10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(12):1437-1447. doi:10.7150/ijbs.16529 This issue Cite
Research Paper
MiR-99a and MiR-491 Regulate Cisplatin Resistance in Human Gastric Cancer Cells by Targeting CAPNS1
1. Department of Cell Biology, School of Basic Medical Sciences, Nanjing Medical University
140 Hanzhong Road, Nanjing 210029, P.R. China.
2. Department of Molecular Cell Biology and Toxicology, School of Public Health, Nanjing Medical University
140 Hanzhong Road, Nanjing 210029, P.R. China.
3. Laboratory of Cancer Biology, Biomedical Research Center, Sir Runrun Shaw Hospital, Zhejiang University, Hangzhou, People's Republic of China.
Received 2016-6-19; Accepted 2016-9-24; Published 2016-11-5
Abstract
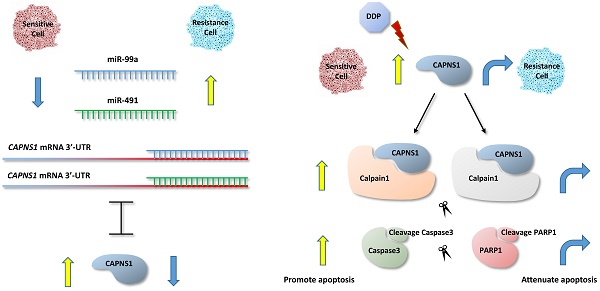
Cisplatin is the first-line agent utilized for the clinical treatment of a wide variety of solid tumors including gastric cancer. However, the intrinsic or acquired cisplatin resistance is often occurred in patients with gastric cancer and resulted in failure of cisplatin therapy. In order to investigate if miRNA involves in cisplatin resistance of human gastric cancer, we first screened and compared the expression of miRNAs between cisplatin resistant gastric cancer cell lines SGC-7901/DDP and BGC-823/DDP and their sensitive parental cells by miRNAs microarray and followed by analysis of 2D-GE/MS to identify their target proteins. We found both miR-99a and miR-491 were upregulated while their target gene calpain small subunit 1 (CAPNS1) was downregulated in resistant gastric cancer cells. Dual-luciferase- reporter assays with wild-type and mutated CAPNS1 3'-UTR confirmed their specificity of targeting. Inhibition of miR-99a and miR-491, or overexpress CAPNS1 can enhance cisplatin sensitivity of the resistant cells while transfection of two miRNAs' mimics or si-CAPNS1 in the sensitive cells can induce their resistance. Moreover, our results demonstrated CAPNS1 positively regulated calpain1 and calpain2, the catalytic subunits of CAPNS1, and cleaved caspase3 which further cleaved PARP1 and directly induced apoptosis. Therefore, miR-99a and miR-491 might be work as novel molecules regulate cisplatin resistance by directly targeting CAPNS1 associated pathway in human gastric cancer cells.
Keywords: miR-99a, miR-491, CAPNS1, cisplatin resistance, gastric cancer.
Introduction
Gastric carcinoma is one of malignancies with third mortality rate in cancers [1]. Surgery combined with radiation and/or chemotherapy is a major clinical treatment for gastric cancer. Cisplatin is an effective DNA-damaging antitumor agent utilized for the treatment of a wide variety of solid tumors including gastric cancer [2]. Despite of a consistent rate of initial responses, cisplatin usually results in the acquisition of chemoresistance and even progresses into multidrug resistance with cross tolerance to diverse agents thus finally leading to chemotherapy failure. The complicated mechanisms of drug resistance in cancer ranging from overexpression of drug efflux pump proteins and multidrug resistance protein family (MDR), mutation or alteration of drug targets, enhancement of the cellular DNA repair system and attenuation of apoptosis, etc. which remains unclearly demonstrated yet [3]. Micro RNAs (miRNAs) widely exist in animals and plants with function by base pairing with target mRNAs' 3'-UTR, to mediate mRNA cleavage or translational repression [4]. MiRNAs are closely associated with drug resistance, which are supported by the markedly differential expression of miRNAs between sensitive and resistant cells, and the regulation of target genes by miRNAs contributing to resistance [5, 6]. The calpain family genes are known to affect proliferation, apoptosis, migration, differentiation, and autophagy in cancer cells and to directly cleave caspase family members then to induce apoptosis [7]. In mammals, calpain family consists of 16 members, 14 large catalysis subunit members, 1 small regulatory subunit member, and 1 endogenous inhibitor [8]. Calpain small subunit 1 (CAPNS1) is the regulatory subunit targeting calpain1 and calpain2. However, it is unclear whether CAPNS1 is involved in the resistance of gastric cancer.
As one of the first line chemotherapy regimens, cisplatin (DDP) is widely applied in postoperative chemotherapy. In the present study, expression of miRNA was screened and anylysed in gastric cancer cell lines resistant to cisplatin [9], SGC-7901/DDP and BGC-823/DDP and their sensitive parent cells by microarray high-throughput assay. Two-dimensional gel electrophoresis combined with mass spectrometric analyses were further used to identify the difference between the differentially expressed proteins. Both MiR-99a and miR-491 were upregulated while CAPNS1 was downregulated in the resistant cells. Our data also revealed that CAPNS1 was negatively regulated by both miR-99a and miR-491. The mechanism of the indicated impact was preliminarily explored, which showed calpain1 and calpain2, regulated by CAPNS1, were implicated in cisplatin resistance in gastric cancer by cleaving the downstream proteins caspase3 and PARP1 to induce apoptosis.
Materials and methods
Cell lines, cultures and reagents
Human gastric cancer cell lines SGC-7901 and BGC-823 were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. Both the cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100U/ml penicillin, and 100μg/mL streptomycin (Life Technologies, Gibco, MD, USA). Cells were grown at 37°C, in a humidified incubator with 5% CO2. Cisplatin resistant gastric cancer cells lines SGC-7901/DDP and BGC-823/DDP were produced from parental SGC-7901 and BGC-823cells by persistence gradient exposure to cisplatin for about 12 months, which were subjected to increasing concentrations from 0.05μg/mL until the cells acquired resistance to 1μg/mL of cisplatin (Sigma-Aldrich, St. Louis, MO, USA). Prior to each experiment, SGC-7901/DDP and BGC-823/DDP cells were cultured in original RPMI 1640 medium for 2 weeks.
Plasmids, siRNA and transfection
MiRNA mimics, inhibitor, negative control, small interfering RNA (siRNA) specific for CAPNS1, and nonspecific control siRNA was synthesized (Ribobio, Guangzhou, China). si-CAPNS1, target sequences CGATCAGGGACCATTTGCA, sense sequences 5'-CGAUCAGGGACCAUUUGCA dTdT-3', antisense sequences 3'-dTdT GCUAGUCCCUGGUAAACGU-5'. HA-CAPNS1 overexpression plasmid was cloned into pcDNA3.0 by PCR amplification and sequenced. (Genechem, Shanghai, China). CAPNS1 3'-UTR-WT/MUT (about 500nt before and after the binding sites) was cloned into Dual-luciferase-plasmid (Ribobio, Guangzhou, China). All the miRNAs mimics, inhibitors, plasmid DNA or siRNA was transfected into cells with Lipofectamine 2000 according to manufacturer's instructions. 50nM of RNA duplex and 100 nM of miRNA inhibitor were used for each transfection.
RNA isolation and qRT-PCR
Total RNA was isolated from cell lines with Trizol reagent (Invitrogen, Carlsbad, CA) and miRNeasy mini kit (Qiagen, CA, USA). For synthesis of cDNAs specific to miR-99a and miR-491, a miScript II RT Kit (Qiagen, CA, USA) was used. The qRT-PCR reactions were performed with miScript SYBR® Green PCR Kit (Qiagen, CA, USA). Expression levels of RNU6 snRNA was used as loading control. The PCR was monitored in real-time using an Applied Biosystems 7900HT sequence detection system (Applied Biosystems). That allowed determination of the threshold cycle (Ct) at the time exponential amplification of the PCR products began. The average Ct, from triplicate assays, was used for further calculations. Primers are synthesized by QIAGEN (miScript Primer Assays Kit). Results are expressed as the mean value of triplicate experiments. Relative expression levels were normalized to controls=2- (Target Ct-Control Ct).
miRNA and Protein expression profiles and data analysis
MiRNA expression profiling of four cell lines was performed by Affymetrix GeneChip miRNA 2.0 Array (CapitalBio Corporation, Beijing, China). After data normalization and filtering, 13 miRNAs were used for unsupervised hierarchical clustering. Cluster and Tree View programs (Stanford University, CA, USA) were used for visualization. Protein expression profiles performed by 2D-MS which was described previously [9].
Luciferase assay
HEK-293 Cells were seeded into 24-well plates one day before transfection, and then co-transfected with 30ng of pmiR-RB-REPORTTM vectors (Ribobio, Guangzhou, China), which harbored CAPNS1 3'UTR wild-type or mutant constructs, and 50nM of miRNAs. After 48h, luciferase activity was measured with a Dual-luciferase assay kit (Promega). Renilla luciferase activity was normalized to firefly luciferase activity.
Clonogenic assay
After 48h, the transfected SGC-7901, BGC-823, SGC7901/DDP, BGC823/DDP cells were trypsinized and treated with cisplatin at indicated doses for 2h; the cells then further cultured in 6-well plates with 300 cells/well for 2 weeks to SGC-7901 and BGC-823 cells or 3 weeks to SGC-7901/DDP and BGC-823/DDP cells, respectively. For scoring colonies, the cells were fixed in 1ml methanol for 15 min and stained with Giemsa for 10 min. The clone numbers were expressed as mean ± SD from at least three independent experiments.
TUNEL assay
Apoptosis was assessed using TUNEL assay apoptotic cell detection (Roche, Basel, Switzerland) according to the manufacturer's instructions. Percentages of apoptotic cells were counted from at least 500 cells. Cell nuclei were stained with DAPI. The confocal images of cells were sequentially acquired with Zeiss AIM software on a Zeiss LSM 510 confocal microscope system.
Western blotting
Western blots were performed as previously described [10]. The antibodies used were the monoclonal anti-anti-β-actin (1:2000, Beyotime, Jiangsu, China); monoclonal anti-CAPNS1; monoclonal anti-calpain1; monoclonal anti-calpain2 (1:2000, Abcam, Shanghai, China); monoclonal anti-PARP1(1:1000, Cell Signaling Technology, Massachusetts, USA); monoclonal anti-caspase-3 (1:1000, Cell Signaling Technology, Massachusetts, USA) antibodies.
Statistical analysis
Data were expressed as the mean ± SD. The statistical significance of the studies was determined by the parametric unpaired Student's t-test. All statistical analyses were performed with SPSS statistical package, version 17.0. Differences were considered significant when P < 0.05 (2 tailed).
Result
MiRNA screening from cisplatin resistant and sensitive gastric cancer cells
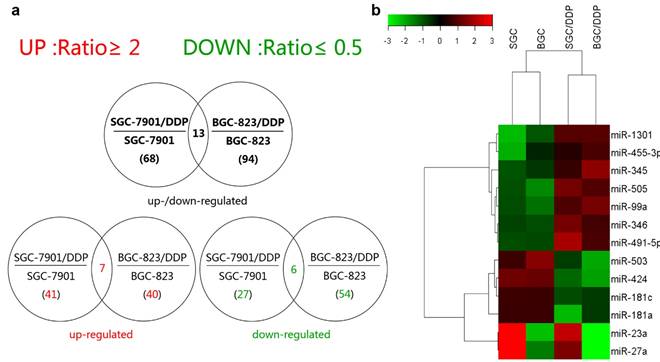
The differential expression profiles in resistant gastric cancer cells and their parental sensitive cells were firstly determined by miRNA microarray analysis. Affymetrix miRNA GeneChip® 2.0 has 15,644 probe sets containing 1,105 human mature miRNAs. Scanning and counting the signal intensity of these probes on the chips of 4 cell lines, a total of 68 miRNAs exhibiting more than 2-fold discrepancy were found in miRNA expression profiling analysis of SGC-7901 and SGC-7901/DDP, including 41 upregulated miRNAs and 27 downregulated ones in SGC-7901/DDP (signal intensity ratio≥2 or ≤0.5) and 94 miRNAs showed 2-fold expression change between BGC-823 and BGC-823/DDP. Among them, 40 miRNAs were upregulated, and 54 downregulated in BGC-823/DDP (Supplementary Tables 1 and 2). Seven miRNAs were simultaneously upregulated whereas six downregulated in both the resistant cells lines (Figure 1a and Figure 1b). The fold change of them detected by microarray was shown in Table 1 (All the detail series matrix files were uploaded to GEO database, GEO accession: GSE86195). Our previously study confirmed that CAPNS1 was downregulated in BGC-823/DDP by 2D-MS and western blot (data was not shown here) [9]. So we predicted many candidate miRNAs which could regulate CAPNS1 by two miRNA databases miRanda (http://www.microrna.org/microrna/home.do) and MicroCosm Targets Version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/). Then we searching for intersection of the prediction list and the co-upregulated miRNAs list, only two miRNAs: miR-99a and miR-491 were found in the intersection. So, we choose them for the further study.
Differential miRNA expressions in both SGC-7901/DDP and BGC-823/DDP cells.
| Probe | Fold-Change (signal ratio≥2or≤0.5) | |
|---|---|---|
| SGC-7901/DDP VS SGC-7901 | BGC-823/DDP VS BGC-823 | |
| hsa-miR-1301 | 6.46 | 3.26 |
| hsa-miR-491-5p | 5.31 | 2.69 |
| hsa-miR-455-3p | 4.27 | 2.14 |
| hsa-miR-505 | 3.87 | 4.39 |
| hsa-miR-346 | 3.47 | 2.67 |
| hsa-miR-99a | 2.89 | 4.82 |
| hsa-miR-345 | 2.58 | 3.84 |
| hsa-miR-23a | 0.48 | 0.43 |
| hsa-miR-503 | 0.44 | 0.13 |
| hsa-miR-181c | 0.39 | 0.45 |
| hsa-miR-27a | 0.34 | 0.4 |
| hsa-miR-424 | 0.23 | 0.16 |
| hsa-miR-181a | 0.19 | 0.46 |
MiRNA expression profile discriminate between cisplatin-resistance and sensitive cells. (a) Venn diagram on total number (in parenthesis) and overlapping number of differentially expressed miRNAs calculated in cell line pairs consisting of the cisplatin-resistant (/DDP added to the paternal cell line's name) relative to the cisplatin-sensitive paternal cell lines (b) Heat map of the 13 intersectional miRNAs deregulated expression in both of resistant cells compared with their parents. Green and red colors indicate relative low and high expression levels across the samples.
MiR-99a and miR-491 were upregulated while CAPNS1 was downregulated in cisplatin-resistant cells
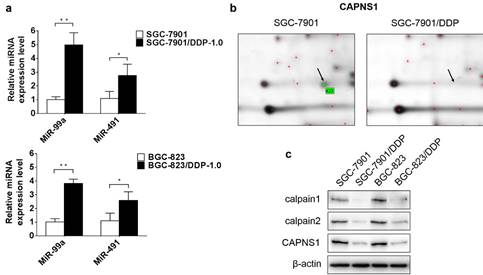
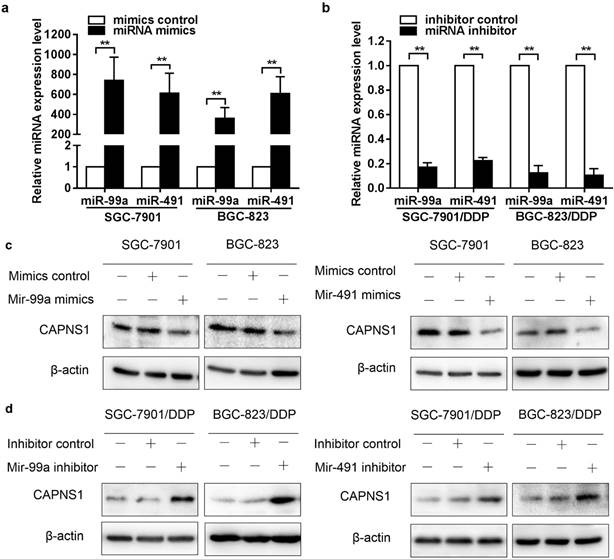
To verify the data obtained from miRNA microarray, miR-99a and miR-491 relative expression levels were detected by qRT-PCR. The expression of miR-99a appreciably higher in cisplatin-resistant cells than in sensitive cells (**P<0.01), and miR-491 were slightly high expression in cisplatin-resistant cells ((*P<0.05) (Figure 2a). In order to find out the target genes of miR-99a and miR-491, different expressed proteins in these two kind cell lines were detected by 2D-MS. The data showed CAPNS1 with the highest expression changes in SGC-7901/DDP compare with SGC-7901 among the proteins analyzed by MS (Figure 2b). Western blot data demonstrated CAPNS1, calpain1 and calpain2 were downregulated dramatically in two resistant cell lines (Figure 2c). Meanwhile, CAPNS1 was a potential target gene of miR-99a and miR-491 which was predicted by miRNA database websites: MicroCosm Targets Version 5 and MiRanda.
The CAPNS1 3'-UTR is a target for both miR-99a and miR-491
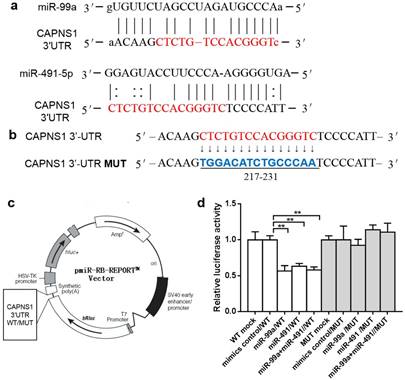
MiRanda predicted both miR-99a and miR-491 matched to the sequence of CAPNS1 mRNA 3'-UTR from 212-239 (Figure 3a). There are 15nt shared by miR-99a and miR-491. We further designed mutated target sequence and constructed the wild type (WT) and mutation type (MUT) report gene plasmid vector (Figure 3b-c), and co-transfecting with miRNA mimics or control into HEK-293 cell lines. The data showed both the mimics and inhibitors of miR-99a or miR-491 worked well and with a significant difference between them in relative luciferase activity (**P<0.01, Figure 3d). However, these mimics and inhibitors did not work on the mutated target.
MiR-99a and miR-491 negatively regulated CAPNS1 expression
As it was shown in figure 2a, miR-99a and miR-491 were low expressed in SGC-7901 and BGC-823 while high expressed in SGC-7901/DDP and BGC-823/DDP. Here, miR-99a and miR-491 mimics and inhibitors were transfected into cisplatin sensitive and resistant cells, respectively. The data of qRT-PCR indicated that transfection of miR-99a and miR-491 mimics resulted in an extremely high expression in two sensitive cells (**P<0.01, Figure 4a). Similarly, transfection of miR-99a and miR-491 inhibitors reduced miRNAs expression in resistant cells (**P<0.01, Figure 4b). CAPNS1 expression was downregulated by miR-99a or miR-491 mimics in sensitive cells (Figure 4c), and was upregulated by miR-99a or miR-491 inhibitor in resistant cells (Figure 4d).
MiR-99a and miR-491 upregulate in SGC-7901/DDP and BGC-823/DDP while CAPNS1, its catalytic subunits calpain1 and calpain2 all downregulate dramatically. (a) Relative expression level of miR-99a and miR-491 in sensitive cells and resistant cells detected by Real-time PCR (n=3, bar, mean ± SD., *P<0.05, **P<0.01) (b) Cropped 2D gel images of selected protein CAPNS1 in SGC-7901 and SGC-7901/DDP, CAPNS1 was detected by mass spectrometry. (c) Expression of CAPNS1, calpain1 and calpain2 were all lower visibly in resistant cells than sensitive cells as detected by western blot analysis.
MiR-99a and miR-491 target with CAPNS1 3'-UTR and regulated CAPNS1 mRNA. (a) Comparison of nucleotides between the miR-99a and miR-491 total sequence and the target CAPNS1 3'-UTR sites. The link lines link two paired nucleotides. The red ones are the share region miR-99a and miR-491 combined with. (b) The wild type CAPNS1 3'-UTR and the mutational 3'-UTR inserted into Dual Luciferase Reporter plasmid. (c) The plasmid profiles of Dual Luciferase Reporter included CAPNS1 3'-UTR WT/MUT fragments. (d) Luciferase activity of HEK-293 co-transfected with either mimics control or miRNA mimics and Dual Luciferase Reporter plasmids detected by luciferase assay. In miR-99a or miR-491 alone or simultaneously combined with wild type plasmid transfection leaded to luciferase activities remarkable decrease, but not occurred in mutational groups. (n=3, bar, mean ± SD, **P<0.01)
MiR-99a and miR-491 inversely regulates CAPNS1 expression in resistant cells and sensitive cells. (a, b) Relative expression level of miR-99a and miR-491 in sensitive cells transfected in control or mimics and resistant cells transfected in control or inhibitors detected by Real-time PCR. (n=3, bar, mean ± SD, **P<0.01) (c) Expression level of CAPNS1 was down regulated by transfected miR-99a (left column) or miR-491(right column) mimics in sensitive cells as detected by western blot analysis. (d) Expression level of CAPNS1 was up regulated by transfected miR-99a (left column) or miR-491 (right column) inhibitors in resistant cells as detected by western blot analysis.
MiR-99a, miR-491 and CAPNS1 regulate cisplatin resistance in human gastric cancer cells
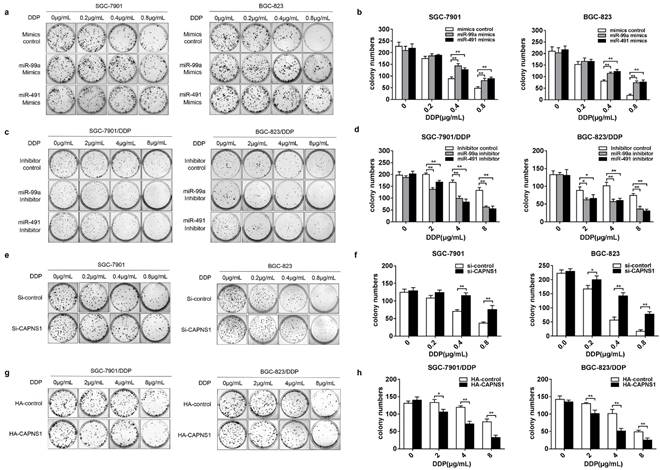
To evaluate whether miR-99a and miR-491 affected cisplatin induced DNA damage and cell death in both sensitive and resistant cells, we performed plate colony formation assay. Transfection of miR-99a or miR-491 mimics or their control into SGC-7901 and BGC-823, respectively, and followed by treatment of the cells with different concentrations of cisplatin medium for 2 h. As a result, more clonogenic units were produced in mimics groups (Figure 5a). Counting down every colony in the plates and compared to control groups, the data showed significance between two groups under 0.4 or 0.8μg/mL cisplatin treatment (Figure 5b, **P<0.01). The similar effects were observed in SGC-7901 and BGC-823 cells when knocked down CAPNS1 (Figure 5e, f, **P<0.01). However, when transfection of miR-99a or miR-491 inhibitor into SGC-7901/DDP and BGC-823/DDP cells, less colony formation than the inhibitor control groups were observed (Figure 5c). Statistical analysis showed that colony numbers were significance between normal saline and cisplatin treatment (2 or 4 or 8μg/mL) (Figure 5d, **P<0.01). Likewise, colony numbers were significantly decreased in resistant cells with high expressed CAPNS1 (Figure 5g, h, **P<0.01). The results demonstrated that miR-99a and miR-491 promoted cisplatin induced colony formation in gastric cells though inhibiting expression of CAPNS1.
MiR-99a and miR-491 regulating their target CAPNS1 and the number of colony induced in cisplatin. (a, c) Plate colony formation assay about four cells under cisplatin treatment. a, SGC-7901 and BGC-823 transfected mimics formed more clonogenic units than which groups transfected mimics control under treated by cisplatin 0.4, 0.8μg/mL for 2 hours. c, SGC-7901/DDP and BGC-823/DDP transfected inhibitors produced less clonogenic units than which groups transfected inhibitor control when them was been treated by cisplatin 4,8μg/mL for 2 hours. (b, d) The statistics data about colony numbers in each group counted two weeks later after the cells was planted in six-well plates. The colony is defined to consist of at least 50 cells. b, SGC-7901, left column; BGC-823, right column. d, SGC-7901/DDP, left column; BGC-823/DDP, right column. bar, mean ± SD, **P<0.01. (e) Transfecting Si-CAPNS1 into SGC-7901 and BGC-823, colony numbers less than transfected Si-control under 0.2(only BGC-823), 0.4, 0.8μg/mL cisplatin treatment for 2 hours. (g) Transfecting HA-CAPNS1 into SGC-7901/DDP and BGC-823/DDP, colony numbers more than transfected HA-control under 2, 4, 8μg/mL cisplatin treatment for 2 hours. (f, h) The statistics of counting colony numbers two weeks later after planting. The colony is defined to consist of at least 50 cells. f, SGC-7901, left column; BGC-823, right column. h, SGC-7901/DDP, left column; BGC-823/DDP, right column. bar, mean ± SD., **P<0.01.
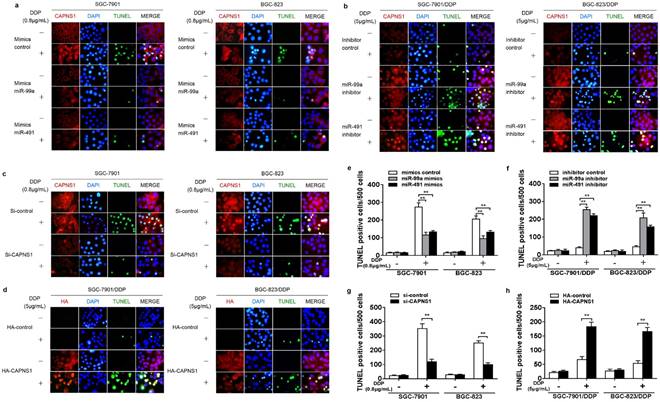
TUNEL assay was used to determine cisplatin-induced apoptosis in cisplatin sensitive and resistant cells. Data showed that both SGC-7901 and BGC-823 cells transfected with miR-99a or miR-491 indicated obviously less apoptotic cells than the cells transfected mimics control when exposed to IC50 of cisplatin (0.8μg/mL) for 24 h (Figure 6a). In contrast, both SGC-7901/DDP and BGC-823/DDP cells transfected with miR-99a and miR-491 showed much more apoptotic cells than their control when exposed to the resistant IC50 of cisplatin for 24 h (Figure 6b). Suggesting that miR-99a and miR-491 regulated cisplatin-triggered apoptosis in both cisplatin sensitive and resistant gastric cancer cells. At the same time, CAPNS1 expression in sensitive cells was inhibited by miR-99a and miR-491 mimics. And, its expression in resistant cells was raised by two miRNA inhibiters. Similarly, knocking down CAPNS1 in cisplatin sensitive cells directly turned them more resistance to cisplatin treatment and high expressed CAPNS1 in resistant cells turned them more sensitive to cisplatin treatment (Figure 6c, d). Consequently, either inhibition of miR-99a and miR-491 or over expression of CAPNS1 promoted apoptosis in resistant cells (Figure 6e-h, **P<0.01). These data indicated that the sensitivity of gastric cancer cells to cisplatin was driven by miR-99a and miR-491 and their targeted CAPNS1expression.
The mechanism of miR-99a, miR-491 and CAPNS1 regulating resistance to cisplatin in human gastric cancer
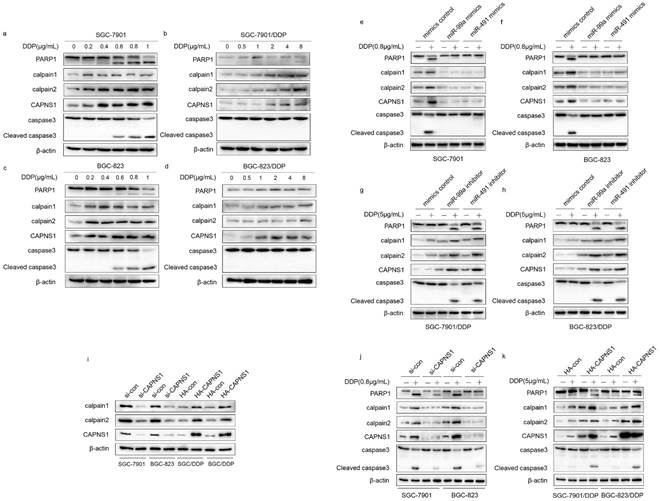
CAPNS1 is a regulatory subunit of calpain1 and calpain2 which are the catalytic subunits. When SGC-7901 and BGC-823 incubated with different concentrations of cisplatin for 24 h, CAPNS1, calpain1 and calpain2 expression levels were increased under very low concentration (0.2μg/mL DDP). Caspase3 and PARP1 were cleaved by high expressed calpain1 and calpain2 (Figure 7a, c). However, in cisplatin resistant cells, the expressions of CAPNS1, calpain1 and calpain2 were not increased until they were exposed under 8μg/mL or higher DDP (Figure 7b, d). As proved above, miR-99a and miR-491 directly negatively regulated CAPNS1. Here, cells transfected mimics or inhibitor of two miRNAs revealed the mechanism of apoptosis inhibition. In sensitive cells, mimics transfection made them down regulate CAPNS1, calpain1 and calpain2, less cleave Caspase3 and PARP1, so them got the ability to resistant apoptosis (Figure 7e, f). On the contrary, resistant cells transfected inhibitors cleave more Caspase3 and PARP1 by upregulation of CAPNS1, calpain1 and calpain2. (Figure 7g, h), which made them accumulate cisplatin induced apoptosis.
Actually, CAPNS1 positively regulated calpain1 and 2 in both sensitive and resistant cells (Figure 7i). In addition, with or without cisplatin, transfection of si-CAPNS1 to SGC-7901 and BGC-823 significantly inhibited expression of calpain1 and calpain2 and resulted in down regulation of cleaved caspase3 and PARP1 (Figure 7j). In resistant cells, high expressed CAPNS1 increased expression of calpain1 and calpain2, which induced to generate more cleavage of caspase3 and PARP1 (Figure 7k). Therefore, CAPNS1 is the key molecular regulated cisplatin resistance in gastric cancer cells, on one hand, it was negatively regulated by miR-99a and miR-491, on the other hand, it positively regulated its catalytic subunits calpain1 and calpain2. These strong evidences indicated the conclusion that gastric cancer cells resistance to cisplatin was regulated via miR-99a/miR-491-CAPNS1-calpain1/calpain2-caspase3-PARP1 pathway.
MiR-99a and miR-491 regulate cisplatin-induced apoptosis though CAPNS1 in both the sensitive and the resistant cells. (a-d) Immunofluorescence assay and TUNEL assay about SGC-7901, BGC-823, SGC-7901/DDP, BGC-823/DDP transfected different fragment or plasmid under 24 hours IC50 cisplatin treatment. Red, CAPNS1 in a, b and c; HA, in d, green, TUNEL, blue, DAPI. (a) SGC-7901 and BGC-823 transfected miR-99a or miR-491 mimics got less TUNEL positive cells after their IC50 of cisplatin treatment (0.8μg/mL) than their compared control. (b) SGC-7901/DDP and BGC-823/DDP transfected miR-99a or miR-491 inhibitors got more TUNEL positive cells after their IC50 of cisplatin treatment (5μg/mL) than their compared control. (c) SGC-7901 and BGC-823 knocked down CAPNS1 decreased the number of TUNEL positive cells after IC50 cisplatin treatment. (d) SGC-7901/DDP and BGC-823/DDP high expressed CAPNS1 increased the number of TUNEL positive cells after their IC50 cisplatin treatment. (e-h) TUNEL positive cells statistics per 500 cells of every groups in every kind of cells. Column, the number of TUNEL positive cells per 500 cell counted randomly. bars, mean ± SD (n=3, **P<0.01).
miR-99a, miR-491and CAPNS1 regulates gastric cancer cells apoptosis though miR-99a/miR-491-CAPNS1-calpain1/calpain2-caspase3-PARP1 pathway. (a, c) SGC-7901 and BGC-823 incubated with 0, 0.2, 0.4, 0.6, 0.8, 1μg/mL cisplatin 24 hours. (b, d) SGC-7901/DDP and BGC-823/DDP incubated with 0, 0.5, 1, 2, 4, 8μg/mL cisplatin 24 hours. (e, f) SGC-7901 and BGC-823 transfected miR-99a or miR-491 mimics 48 hours was unexposed (-) or exposed (+) with 0.8μg/mL cisplatin 24 hours and (g, h) SGC-7901/DDP and BGC-823/DDP transfected miR-99a or miR-491 inhibitors 48 hours was unexposed (-) or exposed (+) with 5μg/mL cisplatin 24 hours. (i) SGC-7901 and BGC-823 knocked down CAPNS1 by siRNA and SGC-7901/DDP and BGC-823/DDP high expressed CAPNS1 by plasmid transfection after 48hours. (j) SGC-7901 and BGC-823 transfected with si-control or si-CAPNS1 48 hours was unexposed (-) or exposed (+) with 0.8μg/mL cisplatin 24 hours. (k) SGC-7901/DDP and BGC-823/DDP transfected with HA-control or HA-CAPNS1 48 hours was unexposed (-) or exposed (+) with 5μg/mL cisplatin 24 hours. All above treatment followed by protein extraction detected CAPNS1, calpain1, calpain2, full length and cleaved caspase3, PARP1 levels by western blot CAPNS1, calpain1, calpain2, caspase3 and PARP1 were analyzed by western blot. β-actin was monitored to ensure equal loading of lanes in all protein levels detections. n=3.
Discussion
Cisplatin resistance in gastric cancer is a great challenge in clinic. However, the detailed molecular mechanisms are still undetermined. As so far, multiple mechanisms for intrinsic or acquired resistance to cisplatin and several related genes had been reported [3, 11, 12]. Among them, previous researches have shown that the suppression of cell death pathway is a crucial mechanism to escape drug induced apoptosis and stay alive in the resistant cells.
MiRNAs have been shown to regulate many biological processes including apoptosis. It has extensively reported that miRNAs regulate drug resistance in human cancers [13]. MiR-503 targeting BCL-2 [14], miR-19a/b targeting PTEN [15], miR-362 targeting NF-κB [16] were identified to regulate resistance in gastric cancer. The connection between apoptosis in resistant cells and miRNAs targeting the genes' mRNA have been revealed, and microRNAs regulate multiple apoptosis pathways in cancer [17]. Both miR-99a and miR-491 may play different roles in various cancers as they regulated different target genes. The miR-99a has been reported to regulate resistance in breast cancer [18], leukemia [19], neck squamous cell carcinoma [20], germ cell tumor [21], pancreatic ductal adenocarcinoma [22], cholangiocarcinoma[23], etc. The relation between miR-491 and liver cancer [24], colorectal cancer [25], ovarian carcinoma [26], pancreatic cancer [27], oral squamous cell carcinoma [28], non-small cell lung cancer [29], etc has also been verified. Recently, miR-99a was reported to regulate vincristine resistance in childhood acute lymphoblastic leukemia [30, 31]. MiR-491-3p was reported modulated chemo- sensitivity in human tongue cancer [32].
In the present study, we found for the first time that miR-99a and miR-491combined with same target in 3'-UTR of CAPNS1. We also confirmed both miR-99a and miR-491 were high expressed in cisplatin resistant gastric cancer cells compared with their parental cells. This situation could maintain low expression of CAPNS1. When the mimics of the both miRNA were transfected into sensitive cells, the resistance of the cells to cisplatin was enhanced significantly. In line with this, transfection of inhibitors of the both miRNA effectively reversed their sensitivity to cisplatin. Interestingly, miR-99a and miR-491 share the same region combined to CAPNS1 3'-UTR. So, we detected the CAPNS1 expression when transfected miR-99a and miR-491 synchronously, but CAPNS1 did not shown significant fold change compared to transfected singly groups (data was not shown here). We speculated that miR-99a and miR-491 did not have synergetic or competitive function to CAPNS1.
CAPNS1, calpain1 and calpain2 had a different basal level between sensitive and resistant cells. CAPNS1 expressed low with high fold-change value in acquired cisplatin resistant cells. The data shown that calpain family members could not be induced by low concentration of DDP in resistant cells but CAPNS1, calpain1 and calpain2 could be induced by low concentration of DDP and participant apoptosis regulation pathway in their parental types. CAPNS1 level in resistant cells is definitely lower than sensitive cells and stably maintained a low expression level under the exposure of IC50 cisplatin (5μg/mL). These differences represented that CAPNS1 was not only a biomarker of resistance but also maintained this acquired characteristic. Therefore, we concluded that CAPNS1 may play an important role in maintenance and development of cellular resistant characteristics to cisplatin in gastric cancer cells. CAPNS1, as a regulatory subunit, positively regulated expression of calpain1 and calpain2[8]. Three of them belongs to calpain family, with function in proliferation, apoptosis [33], migration [34], differentiation [35], and autophagy [36]. CAPNS1 is less reported in cisplatin resistance in gastric cancer study, but its catalytic subunits calpains has been widely reported as key molecules in several apoptosis pathways [37-39]. Del Bello B et al concluded the “crosstalk” between calpain and caspase3/7 in cisplatin-induced apoptosis of melanoma cells [40]. CAPNS1 also involves in many tumor related signal pathways. CAPNS1 mediates apoptosis through PP2A in AKT/FOX3A pathway [39]. And it is also found that CAPNS1 overexpression in hepatocellular carcinoma promoted invasion and metastasis [41]. In addition, Kulkarni S et al found that CAPNS1 promotes HER2 phosphorylation and leads to resistance of breast cancer cells to Trastuzumab [42]. The calpains are a family of cysteine proteases which catalyze a large number of specific substrates with difference in ubiquitin systems [43]. Calpain proteolyzes these substrates in a limited manner rather than digesting them to small peptides, indicating its modulatory functions for the substrate proteins by cutting their interdomain regions [44]. Caspase3 is one of direct substrates cleaved by calpain, this was confirmed in our results and others [40]. In our study, we found that CAPNS1 could affect the expression of calpain1 and calpain2, which suggested that CAPNS1 was a key regulatory subunit which drove catalytic reaction mediated by calpain1 and calpain2. The levels of CAPNS1, calpain1 and calpain2 were synchronous. As a kind of cysteine proteinase, catalytic reaction mediated by calpain need consume intracellular ATP, therefore CAPNS1 becomes the important monitor to decide whether synthesize more catalytic subunits. However, the mechanism of this feedback synthesis is still unknown. We speculated it had some relationship with Ca2+-mediated pathways.
In conclusion, the expressions of CAPNS1, calpain1 and calpain2 were inhibited by increased miR-99a and miR-491 expression in cisplatin resistant gastric cancer cells and the inhibition resulted in resistance to cisplatin induced apoptosis. Both miR-99a and miR-491 might be a potential therapeutic biomarker to reverse cisplatin resistance in gastric cancer. Its clinical significance needs to be further investigated.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported in part by the Foundation of State Key Laboratory of Reproductive Medicine, Nanjing Medical University; the National Natural Science Foundation (grant numbers: 81161120537, 30930080 and 81370078); the Zhejiang provincial natural science foundation of China (LQ15H160004); the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions; and the Natural Science Foundation of Jiangsu Higher Education Institutions (13KJA330001). We are very grateful to Professor Yong Ji (PhD, Dean, School of Pharmacy, Nanjing Medical University) for the English Writing revision.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90
2. Florea A-M, Büsselberg D. Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers. 2011;3:1351-71
3. Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O. et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869-83
4. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376-85
5. Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer gene therapy. 2010;17:523-31
6. Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298-311
7. Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nature reviews Cancer. 2011;11:364-74
8. Sorimachi H, Ishiura S, Suzuki K. Structure and physiological function of calpains. Biochem J. 1997;328( Pt 3):721-32
9. Xu W, Wang S, Chen Q, Zhang Y, Ni P, Wu X. et al. TXNL1-XRCC1 pathway regulates cisplatin-induced cell death and contributes to resistance in human gastric cancer. Cell death & disease. 2014;5:e1055
10. Zhou J, Ye J, Zhao X, Li A, Zhou J. JWA is required for arsenic trioxide induced apoptosis in HeLa and MCF-7 cells via reactive oxygen species and mitochondria linked signal pathway. Toxicology and applied pharmacology. 2008;230:33-40
11. Kang HC, Kim I-J, Park J-H, Shin Y, Ku J-L, Jung MS. et al. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clinical Cancer Research. 2004;10:272-84
12. Kelland L. The resurgence of platinum-based cancer chemotherapy. Nature reviews Cancer. 2007;7:573-84
13. Hong L, Yang Z, Ma J, Fan D. Function of miRNA in controlling drug resistance of human cancers. Current drug targets. 2013;14:1118-27
14. Shu Y. miR-503 modulates drug resistance of human gastric cancer cell lines by targeting BCL2. Translational Gastrointestinal Cancer. 2013;2(S1):AB50
15. Wang F, Li T, Zhang B, Li H, Wu Q, Yang L. et al. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochemical and biophysical research communications. 2013;434:688-94
16. Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL. et al. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-kappaB signaling. Journal of translational medicine. 2014;12:33
17. Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47:163-74
18. Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu C-G. et al. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR 1/P-glycoprotein expression in human cancer cells. Biochemical pharmacology. 2008;76:582-8
19. Bai H, Xu R, Cao Z, Wei D, Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011;585:402-8
20. Dai Y, Xie Ch, Neis JP, Fan CY, Vural E, Spring PM. MicroRNA expression profiles of head and neck squamous cell carcinoma with docetaxel-induced multidrug resistance. Head & neck. 2011;33:786-91
21. Port M, Glaesener S, Ruf C, Riecke A, Bokemeyer C, Meineke V. et al. Micro-RNA expression in cisplatin resistant germ cell tumor cell lines. Mol Cancer. 2011;10:52
22. Dhayat SA, Mardin WA, Seggewiss J, Strose AJ, Matuszcak C, Hummel R. et al. MicroRNA Profiling Implies New Markers of Gemcitabine Chemoresistance in Mutant p53 Pancreatic Ductal Adenocarcinoma. PLoS One. 2015;10:e0143755
23. Lin KY, Ye H, Han BW, Wang WT, Wei PP, He B. et al. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene. 2015
24. Yoon S, Kim TH, Natarajan A, Wang SS, Choi J, Wu J. et al. Acute liver injury upregulates microRNA-491-5p in mice, and its overexpression sensitizes Hep G2 cells for tumour necrosis factor-α-induced apoptosis. Liver International. 2010;30:376-87
25. Nakano H, Miyazawa T, Kinoshita K, Yamada Y, Yoshida T. Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-XL in colorectal cancer cells. International journal of cancer. 2010;127:1072-80
26. Denoyelle C, Lambert B, Meryet-Figuiere M, Vigneron N, Brotin E, Lecerf C. et al. miR-491-5p-induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL-XL and EGFR leading to BIM activation. Cell death & disease. 2014;5:e1445
27. Guo R, Wang Y, Shi WY, Liu B, Hou SQ, Liu L. MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules. 2012;17:14733-47
28. Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC. et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751-64
29. Gong F, Ren P, Zhang Y, Jiang J, Zhang H. MicroRNAs-491-5p suppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res. 2016;8:485-95
30. Akbari Moqadam F, Lange-Turenhout EA, Aries IM, Pieters R, den Boer ML. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk Res. 2013;37:1315-21
31. Kandi R, Gutti U, Saladi RG, Gutti RK. MiR-125b and miR-99a encoded on chromosome 21 co-regulate vincristine resistance in childhood acute megakaryoblastic leukemia. Hematol Oncol Stem Cell Ther. 2015;8:95-7
32. Zheng G, Jia X, Peng C, Deng Y, Yin J, Zhang Z. et al. The miR-491-3p/mTORC2/FOXO1 regulatory loop modulates chemo-sensitivity in human tongue cancer. Oncotarget. 2015;6:6931-43
33. Gafni J, Cong X, Chen SF, Gibson BW, Ellerby LM. Calpain-1 cleaves and activates caspase-7. The Journal of biological chemistry. 2009;284:25441-9
34. Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. Journal of cell science. 2005;118:3829-38
35. Anderton RS, Meloni BP, Mastaglia FL, Greene WK, Boulos S. Survival of motor neuron protein over-expression prevents calpain-mediated cleavage and activation of procaspase-3 in differentiated human SH-SY5Y cells. Neuroscience. 2011;181:226-33
36. Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C. et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell death & disease. 2011;2:e144
37. Tan Y, Wu C, De Veyra T, Greer PA. Ubiquitous calpains promote both apoptosis and survival signals in response to different cell death stimuli. The Journal of biological chemistry. 2006;281:17689-98
38. Sobhan PK, Seervi M, Deb L, Varghese S, Soman A, Joseph J. et al. Calpain and reactive oxygen species targets Bax for mitochondrial permeabilisation and caspase activation in zerumbone induced apoptosis. PLoS One. 2013;8:e59350
39. Bertoli C, Copetti T, Lam EW, Demarchi F, Schneider C. Calpain small-1 modulates Akt/FoxO3A signaling and apoptosis through PP2A. Oncogene. 2009;28:721-33
40. Del Bello B, Moretti D, Gamberucci A, Maellaro E. Cross-talk between calpain and caspase-3/-7 in cisplatin-induced apoptosis of melanoma cells: a major role of calpain inhibition in cell death protection and p53 status. Oncogene. 2007;26:2717-26
41. Bai DS, Dai Z, Zhou J, Liu YK, Qiu SJ, Tan CJ. et al. Capn4 overexpression underlies tumor invasion and metastasis after liver transplantation for hepatocellular carcinoma. Hepatology. 2009;49:460-70
42. Kulkarni S, Saju L, Farver C, Tubbs R. Calpain4 is required for activation of HER2 in breast cancer cells exposed to trastuzumab and its suppression decreases survival and enhances response. International Journal of Cancer. 2012;131:2420-32
43. Ono Y, Sorimachi H. Calpains: an elaborate proteolytic system. Biochimica et biophysica acta. 2012;1824:224-36
44. Saido TC, Sorimachi H, Suzuki K. Calpain: new perspectives in molecular diversity and physiological-pathological involvement. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1994;8:814-22
Author contact
![]() Corresponding author: Professor Shan Xu: xushanedu.cn.
Corresponding author: Professor Shan Xu: xushanedu.cn.

 Global reach, higher impact
Global reach, higher impact