10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(10):1247-1261. doi:10.7150/ijbs.16463 This issue Cite
Review
From Nutrient to MicroRNA: a Novel Insight into Cell Signaling Involved in Skeletal Muscle Development and Disease
1. Institute of Animal Nutrition, Sichuan Agricultural University, Ya'an, Sichuan 625014, P. R. China.
2. Key Laboratory of Animal Disease-Resistance Nutrition, Ministry of Education, China.
Received 2016-6-13; Accepted 2016-8-19; Published 2016-10-17
Abstract
Skeletal muscle is a remarkably complicated organ comprising many different cell types, and it plays an important role in lifelong metabolic health. Nutrients, as an external regulator, potently regulate skeletal muscle development through various internal regulatory factors, such as mammalian target of rapamycin (mTOR) and microRNAs (miRNAs). As a nutrient sensor, mTOR, integrates nutrient availability to regulate myogenesis and directly or indirectly influences microRNA expression. MiRNAs, a class of small non-coding RNAs mediating gene silencing, are implicated in myogenesis and muscle-related diseases. Meanwhile, growing evidence has emerged supporting the notion that the expression of myogenic miRNAs could be regulated by nutrients in an epigenetic mechanism. Therefore, this review presents a novel insight into the cell signaling network underlying nutrient-mTOR-miRNA pathway regulation of skeletal myogenesis and summarizes the epigenetic modifications in myogenic differentiation, which will provide valuable information for potential therapeutic intervention.
Keywords: disease, nutrient, microRNA, mTOR, skeletal myogenesis, signaling pathways
Introduction
Skeletal muscle is the largest component of lean body mass in humans and animals, and its condition is crucial for disease, injury, and regeneration [1]. Skeletal muscle development is a multi-step process that includes the recruitment of myoblasts from myogenic precursors, myoblast proliferation, cell cycle arrest, differentiation and fusion into multinuclear myotubes and then myofibers [2,3]. This is a highly complex process in which environmental factors and signaling pathways integrate to activate muscle-specific regulatory transcription factor expression programs [4,5].
Nutrients are one of the most important epigenetic factors to affect skeletal muscle development. Skeletal muscle has a lower priority in nutrient partitioning compared with other organs during early development [6], leading to the development of skeletal muscle vulnerability to nutrient deficiency. Mammalian target of rapamycin (mTOR) is an evolutionarily conserved protein kinase that regulates a series of biological processes, including cell growth, differentiation and metabolism [7]. During the past several years, many lines of evidence have confirmed that mTOR, located at the heart of a nutrient-sensing signaling network, has emerged as a key regulator of skeletal muscle development [8]. For example, mTOR inactivation in mouse muscle results in severe myopathy, reduced mitochondrial biogenesis, impaired oxidative capacity, and increased glycogen content, leading to premature death [9]. microRNAs (miRNAs) are a group of highly conversed noncoding RNAs that regulate many biological processes by governing target gene expression [10]. Increasing findings have supported a role for miRNAs in the regulation of myogenesis, and changes in miRNA profile of skeletal muscle are associated with various muscular disorders [11-14].
The development of skeletal muscle is a highly complex process in which environmental factors and signaling pathways integrate to activate muscle-specific regulatory transcription factor expression programs [4,5]. Nutrients, as well-established primary environmental factors, activate mTOR signaling to regulate various processes of skeletal muscle development. For instance, leucine supplementation facilitates the proliferation and differentiation of primary preterm rat satellite cells via mTOR complex 1 (mTORC1) signaling [15]. Meanwhile, nutrients are also reported to modulate the expression of miRNAs. Specifically, leucine in C2C12 cells and essential amino acids in human skeletal muscle have both been shown to regulate miRNA expression [16,17]. Interestingly, miRNA expression seems to be directly or indirectly managed by mTOR. Treatment of C3H10T1/2 or C2C12 cells with specific mTOR inhibitor rapamycin was shown to change miRNA profiles [18,19]. Within this context, the regulatory mechanisms of mTOR regulation of miRNA biogenesis were also uncovered during skeletal myogenesis [18-20]. Additionally, Ye et al. (2015) revealed another novel pathway where nutrients dynamically regulate global miRNA biogenesis via mTORC1-Mdm2-Drosha signaling [21]. Here, we present early observations that established epigenetic (nutrient) and endogenetic (mTOR and miRNAs) factors as key myogenic regulators, discuss recent advances concerning the link among nutrients, mTOR and miRNAs in skeletal myogenesis, and summarize our current understanding of cell signalling network involving the nutrient-mTOR-miRNA pathway in skeletal muscle development and implications for skeletal muscle disease.
Nutrient as a key external regulator of skeletal myogenesis
Skeletal muscle is one of the most susceptible tissues to environmental stimuli, such as nutrients and exercise [22]. The role of nutrients in the regulation of skeletal muscle growth and hypertrophy is relatively well established, and the mechanisms are primarily similar to those in protein metabolism in existing muscle fibers [23,24]. However, the role of nutrients in skeletal myogenesis (myofiber formation, including terminal differentiation and fusion) cannot be determined from its requirement for skeletal muscle hypertrophy and growth (increase in protein synthesis that may or may not refer to fusion). In mammals, muscle fibers are primarily formed prenatally, and subsequent muscle development largely depends on muscle fiber hypertrophy [25-27]. Prenatal nutrition mainly influences the number of secondary fibers [26,28]. For instance, maternal undernutrition or a low-protein diet causes a decrease of fast fibers in early stages [29,30], while an increase in fast fibers was observed in later stages [31]. Recently, our results revealed that increased maternal dietary energy intake reduces protein expression of fast-MyHC isoforms and delays differentiation and maturation in foetal skeletal muscle [28]. However, postnatal nutrition largely affects muscle fiber hypertrophy and conversion [32]. Specifically, postnatal undernutrition decreases fiber size and affects glycolytic fast-twitch type II more than other types of myofibers [33-35]. Furthermore, branched-chain amino acid supplementation can prevent skeletal muscle wasting by increasing mitochondrial biogenesis and preserving muscle fiber size in skeletal myocytes and skeletal muscle of middle-aged mice [36].
Multiple studies have also explored the influence of nutrients on myoblast cell proliferation and differentiation in culture. β-Hydroxy-β-methylbutyrate (HMB), a leucine catabolite, has a positive influence on muscle development via the promotion of proliferation and differentiation, the acceleration of fusion, and the reduction of apoptosis in adult myoblast cultures [37]. Fulco et al. (2008) showed that glucose restriction impairs the differentiation of skeletal myoblasts [38]. However, high ambient glucose also suppresses skeletal myogenesis with a two-fold decrease in myoblast fusion [39]. More recently, Averous et al. (2012) demonstrated that the lack of leucine inhibits myoblast differentiation [40]. Meanwhile, our recent results also showed that leucine could promote the proliferation of C2C12 cells [16]. Furthermore, satellite cells are the major muscle stem cells responsible for postnatal skeletal muscle growth and regeneration, which involve several steps including proliferation, migration, and fusion of satellite cells either with an existing fiber or with other satellite cells to form a new muscle fiber [41,42]. However, this process of generating muscle-myogenesis can also be influenced by glucose and essential amino acids (EAAs) [15,43]. For example, in vitro myotube formation of primary preterm rat satellite cells was induced with the administration of the essential amino acid (EAA) leucine, possibly mediated by increased activation of the mTOR signal pathway [15].
mTOR as a crucial internal regulator of skeletal myogenesis
Mammalian target of rapamycin (mTOR) senses and integrates cellular nutrients and energy status to regulate various cellular processes, including cell growth, proliferation, differentiation, metabolism, survival and autophagy [7]. This serine/threonine kinase interacts with several proteins to form two distinct mTOR-containing complexes named mTOR complex 1 (mTORC1) and mTORC2, which are identified by the unique existence of raptor and rictor, mediating rapamycin-sensitive and -insensitive signaling of mTOR, respectively [44,45].
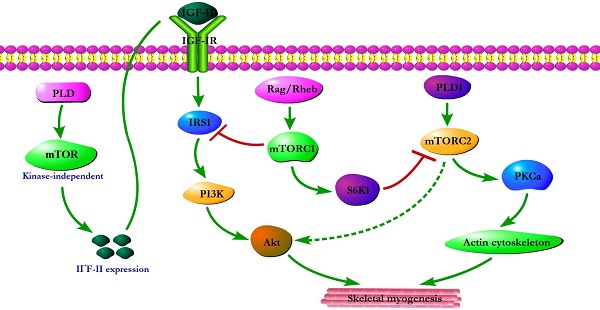
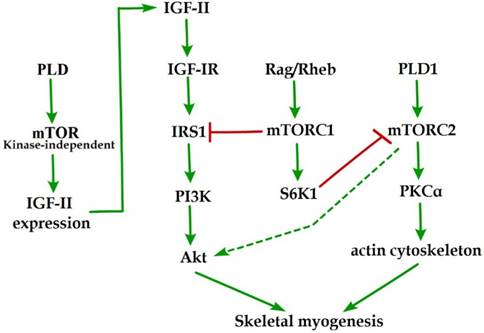
Initial evidence for the involvement of mTOR in skeletal myogenesis originated from the results of Coolican et al. (1997), who uncovered the inhibitory effect of rapamycin on rat L6 myoblast differentiation [46]. Interestingly, results from other teams also revealed that rapamycin repressed C2C12 cell differentiation and skeletal muscle regeneration in rodents [47-50]. Meanwhile, the pharmacological proofs provided strong support for a role of mTOR in skeletal myogenesis by the capacity of a muscle-specific rapamycin-resistant mTOR to rescue C2C12 cell differentiation and skeletal muscle remodeling from rapamycin treatment [48,50,51]. In addition, insulin-like growth factor II (IGF-II) is an important mediator of kinase-independent mTOR in myogenic signaling. During myoblast differentiation, mTOR governs IGF-II transcription through the muscle-specific promoter and enhancer in a kinase-independent manner, and increased IGF-II expression modulates C2C12 cell differentiation via IRS1/Akt pathway [52], which is crucial for skeletal myogenesis [53,54]. Furthermore, data from Yoon and Chen (2008) indicated that phospholipase D1 (PLD1) is placed upstream of mTOR/IGF-II signaling (Figure 1) [55]. Nevertheless, mTORC1 and mTORC2 are involved in distinct signaling pathways and perform distinguishing functions in skeletal myogenesis (Figure 1).
mTOR signaling in skeletal myogenesis. PLD activates kinase-independent mTOR and subsequently modulates myogenic transcription of IGF-II. The Rag GTPases and Rheb activate mTORC1, which subsequently inhibit PI3K-Akt signaling by IRS1 phosphorylation. PLD1 also activates mTORC2 and potentially regulates the phosphorylation of Akt and PKCα. The phosphorylation of S6K1 can repress myogenic function of mTORC2.
mTORC1 signaling in skeletal myogenesis
Reviewing the functions of classical mTORC1 signaling components in skeletal myogenesis has revealed some unexpected discoveries. The key subunit of mTORC1, raptor, is an inhibitor of skeletal myogenesis, as knockdown of raptor promotes and overexpression of raptor inhibits myoblast differentiation [56,57]. Evidence indicates that the inhibitory effects of raptor on C2C12 cell differentiation relies on Ser-307 phosphorylation of insulin receptor substrate 1 (IRS1) by mTORC1 and subsequent suppression of PI3K/Akt signaling (Figure 1) [57]. Within this context, the authors simultaneously investigated the role of Rheb (an activator of mTORC1) in skeletal myogenesis. Likewise, Rheb negatively regulates skeletal myogenesis and most likely exerts an inhibitory function via the negative regulation of IRS1 protein levels by mTOR/raptor [57], which is in contrast to the positive function of Rheb in inducing skeletal muscle hypertrophy [58]. Meanwhile, Yoon and Chen (2013) also proposed Rag GTPases (Rag), another activator of mTORC1, as an inhibitor of myogenic differentiation, and this inhibitory effect of Rag is mediated by mTORC1 inhibition of the IRS1-PI3K-Akt pathway (Figure 1) [59]. Interestingly, ribosomal S6 kinase 1 (S6K1), a major target of mTORC1, is dispensable for myoblast differentiation and nascent myofibers formation in muscle regeneration despite its well-known functions in muscle growth, hypertrophy, and maintenance [50,52,57,60,61]. However, evidence also indicated that S6K1-mediated phosphorylation of Rictor negatively regulates the capacity of mTORC2 to phosphorylate Akt-S473 and persistent activation of S6K1 does not influence IRS1-PI3K signaling during myoblast [62] differentiation [63]. However, in addition, rapamycin-sensitive mTOR signaling governs two distinct stages of skeletal myogenesis: initial myotube/myofiber formation in an mTOR kinase-independent manner and myotube/myofiber maturation in an mTOR kinase-dependent mechanism [50].
mTORC2 signaling in skeletal myogenesis
Rapamycin-insensitive mTORC2 with its important component rictor, phosphorylates Akt at Ser-473 in numerous cellular contexts [64]. Multifunctional kinase Akt is a key nodal point of several pathways to regulate skeletal myogenesis and muscle regeneration [49,53,65]. Results from Shu and Houghton (2009) showed that both rapamycin and rictor downregulation represses the phosphorylation of Akt (S473) and blocks terminal differentiation (fusion) of C2C12 myoblasts [66]. Within this context, the expression of constitutively active Akt in rictor-deficient C2C12 cell rescues myoblast fusion even in the presence of rapamycin [66], indicating that Akt acts as a mediator of rictor/mTORC2 function in myogenesis. Moreover, Jaafar et al. (2011) has shown that PKCα, an identified substrate of mTORC2, is required for skeletal myogenesis and mediates the myogenic function of mTORC2 [56]. These authors have also proposed that PLD1 may function upstream of mTORC2 (Figure 1) [56], and this is in accordance with a report that mTORC2 assembly is regulated by PLD1 and its metabolite phosphatidic acid [67]. Additionally, the role of PLD1 in regulating mTORC2 may be associated with actin cytoskeleton remodeling [68], because it is well ascertained that mTORC2 controls the actin cytoskeleton in various cell types [69,70].
Although mTORC2 is a rapamycin-insensitive mTOR complex, persistent rapamycin administration disrupts mTORC2 complex, leading to the reduced phosphorylation of Akt at Ser-473 [71]. Thus, the negative function of rapamycin treatment in skeletal myogenesis can be ascribed to the inhibition of mTORC2 function. Consistently, the phosphorylation of Akt and PKCα in myoblast cells is sensitive to rapamycin administration [56,66]. Nevertheless, decisive evidence for mTORC2 serving as the correlative target of rapamycin in skeletal myogenesis is insufficient. Although expression of phosphomimetic mutant Akt (S437D) can restore C2C12 cell fusion from rapamycin administration [66], it is possible that Akt exerts its function on myogenesis via mTOR-dependent IGF2 pathway.
MicroRNA biogenesis and function in skeletal muscle development
MicroRNAs (miRNAs) are evolutionarily highly conserved, small non-coding regulatory RNAs that are approximately 21-23 nucleotides in length. They function in gene silencing and translational suppression by binding to the 3'-UTR of their target mRNAs. However, miRNAs have recently been reported to have the potential to activate translation [72]. In various biological processes, miRNAs regulate the protein levels of pivotal regulatory factors or serve as a switch to govern gene expression [73]. Post-transcriptional regulation mediated by miRNAs increases the complicacy of gene expression regulation and promotes its accuracy and sensitivity [74].
Synthesis and regulatory mechanism of microRNAs
Most miRNA genes come from regions of the genome that are quite distant from formerly annotated genes, suggesting that they derive from independent transcription units with their own promoter [75]. Nevertheless, a sizable minority of miRNA genes are derived from the introns of protein coding transcription units, implying that these miRNAs are dependent on the promoter region of their host genes and mRNA splicing patterns [76]. Additionally, other miRNA loci are found in clusters and are transcribed as poly-cistronic primary transcripts [77,78].
miRNA transcription is mainly performed by RNA polymerase II (Pol II) and is regulated by RNA pol II-related transcription factors and epigenetic regulators [79,80]. Following transcription, primary miRNA (pri-miRNA) containing a local hairpin structure undergoes multiple steps of maturation [77]. Specifically, most pri-miRNAs are processed by Drosha and its interacting partner DGCR8 (DiGeorge syndrome critical region gene 8) to form precursor miRNAs (pre-miRNAs), which are then exported from nucleus into cytoplasm by exportin-5. The pre-miRNAs are further processed by Dicer to form mature miRNA, which combines with the Argonaute (Ago) family of proteins within RISC (RNA-induced silencing complex). The function of RISC in translational control is mediated by a key Ago partner GW182, which recruits deadenylating/decapping enzymes to allow exonucleases to attack unprotected mRNA and represses translation by competing with the cap binding protein eIF4E and/or interfering with ribosome scanning in the cytoplasm [81]. In addition to their predominant functions in translational repression, miRNAs have also been involved in translational promotion depending on specific miRNA:mRNA base-pairing and Ago2 in mitochondria [72].
In animals, most of the studied miRNAs interact with specific seed sequences in the 3′-untranslated region (3′-UTR) of target genes to promote mRNA degradation and translational repression. The miRNA 5′-proximal seed sequence (positions 2-8) provides most of the pairing specificity and is generally essential for miRNA function [82-84]. Furthermore, a single target mRNA (3′-UTR in particular) can have more than one different miRNA binding sites, and a single miRNA can also regulate multiple target mRNAs by binding to its corresponding binding site in the 3′-UTRs of target mRNAs [85,86]. Seed sequences are particularly conserved in the 3′-UTR of target mRNAs, implying that the primary regulatory functions of miRNAs are derived from the 3′-UTR [87]. However, several studies have demonstrated that miRNA binding sites can also exist in the coding sequence [88] as well as in the promoter region of the target gene [89,90].
MicroRNAs in skeletal muscle developmental processes
Much like mTOR, miRNAs have also been confirmed as key regulators in various biological processes. An essential role for miRNAs in skeletal muscle development is identified by muscle-specific Dicer knockdown in mice, a key enzyme in the processing of miRNAs, which leads to perinatal lethality with decreased skeletal muscle mass, increased apoptosis of muscle cells and abnormal myofiber morphology [91]. In addition, postnatal deletion of Dicer in cardiomyocytes results in downregulation of miRNAs with a loss of cardiac contractility, cardiac function, and premature death [92,93]. It is significant to understand that Dicer is also involved in the expression of other types of small RNAs, whereas DGCR8, the interacting partner of Drosha, functions specifically in the miRNA biogenesis pathway [94]. Mice with the deletion of striated muscle-specific DGCR8 leads to serious dilated cardiomyopathy and heart failure, while the phenotype in skeletal muscle has not been described [95]. Although the aforementioned studies indicate the importance of miRNA biogenesis for muscle development and function, they do not mention whether this requisite function of Dicer and DGCR8 reflects essential roles of specific miRNAs or non-specific miRNAs in these processes. Here, we present the reported targets of miRNAs with well-known myogenic functions in skeletal muscle (Table 1).
Muscle-specific microRNAs and myogenesis
A range of miRNAs, which are highly expressed in skeletal and/or cardiac muscle (named as myomiRs) has been validated and includes miR-1, miR-133a, miR-133b, miR-206, miR-208b, and miR-499 [96-98]. The functions of these miRNAs during skeletal myogenesis have been widely studied and summarized. Of course, the best-studied myomiRs are miR-1-1/133a-2, miR-1-2/133a-1 and miR-206/133b, which originate from bicistronic transcripts on three different chromosomes [11]. In terms of function, these myomiRs regulate multiple aspects of skeletal muscle development and homeostasis by targeting myogenesis-related genes. In terms of structure, the mature sequences of miR-1 and miR-206 have identical seed regions, and only two mismatched bases exist between miR-133a and miR-133b. The similar origin and sequence of these myomiRs imply that they would also have a close relationship in skeletal myogenesis.
miRNAs and targets associated with skeletal myogenesis in mammals
| Functions | miRNAs | Targets | References |
|---|---|---|---|
| Inhibit proliferation Promote differentiation | miR-1/miR-206/miR-24/ miR-29/miR-128 | HDAC4, Pax3, Pax7, Igfbp5, c-Met, Cx43, YY1, Hand2, PTB, Utrn, Fstl1, TIMP3, Notch3, and pola1 | [18,72,88,101,106,166,171-185] |
| Promote proliferation Inhibit differentiation | miR-133/miR-17-92 | SRF, UCP2, Foxl2, and ENH1 | [101,102,124] |
| Promote proliferation Promote differentiation | miR-27a | MSTN | [16,186,187] |
| Inhibit proliferation Inhibit differentiation | miR-203 | C-Jun and MEF2C | [188] |
| Promote differentiation | miR-26a/miR-27b/miR-146b/miR-148a/miR-181/miR-199a-5p/miR-214/miR-322/miR-378/miR-424/miR-486/miR-503/miR-675 | Ezh2, TGF-β, BMP, HDAC6, Pax3, ROCK1, Smad4, Notch1, Hmga2, HoxA-11, FZD4, JAG1, WNT2, Cdc25A, MyoR, Pax7, Smad1, Smad5 and Cdc6 | [1,108,173,189-198] |
| Inhibit differentiation | miR-23a/miR-30-5p/miR-31/miR-98/miR-124/miR-125b/miR-155/miR-186/miR-199a-3p/miR-221/miR-222/miR-374b/miR-1192 | Myh, MBNL, Myf5, E2F5, Dlx5, IGF-II, MEF2A, MyoG, IGF-I, mTOR, RPS6KA6, P27, Myf6 and HMGB1 | [19,199-210] |
| Promote proliferation | miR-151-3p/miR-2400 | ATP2a2 and MyoG | [211,212] |
| Inhibit proliferation | miR-125a-5p/miR-195/miR-351/ miR-489/miR-497 | E2F3, CDC25, CCND and Dek | [213-216] |
| Myofiber specification | miR-23a/miR-133/miR-151-3p/miR-208b/miR-499/miR-494 | MEF2C, TEAD1, Sp3, ATP2a2, Thrap1, Sox6, HP-1β, and Purβ | [98,100,105,210,212,217] |
Among these myomiRs, miR-1 and miR-206 suppress myoblast proliferation and accelerate differentiation by targeting numerous target genes. Suppression of endogenous miR-1 and miR-206 inhibits downregulation of most target genes in differentiation cells [99], thus indicating that miRNA activity and target interaction are essential for muscle differentiation. Conversely, miR-133 promotes myoblast proliferation and inhibits differentiation via its specific regulation of the expression of several key genes, such as SRF, Foxl2 and TEAD1 [100-102]. Liu et al. (2011) reported that mice lacking both miR-133a-1 and miR-133a-2 developed progressive centronuclear myopathy in fast-twitch myofibers, accompanied by mitochondrial dysfunction and a fast-to-slow myofiber switch [103]. In addition, a local injection of miR-1, miR-133 and miR-206 mimics can act together to induce the expression of myogenic markers (myoD1, Pax7 and myogenin), thus promoting myogenic differentiation in a rat injury model [104]. However, an unexpected finding revealed that muscle-specific miR-1 stimulates mitochondrial translation of mtDNA-encoded transcripts [72]. Furthermore, muscle-restricted miR-208/miR-499 is a family of intronic miRNAs encoded by their host genes, Myh6, Myh7 and Myh7b, governing myofiber type switch [98]. Intriguingly, miR-499 was discovered to be the most remarkably decreased miRNA in response to hindlimb suspension [105], implying its potential significance in muscle maintenance.
Nonmuscle-specific microRNAs and myogenesis
In addition to the muscle-specific and muscle-restricted miRNAs presented above, numerous extensively expressed miRNAs are also associated with skeletal myogenesis, and their expression patterns are altered during myogenic differentiation. It was reported that these nonmuscle-specific miRNAs modulate skeletal muscle development through multiple targets (Table 1). Muscle development-related genes, such as Akt3, MSTN, YY1, MEF2, Pax3 and MyoG, are targeted by these miRNAs. Although these miRNAs are not muscle-specific expressed and some of them may be lacking in the process of subsequent differentiation, their regulatory functions in skeletal myogenesis are irreplaceable. For instance, reduced expression of miR-24 dramatically decreases the myogenic markers, myogenin, MHC and MEF2, and therefore inhibits myotube formation [106]. The inhibition of miR-214 [107], miR-26a [108] can also block this process.
Cross regulation of nutrients, mTOR and microRNAs in skeletal muscle
Regulation of mTOR activation by nutrients
Mammalian target of rapamycin (mTOR) is located at the heart of a nutrient-sensing signaling network, and its activity can be regulated by multiple nutrients, such as amino acids and glucose, thus influencing muscle cell proliferation, differentiation, autophagy, and metabolism [109]. In this point, it is significant to understand the molecular mechanisms underlying how nutrients regulate the activity of mTOR.
Amino acids are one of the most studied nutrients in the regulation of mTOR activation during myogenesis [110]. Raptor-associated mTORC1 aggregates the signaling network that transduces amino acid sufficiency and mitogenic signals. For example, leucine administration promotes the proliferation and differentiation of rat satellite cells partly via mTORC1 signaling pathway [15]. Several mediators of amino acid signals, including Rag GTPase and the class III phosphinositide 3-kinase (PI3K) Vps34, have been reported to lie upstream of mTORC1 [59]. Rag heterodimers, which are regulated by the guanine nucleotide exchange factor activity of the Ragulator protein complex and GTPase-activating protein activity of the GATOR complex, bind raptor and translocate mTORC1 to the lysosomal surface in response to amino acid stimulation, which is necessary for the activation of mTORC1 [111,112]. Vps34 mediates amino acid signals to activate mTORC1 by targeting phospholipase D1 (PLD1) for lysosomal translocation and activation [113,114]. PLD1 and its product, phosphatidic acid (PA), are important for myogenic differentiation, as well as amino acid activation of mTORC1 [114,115]. Additionally, amino acids have also been confirmed to activate mTOR complex 2 (mTORC2) through PI3K/Akt signaling during myoblast differentiation [116].
Regulation of microRNA expression by nutrients
It has been well established that nutrients can modulate protein-coding gene expression [117]. However, increasing evidence has emerged that the major nutrients, such as glucose and amino acids, alter the expression of miRNA [16,17,118,119]. Given the important role of nutrients in the modulation of skeletal muscle development, one could expect that changes in miRNA biogenesis in response to nutrient status may influence, at least in part, the regulation of skeletal myogenesis. Thus, Drummond et al. (2009) showed that acute essential amino acids (EAAs) ingestion elicited robust increases in miR-1, miR-23a, miR-208b, and miR-499 expression, with an accompanied increase in MyoD1 and FSTL1 mRNA expression and decrease in myostatin and MEF2C mRNA expression in human skeletal muscle [17]. Recently, our study reported that miR-27a was induced by leucine and contributed to leucine-induced proliferation promotion in C2C12 cells [16]. It is probably that sustaining consumption of a high-protein diet causes a similar miRNA expression pattern and subsequent functional outcome. Moreover, Zhu et al. (2014) showed that seven miRNAs were significantly upregulated or downregulated within 1 h after refeeding in skeletal muscle of Chinese perch [120], which were involved in a fast-response signaling system in regulating skeletal muscle growth.
Regulation of microRNA expression by mTOR
Given the well-recognized significance of both mTOR and microRNAs in skeletal myogenesis, it would not be surprising that some connections between them exist. Totary-Jain et al. (2013) showed that comprehensive reprogramming of miRNA expression occurred with long-term rapamycin treatment[121]. The miRNAs profiling study by Sun et al. (2010) has uncovered that miR-1 expression is regulated by mTOR control of protein stability of the major myogenic transcription factor MyoD [18], which is located directly upstream enhancer of several other myogenic miRNAs, such as miR-133 and miR-206 [11]. Like that of miR-1, the expression patterns of miR-133 and miR-206 were regulated by MyoD [20], and the acute increase of miR-133 and miR-206 levels was entirely blocked by mTOR-specific inhibitor rapamycin during C2C12 cell differentiation [18]. However, the precise mechanism by which mTORC1 regulates MyoD stability is not clear. Additionally, several other induced miRNAs during myoblast differentiation were sensitive to rapamycin treatment although to lesser degrees. [18]. Furthermore, Ge et al. (2011) showed that rapamycin administration completely blocked the decline of mature miR-125b during differentiation in both C2C12 cells and primary myocytes [19]. Most recently, Ye et al. (2015) proposed another pathway in which mTOR activation markedly downregulated miRNA biogenesis via the upregulation of Mdm2, an essential and sufficient E3 ligase for ubiquitinylation of miRNA-processing enzyme DROSHA [21]. Besides, this analogous result has also been observed in other cell types. For instance, in cancer cells, long-term treatment with rapamycin resulted in remarkable changes in miRNA profiles [121], and these alterations are associated with resistance to rapamycin. These findings stress the influence that mTOR has on miRNA biogenesis and are consistent with the results observed in mouse research, where a maternal low-protein diet was exhibited to alter a subset of miRNAs in offspring via inhibition of mTORC1 signaling [122]. However, miRNA biogenesis regulated by mTORC2 in skeletal muscle has never been reported.
Nutrient regulation of mTOR-microRNA pathways in the context of skeletal muscle development
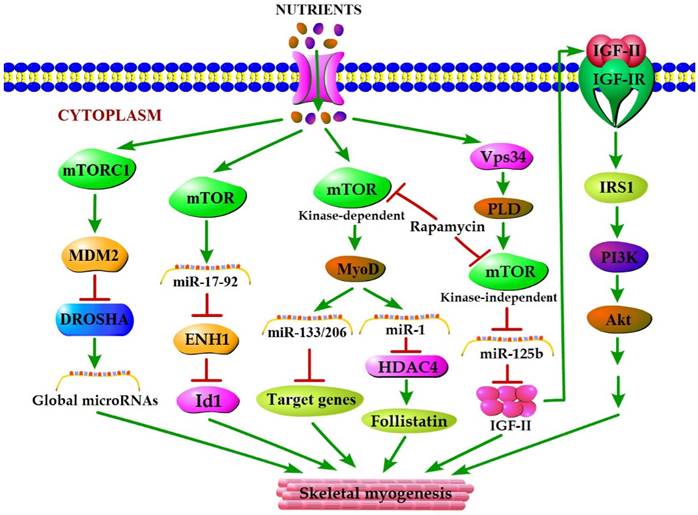
mTOR plays a vital role in the cellular response to nutrient availability and energy status. The link between mTOR activity and miRNA biogenesis, as described above, suggests that the miRNA biogenesis machinery may be responsive to energy and nutrient availability during skeletal myogenesis. Sun et al. (2010) reported that mTOR controlled MyoD-dependent transcription of muscle-specific microRNA miR-1 by regulating MyoD protein stability both in differentiating myoblasts and in mouse regenerating skeletal muscle [18]. Moreover, the functional pathway downstream of mTOR and miR-1 is delineated, in which miR-1 inhibition of histone deacetylase 4 (HADC4) leads to the production of follistatin and subsequent myocyte fusion [18]. Notably, MyoD also functions directly upstream enhancer of miR-133 and miR-206 [20], and the increased levels of miR-133 and miR-206 during myoblast differentiation were repressed with rapamycin treatment [18]. Evidence from Ge et al. (2011) showed that miR-125b biogenesis is negatively modulated by mTOR signaling in a kinase-independent manner both in vitro and in vivo as a part of a dual mechanism by which mTOR regulates the production of IGF-II, a master switch governing the initiation of skeletal myogenesis [19]. It was also confirmed that overexpression of PLD1 relieves the inhibitory effect of rapamycin on L6 cell differentiation [56], which is in line with a competitive mechanism between phosphatidic acid and rapamycin for combining with mTOR [123]. In C2C12 cells, PLD1 can be activated by amino acids [114] and was proven to act on PLD1 upstream of the mTOR/IGF2 pathway [55]. Nevertheless, mTOR markedly regulates the biogenesis of these two miRNAs through distinct manner, one dependent (miR-1) and one independent (miR-125b) of its kinase activity. In addition, numerous miRNAs other than miR-1 and miR-125b have exhibited various degrees of rapamycin sensitivity, indicating that these miRNAs may be directly or indirectly regulated by mTOR [18]. Moreover, Totary-Jain et al. (2013) revealed that long-term rapamycin treatment exhibited comprehensive reprogramming of miRNA expression with the up-regulation of miR-17-92 cluster [121], which could regulate myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis [124]. However, our recent results showed that miRNA-27a was induced by leucine and contributed to leucine-induced proliferation promotion in C2C12 cells [16]. Although increased leucine levels stimulate mTOR signaling and cell growth in C2C12 skeletal muscle cells [125], the role of mTOR in leucine-induced miR-27a biogenesis during myoblast proliferation needs further verification. Recently, another regulatory mechanism in mouse and human cells was proposed. Under high nutrient conditions (amino acid or glucose sufficient), mTORC1 is activated, and it increases the levels of ubiquitin E3 ligase Mdm2 by a p53-dependent transcriptional and P53-independent post-transcriptional pathway. Intriguingly, Mdm2 ubiquitinates and targets the miRNA-processing enzyme Drosha for proteasomal-dependent degradation, leading to a global decrease of miRNA biogenesis [21]. Under low nutrient conditions (amino acid starvation or glucose deprivation), mTORC1 is not active and Mdm2 levels are low. Drosha levels were elevated leading to an increase in global miRNA biogenesis [21]. Using a high-throughput screen of a miRNA mimic library, four miRNA mimics (miR-297, miR-376-3p, miR-567, and miR-627-5p) were identified that were able to increase the resistance of Drosha-silenced cells to nutrient deficiency [21]. Two of the four miRNAs (miR-297 and miR-567) significantly improved Drosha protein levels, implying that these two miRNAs may protect cells from apoptosis by maintaining Drosha level [21]. Although the differences exist in the nutrient regulation of miRNA biogenesis during myogenesis, the common characteristic of nutrient-mTOR-miRNA signaling pathway can further explain the mechanism of nutrient regulation of muscle development (Figure 2).
A model for nutrients-mTOR-miRNA signaling in skeletal myogenesis. MiRNA biogenesis in response to nutrient conditions regulates skeletal myogenesis via an mTORC1-MDM2-Drosha signaling. Meanwhile, mTOR controls skeletal myogenesis through miR-17-92/ENH1/Id1 regulatory axis. Besides, nutrient regulation of certain miRNA expression during skeletal myogenesis depends on two rapamycin-sensitive myogenic mTOR signaling pathways. A kinase-dependent mTOR pathway regulates myogenesis by governing the expressions of various miRNAs (miR-1, miR-133 and miR-206) through stabilizing major myogenic transcription factor MyoD. A kinase-independent mTOR signaling governs IGF-II transcription at a muscle-specific enhancer via its negative regulation of miR-125b level, and IGF-II in turn regulates skeletal myogenesis through activating the IGF-I receptor (IGF-IR) and PI3K/Akt signaling.
Epigenetic modifications in skeletal muscle development
The terminology “epigenetics” refers to mitotically and meiotically heritable alterations of gene expression without changing the DNA sequence [126]. In addition to miRNAs, DNA methylation and histone modifications are known to be the main areas of epigenetics that have profound influences on gene expression control and skeletal myogenesis [127,128]. Additionally, epigenetic modifications can influence the expression of miRNAs in different cell types, such as smooth muscle cells, cancer cells, and adipocytes [129-132].
DNA methylation almost solely occurs at CpG-containing promoters (CpG islands) and is performed by transferring a methyl group from S-adenosylmethionine (SAM) to the fifth carbon of a cytosine ring mediated by DNA methyltransferases (DNMTS) [133]. The initial relationship between DNA methylation and skeletal myogenesis was determined by the pioneering research that genome demethylation with either 5-azacytidine treatment or overexpression of antisense RNA against Dnmt1 induces the transdifferentiation of mouse fibroblasts into myoblasts [134,135]. Later, it was found that demethylation of the distal enhancer in MyoD gene and MyoG promoter is essential for the initiation of differentiation program [136,137]. Furthermore, many studies have also provided direct evidence that DNA methylation and DNA methyltransferases are crucial for skeletal myogenesis [138-142].
Although numerous studies have focused on the effects of DNA methylation, there are proofs emerging on histone modifications in skeletal muscle development [143]. Histone modifications are related to both gene expression activation, including trimethylation of lysine 4 of histone H3 (H3K4me3) and acetylation of histones H3 and H4 (acetyl H3, acetyl H4), and gene expression repression, such as trimethylation of lysine 20 of histone H4 (H4K20me3) trimethylation of lysines 9 and 27 of histone H3 (H3K9me3; H3K27me3) [128]. Tao et al. (2011) revealed that Set7/9, a SET domain-containing histone 3 lysine 4 (H3-K4) methyltransferase, directly interacts with MyoD to promote myoblast differentiation and myofibril assembly [144]. By contrast, lysine methyltransferase G9a mediating histone H3 lysine-9 di-methylation (H3K9me2) on MyoD target promoters inhibits skeletal muscle differentiation [140]. Moreover, numerous studies have revealed that histone acetyltransferases (HATs) and histone deacetylases (HDACs) regulate the acetylation state of histones and certain myogenic transcription factors to govern the activation or repression of skeletal muscle-specific genes [145,146].
It has also been reported that nutrition factors affect the process of epigenetic modifications in skeletal muscle development. Diets deficient in methyl group donors lead to global DNA hypomethylation in rodents [147-149]. Likewise, maternal protein restriction in rats leads to reduced PGC-1α expression through altered DNA methylation in skeletal muscle [150]. Conversely, a high-fat overfeeding diet increases DNA methylation of the PPARGC1A promoter in the skeletal muscle of young healthy men [151]. Additionally, maternal protein restriction during gestation increases the expression of CCAAT/enhancer-binding protein (C/EBPβ) and glucose transporter 4 (GLUT4) in female offspring rat skeletal muscle through the regulation of histone modification at their promoter regions [152,153].
Implications for muscle-related disease
Primary skeletal muscle disorders are induced by defective structural proteins, enzymes or abnormal signaling molecules and involve several different groups of diseases, such as muscular dystrophies, inflammatory myopathies and myopathies [154]. While these diseases can be classified in accordance with their clinical and pathological traits, they comprise Duchenne muscular dystrophy (DMD), Becker muscular dystrophy (BMD), facioscapulohumeral muscular dystrophy (FSHD) and limbgirdle muscular dystrophies type 2A and 2B, as well as other myopathies, such as polymyositis, dermatomyositis, Miyoshi myopathy and inclusion body myositis [155].
With the development of miRNA biology, numerous studies and extensive miRNAs expression profiling have revealed that miRNA dysregulation is a common characteristic of muscle pathology (Table 2) and in some situations individual miRNAs have been shown to cause or relieve disease. Based on our aforementioned understanding, therapeutic manipulation of nutrients, mTOR or miRNAs provide the potentially powerful approaches to treat skeletal muscle diseases. For instance, supplementation of DMD patients with branched-chain amino acids (BCAAs), especially leucine, was a potential and logical approach to reduce the rate of disease progression [156]. Meanwhile, leucine has been shown to increase the proliferating satellite cell number and improve the overall morphology, myofiber size gain and strength recovery in regenerating muscles [157-159]. In this regard, Drummond et al. (2009) showed that essential amino acids (EAAs) increased the expression of miR-499, -208b, -23a, -1, pri-miR-206 and muscle-growth genes MyoD1 and FSTL1, while decreasing the expression of myostatin (an inhibitor of muscle growth) and MEF2C in human skeletal muscle [17]. Luciferase analysis confirmed that miR-499 can effectively target the 3'-UTR of myostatin [160], suggesting that the increase in miR-499 by EAAs inhibited the negative regulatory role of myostatin in skeletal muscle disease. Moreover, skeletal muscle mTOR activation was significantly higher in mdx mice, a model for DMD [161], indicating that the regulation of mTOR activation can alleviate this pathologic condition. Eghtesad et al. (2011) reported treated mdx mouse with the mTOR inhibitor rapamycin both systemically and locally led to a significant decline of muscle fiber necrosis in tibialis anterior and diaphragm muscles 6 weeks post-treatment [162]. Additionally, the sequence-specific actions of miRNAs coupled with recent advances in the delivery of modified oligonucleotides suggested that modulation of miRNA expression may constitute a new therapeutic approach to treat several skeletal muscle diseases. Targeting of miR-31 has been postulated to act to restore dystrophin expression in Duchenne muscular dystrophy (DMD), and inhibition of miR-31 expression in human DMD myoblasts led to the rescue of dystrophin expression [163]. This suggests that strategies that can utilize miRNA-based therapies to recover dystrophin may be feasible therapeutic options to ameliorate clinical symptoms associated with DMD. The development of the muscle-derived rhabdomyosarcoma (RMS) can also be prevented by the re-expression of represented miRNAs, including miR-206, miR-203 and miR-29, which act by reprogramming the profile of RMS cells toward one of terminal differentiation [164-166]. Moreover, the targeting of miR-206 may provide therapeutic in amyotrophic lateral sclerosis (ALS), which is characterized by the loss of motor neuron supply to skeletal muscle and severe skeletal muscle atrophy [167].
Another example is autophagy. Autophagy activation may be critical not only for protein breakdown and muscle atrophy but also for myofiber survival, which can contribute to sarcopenia. Under sufficient nutrients, autophagy is inhibited by mTORC1; on the other hand, lack of nutrients through mTORC1 suppression is a well-established pathway for the activation of autophagy [168], which is initiated to reallocate nutrients from nonessential components to those that are vital to survival [169]. However, Wu et al. (2012) reported that miR-20a and miR-106b negatively regulated autophagy induced by leucine deprivation in C2C12 myoblasts [170]. Therefore, we infer that the nutrient-mTOR-miRNA pathway appears to regulate autophagy dependently of the canonical nutrient-sensing mTORC1 pathway.
miRNA abnormalities associated with disease in skeletal muscle
| Disease | miRNA | References |
|---|---|---|
| Duchenne muscular dystrophy | Increased expression of 37 identified miRNAs and 2 predicted miRNAs, decreased expression of 20 identified miRNAs and 3 predicited miRNAs; increased expression of 8 miRNAs including miR-206, decreased expression of miR-1, miR-135a and miR-29c; increased expression of miR-31 targeting dystrophin; decreased expression of miR-486 | [14,163,218,219] |
| Becker muscular dystophy | Increased expression of miR-221 and miR-146b | [14] |
| Limb-girdle muscular dystrophy type 2A | Increased expression of 80 identified miRNAs and 8 predicted miRNAs; decrease expression of miRR-30a-3p, miR-197 and 2 predicted miRNAs | [14] |
| Limb-girdle muscular dystrophy type 2B | Increased expression of 81 identified miRNAs and 6 predicted miRNAs; decreased expression of miR-30a-3p, miR-510 and 3 predicted miRNAs | [14] |
| Miyoshi myopathy | Increase expression of 64 identified miRNAs and 5 predicted miRNAs; decreased expression of miR-30a-3p, miR-30c, miR-302c and 2 predicted miRNAs | [14] |
| Facioscapulohumeral muscular dystrophy | Increased the expression of 57 identified miRNAs and 5 predicted miRNAs | [14] |
| Polymyositis | Increased expression of 35 identified miRNAs and 2 predicted miRNAs; decreased expression of miRR-30a-3p | [14] |
| Inclusion body myositis | Increased expression of 20 identified miRNAs; decreased expression of miR-197 and 1 predicted miRNA | [14] |
| Dermatomyositis | Increased expression of 33 identified miRNAs and 2 predicted miRNAs; decreased expression of miR-30a-3p and 1 predicted miRNA | [14] |
| Myotonic dystrophy type I | Increased the expression of miR-206; increased the expression of miR-1 and miR-335; reduced the expression of miR-29b, miR-29c and miR-33 | [220,221] |
| Rhabdomyosarcoma | Decreased the expression of miR-1 and miR-133a in Rhabdmyosarcoma tumour samples; elevated levels of miR-1, miR-133a, miR-133b and miR-206 in serum | [222-224] |
| Amyotrophic lateral sclerosis | Increased the expression of miR-206 | [167] |
| Atrophy | miR-1, miR-206, miR-133, miR-23a, miR-128, miR-499 and miR-208b are potentially protective | [105,219,225,226] |
| Hypertrophy | Expression of miR-1, miR-133a, miR-206b, miR-23, miR-26a, miR-29a, miR-378, miR-451 and miR-499 was discovered to be regulated | [97,98,225,227-229] |
Conclusions
The importance of nutrient regulation of mTOR-miRNA pathway provides an exciting opportunity to understand the molecular mechanism that controls skeletal muscle development, growth, adaption, regeneration and function. This signaling pathway also provides an avenue to anatomize the mechanisms that may contribute to genetic and acquired muscle disorders and related complications. Future studies should focus on the possibility of utilizing nutrients or mTOR activity regulators as therapeutic approaches via the regulation of miRNA biogenesis in muscle cell models and in vivo studies. Nevertheless, more physiological significance of our current understanding awaits confirmation by future in vivo studies, such as the generation and examination of skeletal muscle-specific mTOR-null animals, because conventional mTOR knockdown leads to early embryonic lethality in mice.
Acknowledgements
We are grateful to the members of our laboratories for the critical reading of the manuscript and helpful discussion. This work is supported by the National Basic Research Program of China (2012CB124701) and the National Natural Science Foundation of China (no. 31372323).
Competing Interest
The authors have declared that no competing interest exists.
References
1. Alexander M, Kawahara G, Motohashi N, Casar J, Eisenberg I, Myers J. et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 2013;20:1194-208
2. Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11:440-8
3. Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525-32
4. Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683-8
5. Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Elsevier. 2005:585-95
6. Bauman D, Eisemann J, Currie W. Hormonal effects on partitioning of nutrients for tissue growth: role of growth hormone and prolactin. Fed Proc. 1982;41:2538-44
7. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274-93
8. Ge Y, Chen J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J Biol Chem. 2012;287:43928-35
9. Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C. et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187:859-74
10. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102-14
11. Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461-69
12. Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441-8
13. Eisenberg I, Alexander MS, Kunkel LM. miRNAS in normal and diseased skeletal muscle. J Cell Mol Med. 2009;13:2-11
14. Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA. et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016-21
15. Dai JM, Yu MX, Shen ZY, Guo CY, Zhuang SQ, Qiu XS. Leucine promotes proliferation and differentiation of primary preterm rat satellite cells in part through mTORC1 signaling pathway. Nutrients. 2015;7:3387-400
16. Chen X, Huang Z, Chen D, Yang T, Liu G. MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 cells. Int J Mol Sci. 2013;14:14076-84
17. Drummond MJ, Glynn EL, Fry CS, Dhanani S, Volpi E, Rasmussen BB. Essential amino acids increase microRNA-499,-208b, and -23a and downregulate myostatin and myocyte enhancer factor 2C mRNA expression in human skeletal muscle. J Nutr. 2009;139:2279-84
18. Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189:1157-69
19. Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69-81
20. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721-26
21. Ye P, Liu Y, Chen C, Tang F, Wu Q, Wang X. et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol Cell. 2015;57:708-20
22. Liu X, Pan S, Li X, Sun Q, Yang X, Zhao R. Maternal low-protein diet affects myostatin signaling and protein synthesis in skeletal muscle of offspring piglets at weaning stage. Eur J Nutr. 2014:1-9
23. Paddon-Jones D, Sheffield-Moore M, Zhang XJ, Volpi E, Wolf SE, Aarsland A. et al. Amino acid ingestion improves muscle protein synthesis in the young and elderly. Am J Physiol Endocrinol Metab. 2004;286:E321-8
24. Drummond MJ, Rasmussen BB. Leucine-enriched nutrients and the regulation of mammalian target of rapamycin signalling and human skeletal muscle protein synthesis. Curr Opin Clin Nutr Metab Care. 2008;11:222-6
25. Du M, Zhao JX, Yan X, Huang Y, Nicodemus LV, Yue W. et al. Fetal muscle development, mesenchymal multipotent cell differentiation, and associated signaling pathways. J Anim Sci. 2011;89:583-90
26. Zhu MJ, Ford SP, Means WJ, Hess BW, Nathanielsz PW, Du M. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J Physiol. 2006;575:241-50
27. Zhu MJ, Ford SP, Nathanielsz PW, Du M. Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle. Biol Reprod. 2004;71:1968-73
28. Zou T, He D, Yu B, Yu J, Mao X, Zheng P. et al. Moderately increased maternal dietary energy intake delays foetal skeletal muscle differentiation and maturity in pigs. Eur J Nutr. 2015:1-11
29. Fahey A, Brameld J, Parr T, Buttery P. The effect of maternal undernutrition before muscle differentiation on the muscle fiber development of the newborn lamb. J Anim Sci. 2005;83:2564-71
30. Mallinson JE, Sculley DV, Craigon J, Plant R, Langley-Evans SC, Brameld JM. Fetal exposure to a maternal low-protein diet during mid-gestation results in muscle-specific effects on fibre type composition in young rats. Br J Nutr. 2007;98:292-9
31. Daniel Z, Brameld J, Craigon J, Scollan N, Buttery P. Effect of maternal dietary restriction during pregnancy on lamb carcass characteristics and muscle fiber composition. J Anim Sci. 2007;85:1565-76
32. Greenwood P, Hunt A, Hermanson J, Bell A. Effects of birth weight and postnatal nutrition on neonatal sheep: II. Skeletal muscle growth and development. J Anim Sci. 2000;78:50-61
33. Goldspink G, Ward P. Changes in rodent muscle fibre types during post-natal growth, undernutrition and exercise. J Physiol. 1979;296:453-69
34. Stickland N, Widdowson EM, Goldspink G. Effects of severe energy and protein deficiencies on the fibres and nuclei in skeletal muscle of pigs. Br J Nutr. 1975;34:421-8
35. Lefaucheur L, Ecolan P, Barzic Y-M, Marion J, Le Dividich J. Early postnatal food intake alters myofiber maturation in pig skeletal muscle. J Nutr. 2003;133:140-7
36. D'Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F. et al. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 2010;12:362-72
37. Kornasio R, Riederer I, Butler-Browne G, Mouly V, Uni Z, Halevy O. β-hydroxy-β-methylbutyrate (HMB) stimulates myogenic cell proliferation, differentiation and survival via the MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2009;1793:755-63
38. Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA. et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661-73
39. Grzelkowska-Kowalczyk K, Wieteska-Skrzeczyńska W, Grabiec K, Tokarska J. High glucose-mediated alterations of mechanisms important in myogenesis of mouse C2C12 myoblasts. Cell Biol Int. 2013;37:29-35
40. Averous J, Gabillard J, Seiliez I, Dardevet D. Leucine limitation regulates myf5 and myoD expression and inhibits myoblast differentiation. Exp Cell Res. 2012;318:217-27
41. Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628-33
42. Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4:a008342
43. Yue T, Yin J, Li F, Li D, Du M. High glucose induces differentiation and adipogenesis in porcine muscle satellite cells via mTOR. BMB Rep. 2010;43:140-5
44. Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158-66
45. Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596-603
46. Coolican SA, Samuel DS, Ewton DZ, McWade FJ, Florini JR. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem. 1997;272:6653-62
47. Cuenda A, Cohen P. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J Biol Chem. 1999;274:4341-6
48. Erbay E, Chen J. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J Biol Chem. 2001;276:36079-82
49. Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci U S A. 2002;99:9213-8
50. Ge Y, Wu AL, Warnes C, Liu J, Zhang C, Kawasome H. et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am J Physiol Cell Physiol. 2009;297:C1434-44
51. Shu L, Zhang X, Houghton PJ. Myogenic differentiation is dependent on both the kinase function and the N-terminal sequence of mammalian target of rapamycin. J Biol Chem. 2002;277:16726-32
52. Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163:931-6
53. Jiang BH, Aoki M, Zheng JZ, Li J, Vogt PK. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc Natl Acad Sci U S A. 1999;96:2077-81
54. Kaliman P, Canicio J, Shepherd PR, Beeton CA, Testar X, Palacín M. et al. Insulin-like growth factors require phosphatidylinositol 3-kinase to signal myogenesis: dominant negative p85 expression blocks differentiation of L6E9 muscle cells. Mol Endocrinol. 1998;12:66-77
55. Yoon MS, Chen J. PLD regulates myoblast differentiation through the mTOR-IGF2 pathway. J Cell Sci. 2008;121:282-9
56. Jaafar R, Zeiller C, Pirola L, Di Grazia A, Naro F, Vidal H. et al. Phospholipase D regulates myogenic differentiation through the activation of both mTORC1 and mTORC2 complexes. J Biol Chem. 2011;286:22609-21
57. Ge Y, Yoon MS, Chen J. Raptor and Rheb negatively regulate skeletal myogenesis through suppression of insulin receptor substrate 1 (IRS1). J Biol Chem. 2011;286:35675-82
58. Goodman CA, Miu MH, Frey JW, Mabrey DM, Lincoln HC, Ge Y. et al. A phosphatidylinositol 3-kinase/protein kinase B-independent activation of mammalian target of rapamycin signaling is sufficient to induce skeletal muscle hypertrophy. Mol Biol Cell. 2010;21:3258-68
59. Yoon MS, Chen J. Distinct amino acid-sensing mTOR pathways regulate skeletal myogenesis. Mol Biol Cell. 2013;24:3754-63
60. Park I-H, Erbay E, Nuzzi P, Chen J. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp Cell Res. 2005;309:211-9
61. Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E. et al. Atrophy of S6K1-/- skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7:286-94
62. Hamilton DL, Philp A, MacKenzie MG, Baar K. Prolonged activation of S6K1 does not suppress IRS or PI-3 kinase signaling during muscle cell differentiation. BMC Cell Biol. 2010;11:37
63. Julien LA, Carriere A, Moreau J, Roux PP. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol. 2010;30:908-21
64. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098-101
65. Marshall JL, Holmberg J, Chou E, Ocampo AC, Oh J, Lee J. et al. Sarcospan-dependent Akt activation is required for utrophin expression and muscle regeneration. J Cell Biol. 2012;197:1009-27
66. Shu L, Houghton PJ. The mTORC2 complex regulates terminal differentiation of C2C12 myoblasts. Mol Cell Biol. 2009;29:4691-700
67. Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411-20
68. Mebarek S, Komati H, Naro F, Zeiller C, Alvisi M, Lagarde M. et al. Inhibition of de novo ceramide synthesis upregulates phospholipase D and enhances myogenic differentiation. J Cell Sci. 2007;120:407-16
69. Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A. et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122-8
70. Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H. et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296-302
71. Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF. et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol cell. 2006;22:159-68
72. Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y. et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607-19
73. Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209-18
74. Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′ UTR evolution. Cell. 2005;123:1133-46
75. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116:281-97
76. Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165-73
77. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663-70
78. Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S. et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-32
79. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957-66
80. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-60
81. Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19-31
82. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-98
83. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell. 2005;120:15-20
84. Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363
85. Xie X, Lu J, Kulbokas E, Golub TR, Mootha V, Lindblad-Toh K. et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338-45
86. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ. et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500
87. Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105
88. Shi L, Zhou B, Li P, Schinckel AP, Liang T, Wang H. et al. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell Signal. 2015;27:1895-1904
89. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452-60
90. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608-13
91. O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ. et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359-68
92. Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z. et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111-6
93. da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM. et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567-76
94. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126-39
95. Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R. et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585-94
96. Callis TE, Deng Z, Chen JF, Wang DZ. Muscling through the microRNA world. Exp Biol Med. 2008;233:131-8
97. McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306-13
98. van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA. et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662-73
99. Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts. J Biol Chem. 2012;287:40360-70
100. Zhang D, Wang X, Li Y, Zhao L, Lu M, Yao X. et al. Thyroid hormone regulates muscle fiber type conversion via miR-133a1. J Cell Biol. 2014;207:753-66
101. Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-33
102. Luo Y, Wu X, Ling Z, Yuan L, Cheng Y, Chen J. et al. microRNA133a Targets Foxl2 and Promotes Differentiation of C2C12 into Myogenic Progenitor Cells. DNA Cell Biol. 2015;34:29-36
103. Liu N, Bezprozvannaya S, Shelton JM, Frisard MI, Hulver MW, McMillan RP. et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J Clin Invest. 2011;121:3258-68
104. Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495-505
105. McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of β-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39:219-26
106. Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G. et al. Transforming growth factor-β-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690-9
107. Feng Y, Cao JH, Li XY, Zhao SH. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochem Funct. 2011;29:378-83
108. Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836-43
109. Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells. 2013;35:463-73
110. Jewell JL, Russell RC, Guan K-L. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133-9
111. Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290-303
112. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496-501
113. Yoon M-S, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195:435-47
114. Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B. et al. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1). J Biol Chem. 2011;286:25477-86
115. Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118-23
116. Tato I, Bartrons R, Ventura F, Rosa JL. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J Biol Chem. 2011;286:6128-42
117. Clarke S, Abraham S. Gene expression: nutrient control of pre-and posttranscriptional events. FASEB J. 1992;6:3146-52
118. Druz A, Betenbaugh M, Shiloach J. Glucose depletion activates mmu-miR-466h-5p expression through oxidative stress and inhibition of histone deacetylation. Nucleic Acids Res. 2012;40:7291-302
119. Dey N, Das F, Mariappan MM, Mandal CC, Ghosh-Choudhury N, Kasinath BS. et al. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem. 2011;286:25586-603
120. Zhu X, Chen D, Hu Y, Wu P, Wang K, Zhang J. et al. The microRNA Signature in Response to Nutrient Restriction and Refeeding in Skeletal Muscle of Chinese Perch (Siniperca chuatsi). Mar Biotechnol. 2015;17:180-9
121. Totary-Jain H, Sanoudou D, Ben-Dov IZ, Dautriche CN, Guarnieri P, Marx SO. et al. Reprogramming of the microRNA transcriptome mediates resistance to rapamycin. J Biol Chem. 2013;288:6034-44
122. Alejandro EU, Gregg B, Wallen T, Kumusoglu D, Meister D, Chen A. et al. Maternal diet-induced microRNAs and mTOR underlie β cell dysfunction in offspring. J Clin Invest. 2014;124:4395-410
123. Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942-5
124. Qiu H, Liu N, Luo L, Zhong J, Tang Z, Kang K. et al. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death & Differentiation. 2016
125. Areta JL, Hawley JA, Ye JM, Chan MS, Coffey VG. Increasing leucine concentration stimulates mechanistic target of rapamycin signaling and cell growth in C2C12 skeletal muscle cells. Nutr Res. 2014;34:1000-7
126. Wilkins JF. Genomic imprinting and methylation: epigenetic canalization and conflict. Trends Genet. 2005;21:356-65
127. Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R-9
128. Perdiguero E, Sousa-Victor P, Ballestar E, Muñoz-Cánoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541-50
129. Hu W, Wang M, Yin H, Yao C, He Q, Yin L. et al. MicroRNA-1298 is regulated by DNA methylation and affects vascular smooth muscle cell function by targeting connexin 43. Cardiovasc Res. 2015;107:534-45
130. Han L, Witmer PDW, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284-8
131. Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277-81
132. Du J, Cheng X, Shen L, Tan Z, Luo J, Wu X. et al. Methylation of miR-145a-5p promoter mediates adipocytes differentiation. Biochem Biophys Res Commun. 2016;475:140-8
133. Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481-514
134. Taylor SM, Jones PA. Multiple new phenotypes induced in 10T12 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771-779
135. Szyf M, Rouleau J, Theberge J, Bozovic V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. J Biol Chem. 1992;267:12831-12836
136. Palacios D, Puri PL. The epigenetic network regulating muscle development and regeneration. J Cell Physiol. 2006;207:1-11
137. Lucarelli M, Fuso A, Strom R, Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J Biol Chem. 2001;276:7500-7506
138. Hupkes M, Jonsson MK, Scheenen WJ, van Rotterdam W, Sotoca AM, van Someren EP. et al. Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 2011;25:3861-3872
139. Senesi P, Luzi L, Montesano A, Terruzzi I. DNA demethylation enhances myoblasts hypertrophy during the late phase of myogenesis activating the IGF-I pathway. Endocrine. 2014;47:244-254
140. Ling BMT, Bharathy N, Chung TK, Kok WK, Li S, Tan YH. et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2012;109:841-846
141. Zhang RH, Judson RN, Liu DY, Kast J, Rossi FM. The lysine methyltransferase Ehmt2/G9a is dispensable for skeletal muscle development and regeneration. Skelet Muscle. 2016;6:22
142. Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384-94
143. Jin W, Peng J, Jiang S. The epigenetic regulation of embryonic myogenesis and adult muscle regeneration by histone methylation modification. Biochem Biophys Rep. 2016;6:209-19
144. Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R. et al. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194:551-65
145. McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497-504
146. Gurd BJ. Deacetylation of PGC-1α by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl Physiol Nutr Metab. 2011;36:589-97
147. Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43-9
148. Pogribny IP, Karpf AR, James SR, Melnyk S, Han T, Tryndyak VP. Epigenetic alterations in the brains of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain Res. 2008;1237:25-34
149. Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA. et al. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176-86
150. Zeng Y, Gu P, Liu K, Huang P. Maternal protein restriction in rats leads to reduced PGC-1α expression via altered DNA methylation in skeletal muscle. Mol Med Rep. 2013;7:306-12
151. Brøns C, Jacobsen S, Nilsson E, Ronn T, Jensen CB, Storgaard H. et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab. 2010;95:3048-56
152. Zheng S, Rollet M, Pan YX. Maternal protein restriction during pregnancy induces CCAAT/enhancer-binding protein (C/EBPβ) expression through the regulation of histone modification at its promoter region in female offspring rat skeletal muscle. Epigenetics. 2011;6:161-70
153. Zheng S, Rollet M, Pan YX. Protein restriction during gestation alters histone modifications at the glucose transporter 4 (GLUT4) promoter region and induces GLUT4 expression in skeletal muscle of female rat offspring. J Nutr Biochem. 2012;23:1064-71
154. Güller I, Russell AP. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol. 2010;588:4075-87
155. Goljanek-Whysall K, Sweetman D, Münsterberg AE. microRNAs in skeletal muscle differentiation and disease. Clin Sci. 2012;123:611-25
156. Davoodi J, Hutson SM, Grange RW. Branched chain amino acids in inherited muscle disease: The case of Duchenne Muscular Dystrophy. Springer. 2015:277-87
157. Rogulska A, Kurasz S. Regeneration of crushed skeletal muscles in experimental animals and the effect of leucine on the course of this process in white rat. Pol Med Sci Hist Bull. 1974;15:245-8
158. Pereira MG, Baptista IL, Carlassara EO, Moriscot AS, Aoki MS, Miyabara EH. Leucine supplementation improves skeletal muscle regeneration after cryolesion in rats. PLoS One. 2014;9:e85283
159. Pereira MG, Silva MT, da Cunha FM, Moriscot AS, Aoki MS, Miyabara EH. Leucine supplementation improves regeneration of skeletal muscles from old rats. Exp Gerontol. 2015;72:269-77
160. Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol. 2010;30:1937-45
161. Mouisel E, Vignaud A, Hourde C, Butler-Browne G, Ferry A. Muscle weakness and atrophy are associated with decreased regenerative capacity and changes in mTOR signaling in skeletal muscles of venerable (18-24-month-old) dystrophic mdx mice. Muscle Nerve. 2010;41:809-18
162. Eghtesad S, Jhunjhunwala S, Little SR, Clemens PR. Rapamycin ameliorates dystrophic phenotype in mdx mouse skeletal muscle. Mol Med. 2011;17:917
163. Cacchiarelli D, Incitti T, Martone J, Cesana M, Cazzella V, Santini T. et al. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136141
164. Diao Y, Guo X, Jiang L, Wang G, Zhang C, Wan J. et al. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J Biol Chem. 2014;289:529-39
165. Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M. et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366-78
166. Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J. et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer cell. 2008;14:369-81
167. Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL. et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549-54
168. Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287-95
169. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21-35
170. Wu H, Wang F, Hu S, Yin C, Li X, Zhao S. et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179-86
171. Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X. et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867-79
172. Yan D, Dong XDE, Chen X, Wang L, Lu C, Wang J. et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596-604
173. Dey BK, Gagan J, Dutta A. miR-206 and-486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203-14
174. Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P. et al. TGF-β regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805-14
175. Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677-87
176. Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863-71
177. Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347-65
178. Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77-85
179. Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71-84
180. Liu H, Chen SE, Jin B, Carson JA, Niu A, Durham W. et al. TIMP3: a physiological regulator of adult myogenesis. J Cell Sci. 2010;123:2914-21
181. Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA. et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122:2054-65
182. Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E. et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′ UTR microRNA recognition elements. Mol Cell. 2009;35:610-25
183. Wei W, He H, Zhang W, Zhang H, Bai J, Liu H. et al. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668
184. Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J Biol Chem. 2012;287:25255-65
185. Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126:2678-91
186. Chen X, Huang Z, Chen D, Yang T, Liu G. Role of microRNA-27a in myoblast differentiation. Cell Biol Int. 2014;38:266-71
187. Huang Z, Chen X, Yu B, He J, Chen D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem Biophys Res Commun. 2012;423:265-9
188. Luo W, Wu H, Ye Y, Li Z, Hao S, Kong L. et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014;5:e1347
189. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes & development. 2014;28:491-501
190. Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286:19431-8
191. Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61-74
192. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M. et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278-84
193. Khanna N, Ge Y, Chen J. MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. Plos One. 2014;9:e100657
194. Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem. 2012;287:21093-101
195. Sarkar S, Dey BK, Dutta A. MiR-322/424 and-503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138-49
196. Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ. et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci U S A. 2009;106:13383-7
197. Lee SW, Yang J, Kim SY, Jeong HK, Lee J, Kim WJ. et al. MicroRNA-26a induced by hypoxia targets HDAC6 in myogenic differentiation of embryonic stem cells. Nucleic Acids Res. 2015 gkv088
198. Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180-91
199. Dormoy-Raclet V, Cammas A, Celona B, Lian XJ, van der Giessen K, Zivojnovic M. et al. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA. Nat Commun. 2013;4:2388
200. Ma Z, Sun X, Xu D, Xiong Y, Zuo B. MicroRNA, miR-374b, directly targets Myf6 and negatively regulates C2C12 myoblasts differentiation. Biochem Biophys Res Commun. 2015;467:670-5
201. Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F. et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PloS One. 2009;4:e7607
202. Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song CC. et al. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int J Mol Sci. 2013;15:296-308
203. Antoniou A, Mastroyiannopoulos NP, Uney JB, Phylactou LA. miR-186 inhibits muscle cell differentiation through myogenin regulation. J Biol Chem. 2014;289:3923-35
204. Seok HY, Tatsuguchi M, Callis TE, He A, Pu WT, Wang DZ. miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J Biol Chem. 2011;286:35339-46
205. Qadir AS, Woo KM, Ryoo HM, Yi T, Song SU, Baek JH. MiR-124 Inhibits Myogenic Differentiation of Mesenchymal Stem Cells Via Targeting Dlx5. J Cell Biochem. 2014;115:1572-81
206. Jeremie K, Cindy D, Nadezda M, Julien P, Annick HB, Anna P. miR-98 delays skeletal muscle differentiation by down-regulating E2F5. Biochem J. 2015;466:85-93
207. Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell stem cell. 2012;11:118-26
208. Zhang BW, Cai HF, Wei XF, Sun JJ, Lan XY, Lei CZ. et al. miR-30-5p Regulates Muscle differentiation and alternative splicing of muscle-related genes by targeting MBNL. Int J Mol Sci. 2016;17:182
209. Wang L, Chen X, Zheng Y, Li F, Lu Z, Chen C. et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp Cell Res. 2012;318:2324-34
210. Shen L, Chen L, Zhang S, Zhang Y, Wang J, Zhu L. MicroRNA-23a reduces slow myosin heavy chain isoforms composition through myocyte enhancer factor 2C (MEF2C) and potentially influences meat quality. Meat Sci. 2016;116:201-6
211. Zhang WW, Tong HL, Sun XF, Hu Q, Yang Y, Li SF. et al. Identification of miR-2400 gene as a novel regulator in skeletal muscle satellite cells proliferation by targeting MYOG gene. Biochem Biophys Res Commun. 2015;463:624-31
212. Wei H, Li Z, Wang X, Wang J, Pang W, Yang G. et al. microRNA-151-3p regulates slow muscle gene expression by targeting ATP2a2 in skeletal muscle cells. J Cell Physiol. 2015;230:1003-12
213. Song C, Wu G, Xiang A, Zhang Q, Li W, Yang G. et al. Over-expression of miR-125a-5p inhibits proliferation in C2C12 myoblasts by targeting E2F3. Acta Biochim Biophys Sin. 2015;47:244-9
214. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat Commun. 2014;5:4597
215. Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A. et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524-8
216. Chen Y, Melton DW, Gelfond JA, McManus LM, Shireman PK. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol Genomics. 2012;44:1042-51
217. Yamamoto H, Morino K, Nishio Y, Ugi S, Yoshizaki T, Kashiwagi A. et al. MicroRNA-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and Forkhead box j3. Am J Physiol Endocrinol Metab. 2012;303:E1419-27
218. Alexander MS, Casar JC, Motohashi N, Myers JA, Eisenberg I, Gonzalez RT. et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet muscle. 2011;1:27
219. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-4
220. Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C. et al. Research Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48
221. Perbellini R, Greco S, Sarra-Ferraris G, Cardani R, Capogrossi MC, Meola G. et al. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul Disord. 2011;21:81-8
222. Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A. et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun. 2010;400:89-93
223. Missiaglia E, Shepherd C, Patel S, Thway K, Pierron G, Pritchard-Jones K. et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769-77
224. Rao PK, Missiaglia E, Shields L, Hyde G, Yuan B, Shepherd CJ. et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 2010;24:3427-37
225. Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PloS One. 2009;4:e5610
226. Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~ 27a~ 24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:1
227. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295:E1333-40
228. Nielsen S, Scheele C, Yfanti C, Åkerström T, Nielsen AR, Pedersen BK. et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J Physiol. 2010;588:4029-37
229. Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW. et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol. 2011;110:309-17
Author contact
![]() Corresponding authors: E-mail: hejun8067com (J. He) or dwchenedu.cn (D. Chen). Tel & Fax: +86 835 2882088
Corresponding authors: E-mail: hejun8067com (J. He) or dwchenedu.cn (D. Chen). Tel & Fax: +86 835 2882088

 Global reach, higher impact
Global reach, higher impact