription factor WRKY33 regulates the expression of several different defense-related genes [52] and acts as a downstream target of MPK4 [53]. In Arabidopsis, WRKY22 is an important downstream target of the MAPK-mediated signal transduction pathway that is up-regulated by pathogen infection [54] and certain pathogen-associated molecular patterns (PAMP), including those induced by flagellin and chitin [55,56]. CML38 is a Ca2+ sensor, and WRKY22 and WRKY33 may be indirect downstream targets of Ca2+ that are linked by MPK4 and MPK3/6, as shown in the KEGG pathway ath04626 (http://www.kegg.jp/kegg-bin/show_pathway?ath04626). This pathway was significantly enriched (p = 0.00989 < 0.01) in our DGE results. Both the DGE and the quantitative PCR results showed that CML38, WRKY22 and WRKY33 were up-regulated in the Arabidopsis L3 line (Table 1). Whether directly or indirectly regulated by PevD1, these three genes showed high expression levels but did not result in a significantly different concentration of Ca2+ in four-week-old transgenic Arabidopsis and WT Arabidopsis (results not shown). The high expression level of these three genes may promote a stronger response to the Ca2+ influx that is induced by pathogen infection.
The ABA-responsive pathway may be suppressed by PevD1
ABA is another important phytohormone that regulates a variety of physiological aspects in plants, such as seed maturation, dormancy, seedling growth, and stomatal behavior [6]. ABA is thought to function by inhibiting seed germination and arresting seedling growth [7]. In recent years, ABA has also been shown to be an essential signal that regulates plant defenses against pathogens [57], but the functions that ABA performs against biotic stressors are complicated [58]. Generally, ABA has been shown to act as a negative regulator of plant defense against various pathogens [59]. WRKY40, PP2CA and HAI2 are three important negative regulators that act as suppressers in the ABA-responsive pathway [6]. In transgenic Arabidopsis that overproduce PevD1, the factors WRKY40, PP2CA and HAI2 were expressed at markedly higher levels than were observed in the WT, as shown in both the DGE and the qRT-PCR results (Table 1). All three of these suppressers were activated, indicating that the ABA-responsive pathway may be suppressed by PevD1 and that the ABA-mediated maintenance of seed dormancy and repression of seedling growth were attenuated, which provided the transgenic Arabidopsis with increased resistance to B. cinerea and Pst DC3000 (Figure 2) in addition to promoting earlier germination and enhanced growth (Figure 3).
In conclusion, PevD1 overexpression in Arabidopsis not only enhanced resistance to B. cinerea and Pst DC3000 but also promoted growth and development by reducing the required germination time, increasing seedling growth, advancing bolting time and increasing plant height. PevD1 impacted the expression of Arabidopsis genes known to be involved in Ca2+- and ABA-responsive pathways and the JA biosynthesis pathway and WRKY TFs. PevD1 may therefore act as an inducer of plant defenses and a promoter of plant growth. Further investigations should focus on the details of the crosstalk between the diverse signaling pathways and regulatory webs that are affected by PevD1 to improve our understanding of this protein elicitor's functions in triggering plant innate immunity and regulating plant growth.
Supplementary Material
Supplementary tables and figures.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (No. 31272086).
Additional Information
Sequence data are available in the Sequence Read Archive (SRA) database at the National Center for Biotechnology Information (NCBI) with access number SRA275647.
Competing interests
The authors have declared that no competing interest exists.
References
1. Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742-744
2. Quirino B, Normanly J, Amasino R. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol. 1999;40:267-278
3. Wasternack C. Jasmonates in Plant Growth and Stress Responses. Tran L-SP, Pal S, eds. Phytohormones: a window to metabolism, signaling and biotechnological applications: Springer New York. 2014:221-263
4. Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111:1021-1058
5. Cutler SR, Rodriguez PL, Finkelstein RR. et al. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651-679
6. Fujii H. Abscisic acid implication in plant growth and stress responses. Tran L-SP, Pal S, eds. Phytohormones: a window to metabolism, signaling and biotechnological applications: Springer New York. 2014:37-54
7. Shang Y, Yan L, Liu ZQ. et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909-1935
8. Chen H, Lai Z, Shi J. et al. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010;10:281
9. Gómez-Cadenas A, Ollas C, Manzi M. et al. Phytohormonal Crosstalk Under Abiotic Stress. In: (ed.) Tran PL-S, Pal S. Phytohormones: a window to metabolism, signaling and biotechnological applications. New York, NY: Springer New York. 2014:289-321
10. Derksen H, Rampitsch C, Daayf F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013;207:79-87
11. Kudla J, Batistič O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541-563
12. Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochem Biophys Acta. 2012;1820:1283-1293
13. Takahashi F, Mizoguchi T, Yoshida R. et al. Calmodulin-dependent activation of MAP kinase for ROS homeostasis in arabidopsis. Mol Cell. 2011;41:649-660
14. Yang T, Shad Ali G, Yang L. et al. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal Behav. 2010;5:991-994
15. Kim MC, Chung WS, Yun D-J. et al. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant. 2009;2:13-21
16. Agarwal P, Reddy MP, Chikara J. WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep. 2011;38:3883-3896
17. Rushton PJ, Somssich IE, Ringler P. et al. WRKY transcription factors. Trends Plant Sci. 2010;15:247-258
18. Bakshi M, Oelmüller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9:e27700
19. Zhao M. WRKY transcription factor superfamily: structure, origin and functions. Afr J Biotechnol. 2012:11
20. Adam AL, Pike S, Hoyos ME. et al. Rapid and transient activation of a myelin basic protein kinase in tobacco leaves treated with harpin from erwinia amylovora. Plant Physiol. 1997;115:853-861
21. Shao M, Wang J, Dean RA. et al. Expression of a harpin-encoding gene in rice confers durable nonspecific resistance to Magnaporthe grisea. Plant Biotechnol J. 2008;6:73-81
22. Peng J-L, Bao Z-L, Ren H-Y. et al. Expression of harpinxoo in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology. 2004;94:1048-1055
23. Chuang H-w, Chang P-Y, Syu Y-y. Harpin protein, an elicitor of disease resistance, acts as a growth promoter in Phalaenopsis orchids. J Plant Growth Regul. 2014;33:788-797
24. Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150:1638-1647
25. Takakura Y, Che F-S, Ishida Y. et al. Expression of a bacterial flagellin gene triggers plant immune responses and confers disease resistance in transgenic rice plants. Mol Plant Pathol. 2008;9:525-529
26. Kulye M, Liu HUA, Zhang Y. et al. Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant Cell Environ. 2012;35:2104-2120
27. Peng X-C, Qiu D-W, Zeng H-M. et al. Inducible and constitutive expression of an elicitor gene Hrip1 from Alternaria tenuissima enhances stress tolerance in Arabidopsis. Transgenic Res. 2015;24:135-145
28. Nge KL, Nwe N, Chandrkrachang S. et al. Chitosan as a growth stimulator in orchid tissue culture. Plant Sci. 2006;170:1185-1190
29. Dong H-P, Peng J, Bao Z. et al. Downstream divergence of the ethylene signaling pathway for harpin-stimulated arabidopsis growth and insect defense. Plant Physiol. 2004;136:3628-3638
30. Wang B, Yang X, Zeng H. et al. The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl Microbiol Biot. 2012;93:191-201
31. Bu B, Qiu D, Zeng H. et al. A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 2014;33:461-470
32. Clough SJ, Bent AF. Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16:735-743
33. Brini F, Hanin M, Lumbreras V. et al. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 2007;26:2017-2026
34. Ferrari S, Plotnikova JM, De Lorenzo G. et al. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003;35:193-205
35. Xiao S, Chye ML. Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 2011;156:2069-2081
36. King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301-307
37. Jia Z, Zou B, Wang X. et al. Quercetin-induced H(2)O(2) mediates the pathogen resistance against Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis thaliana. Biochem Biophys Res Commun. 2010;396:522-527
38. Mengiste T, Chen X, Salmeron J. et al. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15:2551-2565
39. Chuang HW, Harnrak A, Chen YC. et al. A harpin-induced ethylene-responsive factor regulates plant growth and responses to biotic and abiotic stresses. Biochem Biophys Res Commun. 2010;402:414-420
40. Mortazavi A, Williams BA, McCue K. et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621-628
41. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106-R106
42. Ferreira JA, Zwinderman AH. On the Benjamini-Hochberg method. Ann Stat. 2006;34:1827-1849
43. Young MD, Wakefield MJ, Smyth GK. et al. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14-R14
44. Brown RL, Kazan K, McGrath KC. et al. A role for the GCC-Box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020-1032
45. Ravichandran S, Stone SL, Benkel B. et al. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol. 2013;13:1-12
46. Liu R, Xu Y-H, Jiang S-C. et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J Exp Bot. 2013;64:5443-5456
47. Liu W, Zeng H, Liu Z. et al. Mutational analysis of the Verticillium dahliae protein elicitor PevD1 identifies distinctive regions responsible for hypersensitive response and systemic acquired resistance in tobacco. Microbiological Res. 2014;169:476-482
48. Hoepflinger MC, Pieslinger AM, Tenhaken R. Investigations on N-rich protein (NRP) of Arabidopsis thaliana under different stress conditions. Plant Physiol Bioch. 2011;49:293-302
49. Tenhaken R, Doerks T, Bork P. DCD - a novel plant specific domain in proteins involved in development and programmed cell death. BMC Bioinformatics. 2005;6:1-6
50. Uthairatanakij A, Silva JATD, Obsuwan BK. Chitosan for improving orchid production and quality. Science. 2007;1:1-5
51. Vanderbeld B, Snedden W. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol Biol. 2007;64:683-697
52. Petersen K, Fiil BK, Mundy J. et al. Downstream targets of WRKY33. Plant Signal Behav. 2008;3:1033-1034
53. Andreasson E, Jenkins T, Brodersen P. et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579-2589
54. Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21-37
55. Wan J, Zhang S, Stacey G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol. 2004;5:125-135
56. Asai T, Tena G, Plotnikova J. et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977-983
57. Adie BAT, Pérez-Pérez J, Pérez-Pérez MM. et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in arabidopsis. Plant Cell. 2007;19:1665-1681
58. Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14:310-317
59. Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473-488
Author contact
![]() Corresponding author: Dewen Qiu, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, No.12 zhongguancun south street, Haidian district, Beijing, 100081, China. Phone: 86-10-62815924, email: qiudewencn.
Corresponding author: Dewen Qiu, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, No.12 zhongguancun south street, Haidian district, Beijing, 100081, China. Phone: 86-10-62815924, email: qiudewencn.

 Global reach, higher impact
Global reach, higher impact