10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(1):85-99. doi:10.7150/ijbs.17390 This issue Cite
Research Paper
Metadata Analysis of Phanerochaete chrysosporium Gene Expression Data Identified Common CAZymes Encoding Gene Expression Profiles Involved in Cellulose and Hemicellulose Degradation
Department of Biology, Lakehead University, 955 Oliver Road, Thunder Bay, Ontario, P7B 5E1, Canada.
Received 2016-8-29; Accepted 2016-10-10; Published 2017-1-1
Abstract
In literature, extensive studies have been conducted on popular wood degrading white rot fungus, Phanerochaete chrysosporium about its lignin degrading mechanisms compared to the cellulose and hemicellulose degrading abilities. This study delineates cellulose and hemicellulose degrading mechanisms through large scale metadata analysis of P. chrysosporium gene expression data (retrieved from NCBI GEO) to understand the common expression patterns of differentially expressed genes when cultured on different growth substrates. Genes encoding glycoside hydrolase classes commonly expressed during breakdown of cellulose such as GH-5,6,7,9,44,45,48 and hemicellulose are GH-2,8,10,11,26,30,43,47 were found to be highly expressed among varied growth conditions including simple customized and complex natural plant biomass growth mediums. Genes encoding carbohydrate esterase class enzymes CE (1,4,8,9,15,16) polysaccharide lyase class enzymes PL-8 and PL-14, and glycosyl transferases classes GT (1,2,4,8,15,20,35,39,48) were differentially expressed in natural plant biomass growth mediums. Based on these results, P. chrysosporium, on natural plant biomass substrates was found to express lignin and hemicellulose degrading enzymes more than cellulolytic enzymes except GH-61 (LPMO) class enzymes, in early stages. It was observed that the fate of P. chrysosporium transcriptome is significantly affected by the wood substrate provided. We believe, the gene expression findings in this study plays crucial role in developing genetically efficient microbe with effective cellulose and hemicellulose degradation abilities.
Keywords: Phanerochaete chrysosporium, Transcriptome, Lignocellulose, Gene Expression Omnibus (GEO), GEO2R, Bioconductor, Carbohydrate Active Enzyme database (CAZy).
Introduction
Recently, production of second generation fuel substrate (lignocellulose) gained much attention as the first generation fuel substrate (corn starch) faces the food vs fuel challenge [1]. Abundant availability of lignocellulosic biomass and its potential for fuel production, encourage the research interest in biorefining scientists to substitute renewable substrates for fossil based products [1]. However, major bottlenecks include the highly recalcitrant nature of lignocellulosics due to which various steps such as thermochemical pretreatment followed by enzymatic hydrolysis and fermentation in upstream processing are required [2-4]. These tedious process increases the costs involved in the production of fuels [2-4]. Though chemical pretreatment methods of polysaccharide degradation do exist, but world-wide biorefining strategies are enzyme based [5]. Usually the pretreated biomass is depolymerized by supplementing the enzyme cocktail which breakdowns the polysaccharides to pentoses and hexoses. Enzyme cocktails produced from the Trichoderma reesei are highly used for the degradation of polysaccharides [6].
Structurally plant cell wall components can be majorly divided into polysaccharide units and polyphenolic lignin units. Plant polysaccharide units are divided into cellulose, hemicellulosic polysaccharides (xyloglucans, xylans, glucomannans and mixed linkage glucans) [7] and pectic polysaccharides (homogalacturonan and rhamnogalacturonan I, II) [8, 9]. Cellulose is the characteristic chemical constituent in all the plant cell walls. Structurally, cellulose is comprised of glucose molecules with β-1,4 glycosidic linkages [9, 10]. These cellulose chains are bound by hydrogen bonds and van der Waals interactions resulting in microfibrils which are crystalline in nature and poses difficulty in enzymatic saccharification [11]. Naturally cellulose exists in crystalline (well-ordered) and amorphous (disordered) forms [11]. Hemicelluloses are hetero polysaccharides including xylans, glucans, mannans and glucomannans with (β-1,4) and (β-1,3) glycosidic linkages. Enzymatic breakdown of hemicellulose was considered easy and simple when compared to cellulose. However, due to the presence of certain recalcitrant oligomeric structures and its complex branching and acetylation patterns make the depolymerization difficult [12]. The enzymatic breakdown of cellulose results only in glucose whereas hemicellulose degradation results in a mixture of different sugars which significantly contain pentoses which are difficult to ferment further [13, 14].
Fermentation of cellulose and hemicellulose monomeric units of plant biomass using microorganisms for the production of bioethanol has gained significance in the recent years. Among different microorganism's, fungi are the efficient plant biomass degraders, especially Basidiomycota phylum. Based on their wood decaying properties, Basidiomycetes fungi are classified into white, brown and soft rot. Phanerochaete chrysosporium, a white rot fungi, can efficiently degrade all the components of plant biomass cellulose, hemicellulose and particularly lignin [15]. Complex structure of lignin enables its degradation by specific microorganisms, thus several studies conducted on P. chrysosporium were based on lignin degradation while few studies were based on cellulose and hemicellulose degradation mechanisms. Although, whole genome sequence of P. chrysosporium showed that it harbors around 240 carbohydrate active enzymes (CAZymes) [15]. Secretome and computational analysis performed by Wymelenberg et al. (2006) has revealed eighteen putative peptide sequences which are allotted to eight specific glycosyl hydrolases [16]. Glycosyl hydrolases were found to be associated with hemicellulose and pectin degradation, putative xylanase and exo glucanase encoding genes xyn10D, exg55A were detected in ground wood submerged cultures [17, 18]. Three unidentified peptides encoding for glycosyl hydrolases were noticed in carbon limited growth conditions [18]. Availability of annotated whole genome sequence of P. chrysosporium have led to the current understanding of its degradative patterns. Based on the PubMed results, 12 large scale gene expression studies were conducted on P. chrysosporium, out of which 10 microarray studies, one RNA-Seq and one Long SAGE studies. These studies have revealed various significant facts about the genes and enzymes involved in cellulose, hemicellulose and lignin degradation mechanisms.
Development of online based genome and enzyme databases like CAZy (Carbohydrate Active Enzyme database) [19], FOLy (Fungal Lignin Oxidizing Enzymes) [20], DOE-JGI (Joint Genome Institute) [21] and FungiDB [22] etc., has significantly influenced the present understanding of fungal genomic and proteomic studies. The CAZy database was developed by Vincent et al. (2014), to consolidate various enzymes that are involved in synthesis and breakdown of carbohydrates and other glycoconjugates [19]. CAZy database was divided into five major classes and distributed into 135 glycoside hydrolases, 99 glycosyl transferases, 24 polysaccharide lyases, 16 carbohydrate esterases and 13 auxiliary activities, it also consists of carbohydrate binding module (CBM) [19]. Levasseur, A et al. (2008) has developed FOLy based on the CAZy database structure and is dependent on family based management of sequence information and their respective accession from different public repositories and its descriptions [20]. FOLy database is divided into two major sections as LO's (Lignin Oxidizing enzymes) and LDA's (Lignin Degrading Auxiliary enzymes) [20].
The availability of whole genome sequences and recently performed genome wide transcriptome studies on P. chrysosporium have inspired us for the current metadata analysis. In the last decade, several studies were conducted to understand the molecular mechanisms underlying lignocellulose degradation by P. chrysosporium. Although, these gene expression studies have explained about the expression of several cellulolytic and hemicellulolytic enzymes, understanding the common significant genes expressed under varied growth conditions will play a crucial role in biofuel production. In our current study, we have rigorously analyzed the whole transcriptome metadata retrieved from NCBI GEO using GEO2R and Bioconductor packages. To the best of our knowledge this is the first metadata analysis report on P. chrysosporium to understand the common gene expression patterns employed for cellulose and hemicellulose degradation.
Data Analysis Methodology
Data collection: Microarray datasets were retrieved from Gene Expression Omnibus NCBI-GEO (https://www.ncbi.nlm.nih.gov/geo/), a public repository for gene expression datasets. All the microarray datasets collected were based on white rot fungus P. chrysosporium cultured on different growth substrates. Till date there are 6 Microarray, 1 RNA sequencing and 1 Long-SAGE (Serial Analysis for Gene Expression) studies based on P. chrysosporium, out of which we have analyzed the microarray and RNA sequencing studies to understand the cellulolytic and hemicellulolytic degradation mechanisms. Accession IDs of microarray datasets retrieved from NCBI GEO are GSE14734 [23, 24], GSE14735 [23, 24] GSE54542 [25], GSE27941[26], GSE52922 [27], GSE69008 [28] GSE69461[29] details of these gene expression datasets were shown in the Table 1.
Details of the P. chrysosporium transcriptome metadata retrieved from NCBI GEO and NCBI SRA
| GEO-ID's | Platform and Technology | Substrate | # of Samples | References |
|---|---|---|---|---|
| GSE54542 | NimbleGen Phanerochaete chrysosporium arrays | Oak acetonic extractives | 6 | [25] |
| GSE27941 | NimbleGen Phanerochaete chrysosporium arrays | Ball milled aspen, Ball milled pine | 6 | [26] |
| GSE52922 | NimbleGen Phanerochaete chrysosporium arrays | P717 hybrid line, Transgenic line 82 Transgenic line 64 | 9 | [27] |
| GSE14734 | NimbleGen Phanerochaete chrysosporium arrays | Cellulose, Glucose, Ball milled aspen | 9 | [23, 24] |
| GSE14735 | NimbleGen Phanerochaete chrysosporium arrays | Replete medium, Carbon limited Nitrogen limited | 9 | [23, 24] |
| GSE69008 | NimbleGen Phanerochaete chrysosporium arrays | Poplar wood substrates | 24 | [28] |
| GSE69461 | Illumina HiSeq 2000 | Picea glauca (spruce sapwood) | 18 | [29] |
Data Analysis: The microarray datasets were analyzed using GEO2R an interactive online tool (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html) and using Bioconductor packages GEOquery and limma based on R software version 3.2.2 and. Following settings were used for analyzing the microarray datasets using GEO2R data analysis tool a) auto detect option used for the log transformation of the data, b) samples and value distribution were obtained using the box-whisker plot c) submitter provided annotations were used for the current analysis. The samples were grouped based on their experimental conditions and further differentially expressed genes were obtained using “Top250” function which internally uses limma (linear models for microarray data). The top differentially expressed genes are obtained after performing multiple testing correction using Benjamini and Hochberg false discovery rate (FDR) method with a p-value 0.05. The supplier provided annotations for the microarray platform were used for analyzing the obtained differentially expressed genes. Gene annotations mainly InterPro Hits, protein ID and genome position were retrieved for the current analysis. The profile graph for the individual genes were obtained from the GEO2R tool using the profile graph option, the profile graphs obtained were reported in Supplementary Material (Figure S1). The P. chrysosporium RP-78 v2.2 genome annotations were obtained from the MycoCosm (fungal genome repository) [30, 31]. Different analysis tool options such as Gene Ontology (GO), EuKaryotic Orthologous Groups (KOG) and CAZy were used for understanding and biological contextualization of the results. The expression values of differentially expressed genes were used to develop hierarchical clusters using the Cluster 3.0 software [32] (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) with options selected cluster for both genes and arrays using the complete linkage. The obtained cluster output files were used as input for the Java Treeview [33] software to develop the dendrograms using the standard conditions. The hierarchical clusters of the differentially expressed genes from each dataset were listed in the Supplementary Material (Figure S2).
The differentially expressed genes obtained from each datasets were compared using Venny 2.1 [34] (http://bioinfogp.cnb.csic.es/tools/venny/) online software to obtain the common genes list. The common gene lists obtained from the Venn diagrams were listed in the Supplementary Material: (Figure S3).
We have retrieved RPKM (Reads Per Kilobase Million sample value for the conditions 96-hour and 40-hour were obtained from the supplementary data provided for GSE69461[29]. The RPKM values from all the samples were further subjected to statistical analysis using limma [35], Glimma (http://bioconductor.org/packages/release/bioc/html/Glimma.html) and edgeR [36-39] Bioconductor packages. The statistically significant genes among different samples were obtained based on the fold change values more than 2.0 also subjected to false discovery rate correction at a p-value 0.05. From the obtained gene list, genes encoding for cellulolytic and hemicellulolytic enzymes and CAZymes were specifically recovered based on the InterPro annotations provided. The results obtained from the statistical analysis was reported in the Table S3 (Supplementary material) these results contain the differentially expressed genes along with their logFC and adjusted P-values (multiple testing correction, FDR) values. The fold change values were used for the clustering analysis and the differentially expressed gene list was further compared with other datasets to obtain commonly expressed genes among the above mentioned gene expression datasets.
Overview of Data analysis
The whole transcriptome datasets considered for our present study can be divided into customized growth medium (Highley's basal medium supplemented with cellulose, glucose or other commercially available nutrients) and complex natural plant biomass medium (ball milled aspen, ball milled pine, spruce wood and poplar wood substrates) based on the media composition used for culturing P. chrysosporium. The customized growth mediums were used in gene expression studies with accession Ids GSE14734 and GSE14735. Growth medium consisting HBM supplemented with 0.5 % (wt/vol) of BMA or cellulose or glucose as sole carbon source was used for the culturing of P. chrysosporium (GSE14734) [23, 24]. For GSE14735 dataset P. chrysosporium was cultured on three different growth mediums a) replete B3 medium (with adequate carbon and nitrogen source), b) carbon limited medium c) nitrogen limited medium [23, 24]. The complex natural plant biomass growth medium was used in gene expression studies with accession numbers GSE27941, GSE52922, GSE54542, GSE69008 and GSE69461. Growth medium used for GSE27941 consists of 0.5% of ball milled aspen and ball milled pine as the sole carbon source supplemented with HBM [26]. Similarly, growth medium used for GSE52922 consisted Wiley milled chemically distinct wood substrates of Populus trichocarpa P717 (parental hybrid clone line) with 65 mol% of syringyl units and the two transgenic lines 64 and 82 with 94 and 85 mol % of syringyl units, supplemented with HBM [27]. GSE69461 medium consists of microtomed tangential sections of Picea glauca (40mm long, 10mm wide and 40 mm thick with a dry weight of 7mg) covered with 90µl of agar supplemented with nitrogen mineral salt medium [29]. GSE54542 medium contained Oak heartwood fine powdered samples extracted using acetone and resuspended further in DMSO followed by a set of extraction processes [25]. Finally, GSE69008 growth medium consisted of chemically distinct Populus trichocarpa wood substrates, which can be divided in to A (high lignin-low glucose) B (low lignin-high glucose) and C (average lignin-average glucose) conditions [28].
Results
Distribution of CAZymes in P. chrysosporium Genome
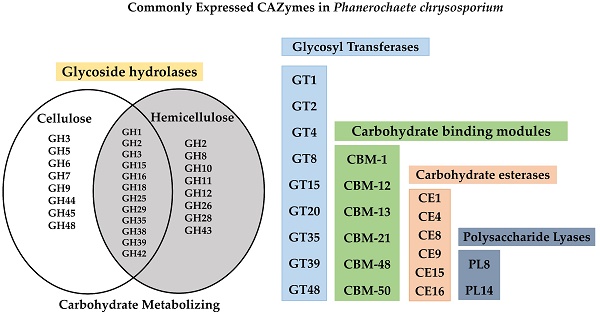
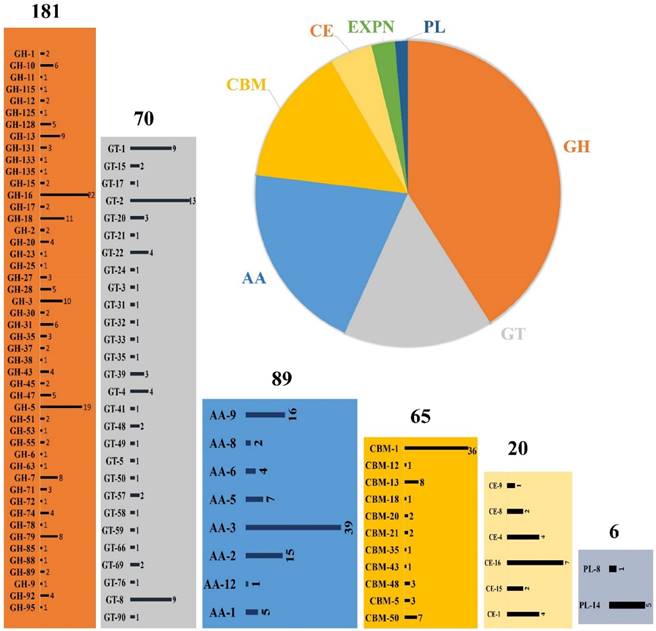
Several studies have already proved the eccentric lignin degrading abilities of P. chrysosporium. However, very few studies were conducted till today to understand the cellulose and hemicellulose degradation mechanisms employed by this organism. Whole genome studies of P. chrysosporium conducted in 2004 by Martinez et.al, have reported that it encodes around 240 putative carbohydrate active enzymes, which mainly encodes 66 glycoside hydrolases, 14 carbohydrate esterases and 57 glycosyltransferases [15]. Present day annotated genome of P. chrysosporium RP-78 version 2.2, genome codes for around 440 putative carbohydrate active enzymes divided into 89 Auxiliary activity enzymes, 65 carbohydrate binding modules, 20 carbohydrate esterases, 181 glycoside hydrolases, 70 glycosyl transferases and 6 polysaccharide lyases (Figure 1). Extracellular cellulases and hemicellulases secreted by fungi can be grouped under glycoside hydrolases [40]. According to Martinez et al. (2004), the genome of P. chrysosporium encodes at least 40 genes for putative endoglucanases (GH5, GH9, GH12, GH61, GH74) 7 exocellobiohydrolases (GH6, GH7) 9 β-glucosidases (GH1, GH3) and 5 polygalacturonase (GH28) [15]. Pedro M et al. (2003) have reported the global correlation studies of CAZymes and their corresponding total number of open reading frames among the bacterial and eukaryotic genomes [41]. The number of genes encoding glycosyl transferases exceeds occasionally by large factor compared to glycoside hydrolases encoding genes observed in eukaryote genome sequences such as Saccharomyces cerevisiae, Saccharomyces pombe, Caenorhabditis elegans, Arabidopsis thaliana, Homo sapiens and Drosophila melanogaster. Contrastingly, the genomic studies of P. chrysosporium revealed large number of glycosyl hydrolases encoding genes rather than glycosyl transferases [15].
Schematic representation of CAZymes distribution in Phanerochaete chrysosporium genome, GH (glycoside hydrolase), GT (glycosyl transferases), AA (Auxiliary activities), CBM (Carbohydrate binding modules), CE (Carbohydrate esterases) and PL (Polysaccharide lyases); the numbers represented on top of each box represents the number of genes encoding for that particular class of enzymes respectively.
Some of the significant findings from the P. chrysosporium gene expression studies with respect to CAZymes were discussed as follows. Wymelenberg et al. (2009) have reported the gene expression data of P. chrysosporium cultured on cellulose, glucose and ball milled aspen (GSE14736) [23, 24]. It revealed several transcripts encoding various cellulases belonging to GH-5, 6, 7 and hemicellulose depolymerizing enzymes such as xylanases, mannases, α-β-galactosidases, xyloglucanase, arabinofuranosidase, polygalacturonase and feruloyl esterases found to be highly upregulated in ball milled aspen along with GH-61 proteins. CAZy encoding genes were not expressed in nitrogen limited medium [23, 24]. Wymelenberg et al. (2011) have conducted the P. chrysosporium gene expression study by culturing it on ball milled aspen and ball milled pine (GSE27941) [26]. The obtained results have supported earlier investigations on the cellulolytic system of P. chrysosporium, various transcripts coding for endo and exoglucanases (CBH1, CEL7D, CBH2, CEL5A) and GH-61 were significantly expressed in BMA samples. While genes encoding cellobiose dehydrogenase (CDH) and aldose-1-epimerase transcripts were highly expressed in BMP but not in BMA [26]. Thuillier, A et al. (2014) have performed the whole transcriptomic study of P. chrysosporium cultured on oak acetonic extractives (GSE54542) [25]. Genes encoding various CAZymes such as cellobiohydrolases, endoglucanases, β-glucosidase, endoxylanases, endo glucanases and mannases were found to be expressed in control conditions and the same genes were downregulated in oak extractives [25]. In 2014, Gaskell et al. has performed gene expression studies on P. chrysosporium to understand the influence of Populus genotype (P717, 82 and 64 transgenic lines) on its gene expression (GSE52922) [27]. Results from this study showed that genes coding for GH-6 (CBH2), AA-3 (CDH), AA9 (LPMO), GH5 (endoglucanases) were highly upregulated in P717. It was also reported that transcripts coding for LPMO's, cellobiose dehydrogenase and aldose-1-epimerase were highly expressed in all the three cultures. In order to understand the lignocellulolytic abilities of P. chrysosporium on spruce wood samples, Korripally et al. (2015) have conducted a gene expression study (GSE69461) [29] and reported that 23 transcripts coding for putative hemicellulases showed more than 4-fold expression. Along with the hemicellulases, genes coding for carbohydrate esterases were also found to be highly expressed in 40 hour samples, which show that these hemicellulases and carbohydrate esterases are mutually involved in depolymerization [29]. Recently, Skyba et al. (2016) has conducted a gene expression study to understand the involvement of specific genes and enzymes involved in lignocellulose degradation by culturing P. chrysosporium on chemically distinct P. trichocarpa wood substrates [28]. Results from this study revealed the genes coding for AA9 (LPMO), GH13 were down regulated when cultured on low glucose substrates [28]. The metadata analysis results from the above mentioned gene expression studies have resulted in differentially expressed common gene lists involved in cellulose and hemicellulose degradation (Figure 2). The common significant gene lists obtained, shown in Venn diagrams were reported in the supplementary data-S2.
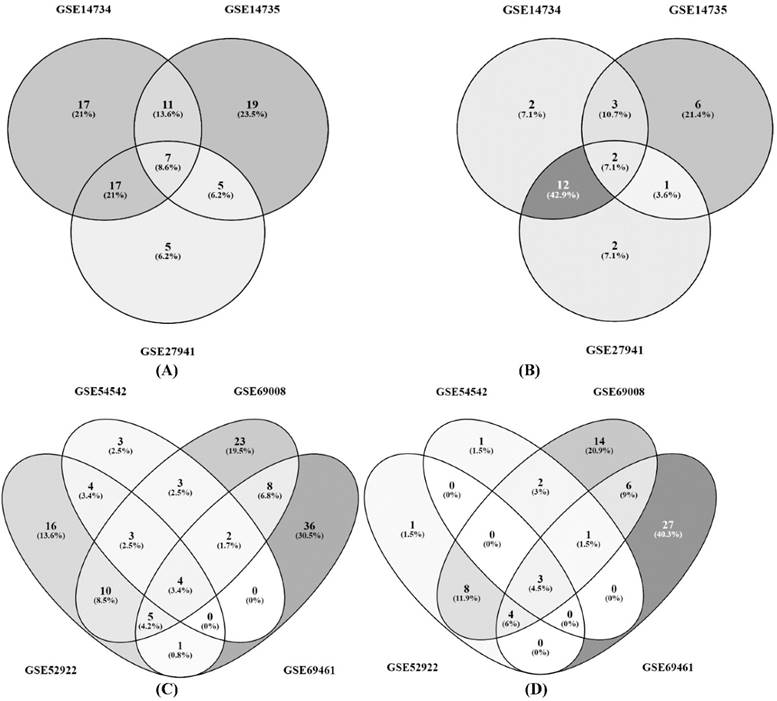
Three way and four way Venn diagrams showing the common differentially expressed genes of P. chrysosporium involved in cellulose and hemicellulose degradation; (A) Differentially expressed genes and (B) Differentially expressed CAZymes, resulted from the datasets GSE14734, GSE14735 and GSE27941; (C) Differentially expressed genes and (D) Differentially expressed CAZymes resulted from the datasets GSE54542, GSE52922, GSE69008 and GSE69461.
Expression of cellulose degrading glycoside hydrolases
Glycoside hydrolases are present in large numbers among carbohydrate active enzymes with wide variety of functional properties. Enzymes belonging to this class cleaves the glycosidic bond linkages of glycosides, glycans and other glycoconjugates [42]. Metadata analysis of P. chrysosporium gene expression studies have resulted in common differentially expressed glycoside hydrolase classes involved in cellulose degradation such as GH-5, GH-6, GH-7, GH-9, GH-44, GH-45, GH-48 and GH-61 [Table 2]. Genes encoding GH-5, GH-6, GH-7 and GH-61 (AA9) were found to be differentially expressed by P. chrysosporium cultured on cellulose (GSE14734), ball milled aspen (GSE14734, GSE27941). It was reported that, in Trichoderma reesei genes coding for cellulolytic and xylanolytic enzymes are regulated by XYR1 and CRE1 transcriptional factors corresponding to the carbon source used for its growth [43]. It was also seen that, presence of glucose in the growth media represses the expression of cellulolytic and xylanolytic genes [44-47]. Above mentioned glycoside hydrolases significantly down regulated in nitrogen limited, carbon limited growth mediums (GSE14735). When P. chrysosporium was cultured on (P. trichocarpa) P717, 82 transgenic lines (GSE52922), genes encoding for GH-2, GH-3, GH-5 were differentially expressed when compared to 64 transgenic line. In growth mediums containing chemically distinct P. trichocarpa, glycoside hydrolase encoding genes GH-5, GH-6, GH-7, GH-61 were found to be highly expressed in high glucose-low lignin and average lignin-average glucose (GSE69008) samples. Most of the cellulose degrading glycoside hydrolase encoding genes were found to be down-regulated in oak acetonic extracts (GSE54542). Glycoside hydrolases involved in cellulose breakdown such as GH-2, 5, 6, 7, 45, 48, 61, were significantly expressed in 96-hour spruce wood samples (GSE69461).
Glycoside hydrolases differentially expressed among different gene expression datasets
| Glycoside Hydrolases | Representing enzyme (Cellulose degrading) | Glycoside Hydrolases | Representing enzyme (Hemicellulose degrading) | Glycoside Hydrolases | Representing enzyme (Oligosaccharide) | |
|---|---|---|---|---|---|---|
| GH-1 | β-Glucosidase β-Galactosidase | GH-1 | β-glucosidase β-galactosidase β-mannosidase β-glucuronidase | GH-1 | β-glucosidase β-galactosidase β-mannosidase β-glucuronidase | |
| GH-2 | β-Galactosidase β-Mannosidase | |||||
| GH-5 | Endo-β-1,4-glucanase, β-glucosidase, Endo-β-1,4-xylanase | |||||
| GH-8 | Endo-1,4-β-xylanase Chitosanase Cellulase | |||||
| GH-3 | β-glucosidase xylan 1,4-β-xylosidase | |||||
| GH-5 | Endo-β-1,4-glucanase, β-glucosidase, Endo-β-1,4-xylanase | |||||
| GH-6 | Endoglucanase Cellobiohydrolase | GH-10 | Endo-1,4-β-xylanase Endo-1,3-β-xylanase | GH-2 | β-galactosidase β-mannosidase β-glucuronidase | |
| GH-7 | Endo-β-1,4-glucanase, endo-β-1,3-1,4-glucanase | GH-11 | Endo-1,4-β-xylanase Endo-1,3-β-xylanase | GH-3 | β-glucosidase xylan-1,4-β-xylosidase α-L-arabinofuranosidase | |
| GH-9 | Endoglucanase Endo-β-1,4-glucanase β-glucosidase | GH-12 | Endoglucanase Xyloglucan hydrolase | GH-28 | Polygalacturonase Exo-polygalacturonase Rhamnogalacturonase | |
| GH-44 | Endoglucanase Xyloglucanase | GH-26 | β-Mannanase exo-β-1,4mannobiohydrolase β-1,3-xylanase Mannobiose producing exo-β-mannanase | GH-29 | α-L-fucosidase α-1,3-1,4-L-fucosidase | |
| GH-45 | Endoglucanase | GH-38 | α-mannosidase mannosyl-oligosaccharide, α-1,2-mannosidase | GH-35 | β-galactosidase exo-β-glucosaminidase exo-β-1,4-galactanase | |
| GH-48 | Reducing end-acting cellobiohydrolase, endo-β-1,4-glucanase | GH-43 | β-xylosidase α-L-arabinofuranosidase | |||
| GH-47 | α-mannosidase | |||||
| GH-61 | Lytic Polysaccharide Monoxygenase (LPMO) | GH-39 | β-xylosidase α-L-iduronidase | |||
| GH-61 | Lytic Polysaccharide Monoxygenase (LPMO) | GH-42 | β-galactosidase, α-L-arabinopyranosidase |
Expression of hemicellulose degrading CAZymes
Most of the naturally occurring plant polysaccharides are partly esterified by acetic acid to protect from microbial glycoside hydrolases as they cannot breakdown acetylated glycosyl units. As a result, microorganisms secrete a set of enzymes known as carbohydrate esterases which targets the acetyl groups of plant polysaccharides [48]. Carbohydrate esterases (CE) catalyze O-de or N-deacetylation of saccharides substituted with either esters or amides, where these sugars play the role of alcohol or amine [48]. Substrates for CEs can be majorly classified into pectin methyl esters (acid form of sugar) and acetylated xylan (sugar alcohol). Presently carbohydrate esterases are classified into 16 different classes in the CAZy database. Therefore, in contrast to cellulose, hemicellulose degradation is commenced by a set of glycoside hydrolase and carbohydrate esterase class enzymes. P. chrysosporium secretes several hemicellulose degrading glycoside hydrolase class enzymes such as endo-1,4-β-xylanase, β-mannosidase, β-xylosidase and several other enzymes. Metadata analysis of the above mentioned publicly available gene expression datasets have resulted in following hemicellulose depolymerizing glycoside hydrolase classes GH-5, GH-8, GH-10, GH-11, GH-12, GH-26, GH-38, GH-43, GH-47 [Table 2]. When P. chrysosporium was cultured on ball milled aspen (GSE14734, GSE27941), genes encoding glycoside hydrolase such as GH-1, GH-2, GH-39, GH-43 which was also expressed in cellulose, and GH-47 were differentially expressed along with acetyl xylan esterases (CE-1). When cultured on replete nutrient medium (GSE14735) genes encoding GH-1, GH-5, GH-7, GH-39, GH-61 glycoside hydrolases were highly expressed. Glycoside hydrolase encoding genes GH-2, GH-16, GH-20 were found to be highly expressed in oak acetonic extractives and GH-3, GH-43, GH-47 were highly expressed in control samples (GSE54542). In GSE52922, P. chrysosporium differentially expressed glycoside hydrolase classes such as GH-2, GH-10, GH-39 in transgenic line 82 and P717, whereas GH-43 encoding genes were expressed in transgenic line 64 growth substrates. We have also observed that hemicellulose degrading glycoside hydrolase GH-5, GH-10, GH-13, GH-43, GH-47 were expressed when P. chrysosporium cultured on high lignin and low glucose samples of P. trichocarpa wood substrates (GSE69008). In GSE69461, when P. chrysosporium cultured on spruce wood samples glycoside hydrolase encoding genes such as GH-5, GH-10, GH-47, GH-53, GH-115, GH-125 and GH-131 were highly upregulated in 96-hour growth samples. Along with these glycoside hydrolases, genes encoding carbohydrate esterases such as acetyl xylan esterase (CE-1) (CE-4), pectin methyl esterase (CE-8), 4-O-methyl-glucuronoyl methylesterase (CE-15) and acetylesterase (CE-16) were also highly expressed in 96-hours samples. At the same time, acetyl xylan esterase (CE-4) and N-acetylglucosamine 6-phosphate deacetylase (CE-9) were highly expressed by P. chrysosporium in 40-hours spruce wood samples.
Glycoside hydrolases involved in fungal cell wall synthesis
Composition of fungal cell walls are dynamic and differs significantly from that of cellulose based plant cell walls [49]. Fungal cell wall majorly consists of polysaccharide units and glycoproteins in which chitin and glucan chains are significantly high. However, other cell wall components vary among the fungal species. Principally, fungal glucan component occurs in long chains of β-1,3-linked glucose units and chitin is synthesized as chains of beta-1,4-linked N-acetylglucosamine units, however chitin is less abundant in fungal cell wall when compared to glucan chains or the glycoprotein portion [49]. Thus, several genes encoding for the enzymes involved in synthesis of β-1,3-glucans and chitin are constantly expressed in fungal cell by protecting the fungi from changes in osmotic pressure and various environmental stress [49]. Glycoside hydrolases encoding genes such as GH-16 (chitin β-1,6-glucanosyltransferase), GH-18 (chitinase), GH-37 (α,α-trehalose), GH-55 (exo-endo-β-1,3-glucanases), GH-71 (α-1,3-glucanse) GH-128 (β-1,3-glucanase). Genes coding for the GH-18 was found to be differentially expressed by P. chrysosporium in all the datasets among various growth conditions. We have also found that GH-16 encoding genes were highly upregulated in oak acetonic extractives. While genes coding for glycoside hydrolase classes such as GH-16, GH-37, GH-55, GH-71, GH-128 were found to be highly expressed in complex natural plant biomass growth substrates (40-hours spruce wood, high lignin-low glucose, average lignin-average glucose), which might be due to the exposure of fungal cell to the various stressful conditions.
Expression of glycosyl transferases encoding genes
Glycosyl transferase enzymes catalyze glycosidic linkages utilizing activated sugar donor containing a phosphate group (mostly nucleoside diphosphate sugars e.g. UDP Gal, GDP Man). In simple terms GT's catalyzes the reactions for joining sugars to a glycone, thus playing a significant role in synthesis of oligosaccharides [50]. From the current metadata analysis, we have found that differentially expressed common glycosyl transferase classes were GT-1, GT-2, GT-4, GT-8, GT-15, GT-20, GT-35, GT-39, GT-48. The type of growth mediums on which P. chrysosporium was cultured, have significant effect on the expression of glycosyl transferase classes. Genes encoding GT-4, GT-8, GT-48 enzyme classes were highly expressed on cellulose growth medium whereas GT-1, GT-2, GT-20, GT-39 were expressed on ball milled aspen, and GT-8, GT-48 genes were upregulated on glucose and replete growth mediums. GT-2 class of enzymes were highly expressed on Oak acetonic extractives and on control conditions GT-8 encoding genes were upregulated. P. chrysosporium when cultured on chemically distinct P. trichocarpa medium, discrete glycosyl transferase classes were highly expressed, they are a) GT-1, GT-4, GT-20 on high lignin- low glucose conditions b), GT-2, GT-8, GT-15, GT-35, GT-39, GT-48 on low lignin-high glucose mediums c) GT-2, GT-8, GT-15, GT-20, GT-35, GT-39, GT-48 on average lignin-average glucose samples. Similarly, class of GT-39 encoding genes were expressed on P717 and 82 transgenic lines. Genes encoding glycosyl transferases such as GT-1, GT-2, GT-15, GT-48 were highly upregulated in 40-hour spruce wood samples.
Gene expression of polysaccharide lyases
Polysaccharide lyases are class of enzymes which are involved in breakdown of activated glycosidic bonds involved in joining certain acidic polysaccharide units [51]. Polysaccharide lyases cleave the polysaccharide units through a eliminase mechanism rather than a hydrolysis resulting in oligosaccharide units [51]. Present day CAZy database has classified polysaccharide lyases into 24 different classes. The metadata analysis of gene expression datasets has shown that, hyaluronate lyase or chondroitin AC lyase (PL-8) encoding genes were down regulated in oak acetonic extracts and upregulated in 96-hour spruce wood, ball milled aspen, nitrogen and carbon limited and 64, 82 transgenic line samples. Genes coding for β-1,4-glucuronan lyase or alginate lyase (PL-14) is highly upregulated in 40-hour spruce wood samples.
Major Facilitator Superfamily encoding genes
Major facilitator superfamily is one of the largest membrane transporter proteins involved in the intracellular transport of a wide variety of chemical compound by solute uniport, solute/solute antiport, solute/cation antiport and solute/cation symport [52]. MFS proteins are involved in transport of simple sugars, oligosaccharides, aminoacids, nucleosides, organophosphate esters and many other compounds [52]. Sugar transporters are class of membrane proteins involved in binding and transport of different carbohydrates, alcohols and acid compounds [53]. Efficient degradation and metabolism of plant cell wall polysaccharides by P. chrysosporium requires sugar transporters along with extracellular glycoside hydrolases [54]. Genes encoding for sugar transporters were highly expressed in cellulose, glucose, replete, ball milled aspen, high glucose-low lignin, average lignin-average glucose, 96-hours culture samples.
Glycosyl transferases and carbohydrate esterases differentially expressed among different gene expression datasets
| Glycosyl Transferases | Representing enzyme | Carbohydrate Esterases | Representing enzymes | Polysaccharide Lyases | Representing enzyme |
|---|---|---|---|---|---|
| GT-1 | UDP-glucuronosyltransferase, zeatin O-β-xylosyltransferase, indole-3-acetate β-glucosyltransferase | CE-1 | acetyl xylan esterase, cinnamoyl esterase, feruloyl esterase, carboxylesterase, S-formylglutathione hydrolase | PL-8 | Hyaluronate lyase, Chondroitin AC lyase, Xanthan lyase |
| GT-2 | cellulose synthase, chitin synthase, dolichyl-phosphate β-D-mannosyltransferase, | CE-4 | acetyl xylan esterase, chitin deacetylase, chitooligosaccharide deacetylase | ||
| GT-4 | sucrose synthase, sucrose-phosphate synthase, α-glucosyltransferase, lipopolysaccharide N-acetylglucosaminyltransferase | CE-8 | Pectin methylesterase | ||
| GT-8 | lipopolysaccharide α-1,3-galactosyltransferase, UDP-lipopolysaccharide α-1,2-glucosyltransferase, lipopolysaccharide glucosyltransferase 1, | CE-9 | N-acetylglucosamine 6-phosphate deacetylase | PL-14 | Alginate lyase, exo-oligo alginate lyase, β-1,4-glucuronan lyase |
| GT-15 | glycolipid 2-α-mannosyltransferase, GDP-Man: α-1,2-mannosyltransferase, | CE-15 | 4-O-methyl-glucuronoyl methylesterase | ||
| GT-20 | α, α-trehalose-phosphate synthase, Glucosylglycerol-phosphate synthase, trehalose-6-P phosphatase, | CE-16 | Acetylesterase | ||
| GT-35 | glycogen or starch phosphorylase | ||||
| GT-39 | protein α-mannosyltransferase | ||||
| GT-48 | 1,3-β-glucan synthase |
Genes encoding for carbohydrate metabolism
Several genes involved in expression and regulation of glycolysis were also highly expressed in the actively growing cultures of P. chrysosporium. Genes encoding for aldose-1-epimerase were highly expressed in cellulose, glucose, control, high glucose-low lignin, average lignin-average glucose, 40-hour spruce wood cultures. Genes coding for enzymes involved in regulation of the glycolysis such as hexokinase, phosphofructokinase and pyruvate kinase were differentially expressed in cellulose, carbon-limited, high glucose-low lignin and average glucose-average lignin, 40-hour spruce wood samples. Fructose-2,6 bisphosphatase, phosphoglucomutase genes were upregulated in glucose, replete, ball milled aspen samples. Enzymes involved in pentose phosphate pathway such as fructose-6-phosphoketolase, transketolase, transaldolase encoding genes were expressed in replete, high glucose-low lignin, average lignin-average glucose and 40-hour spruce wood growth samples. Alpha amylase, aldo/keto reductase, aldehyde dehydrogenase, zinc-containing alcohol dehydrogenase encoding genes were expressed in cellulose, glucose, nitrogen limited, carbon-limited, ball milled aspen, control, 64 transgenic line, high glucose-low lignin, average lignin-average glucose, 40-hours samples. Genes coding for pyruvate decarboxylase, pyruvate kinase was highly upregulated in carbon-limited, nitrogen limited, oak acetonic extractive and 40-hour spruce wood samples.
Genes encoding for carbohydrate binding modules (CBM)
The cellulose and hemicellulose degrading hydrolytic enzymes secreted by polysaccharide degrading microorganisms contains a discrete module with composite molecular architecture linked by unstructured sequences, which consists of a catalytic module and a carbohydrate binding module (CBM) [55]. Carbohydrate binding modules were firstly classified as cellulose binding modules (CBD) as various modules were found to bind with cellulose [56-58]. However, several studies have reported the modules binding to the carbohydrates other than cellulose, which was the main reason behind renaming these modules as CBMs [55]. Currently, the carbohydrate binding modules (CBMs) are divided into 80 major classes in CAZy database. CBM-12 encoding genes were highly expressed in cellulose (GSE14734) ball milled aspen (GSE27941). Genes encoding CBM-1 and CBM-13 were expressed in 96-hour samples and CBM12, CBM-21, CBM-48 and CBM-50 were found to be expressed in 40-hours samples respectively (GSE69461). Similarly, genes encoding CBM-12, CBM-48, were expressed in high glucose-low lignin and average lignin-average glucose growth conditions. The genes coding for carbohydrate binding WSC domain were differentially expressed in cellulose, nitrogen limited, ball milled aspen, control, high lignin-low glucose growth conditions. According to Wymelenberg et al (2006), careful inspection of cro3, cro4 and cro5 cDNA sequences revealed 2 to 4 tandem copies of WSC domain which might be having a role in cell wall integrity and stress component [59]. The exact function of WSC-domain containing proteins in P. chrysosporium is not clear till today [59]. In Trichoderma harzianum, β-1,3- exoglucanase protein which is associated with host cell wall degradation was found to contain 2 copies of WSC domain [60], contrastingly in Saccharomyces cerevisiae, the WSC-containing proteins are involved in maintaining cell wall integrity and heat shock response [61].
Discussion
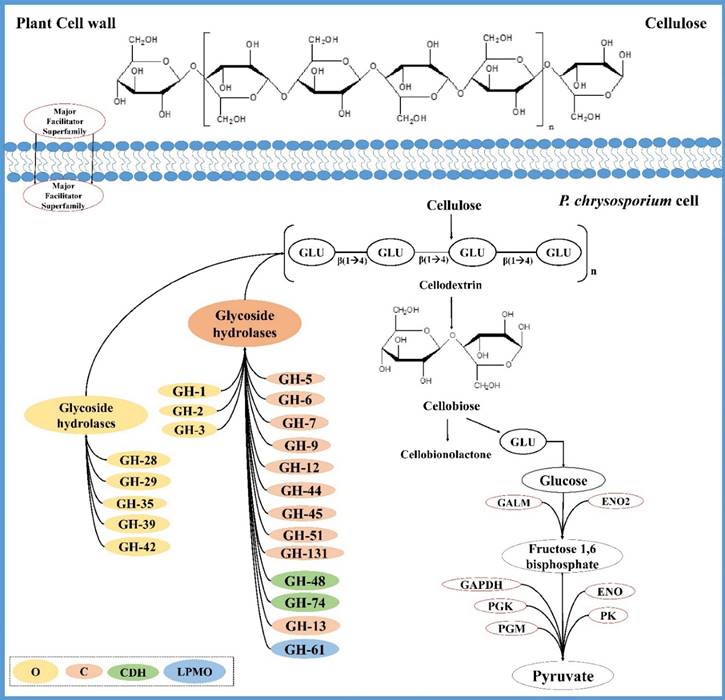
Analyzing the common gene expression patterns of P. chrysosporium involved in degradation of plant biomass will considerably enhance our current understanding. We have analyzed the gene expression datasets retrieved from public repositories based on P. chrysosporium growth conditions by using GEO2R and Bioconductor packages, to find the common differentially expressed genes among various datasets. Several genes encoding for carbohydrate active enzymes were found to be commonly expressed in P. chrysosporium gene expression datasets (Figure 3). Classically, it was well known that the process of cellulose degradation by P. chrysosporium (filamentous fungus) occurs through combination of hydrolytic reactions caused by a) endo-1,4-β-glucanases b) exo-1,4-β-glucanases (cellobiohydrolases) c) β-glucosidases [6]. Initial cellulose degradation by the filamentous fungus occurs through cellulases which are majorly classified under glycoside hydrolases, resulting in cellobiose [62]. Cellobiose, the major product obtained during the cellulose biodegradation is hydrolyzed into two molecules of glucose by cellobiose dehydrogenase (CDH) and β-glucosidase (BGL) [62]. It was reported that P. chrysosporium secretes a single isozyme coding for both BGL and CDH, initially CDH acts on cellobiose resulting in glucose and gluconolactone which are hydrolyzed by BGL [63]. Gene expression studies of P. chrysosporium conducted in the recent years have revealed several significant facts on expression patterns of cellulose degrading glycoside hydrolases and other carbohydrate active enzymes. Endoglucanase (endo-1,4-β-glucanase or 1,4-β-D-glucan 4-glucanohydrolases) is required for the initial hydrolysis of cellulose for breaking internal glycosidic bonds. Majority of the genes encoding for endo and exo glucanases belonging to GH-5, GH-6, GH-7, GH-9, GH-44, GH-45 and GH48 glycoside hydrolase classes were differentially expressed when cultured in cellulose, ball milled aspen as the carbon source (GSE14734), average lignin- average glucose (B10, C10, B20 and C20) (GSE69008) ball milled aspen (GSE27941). While these genes were down regulated in nitrogen limited, carbon limited (GSE14735), ball milled pine (GSE27941) transgenic line 64 (GSE52922), oak acetonic extracts (GSE54542) spruce wood 40 hour samples. In fact, several genes encoding cellulolytic and hemicellulolytic enzymes were not expressed in nitrogen and carbon limited conditions (GSE14735). Lytic polysaccharide monoxygenases (LPMO) are class of enzymes which can potentially breakdown the recalcitrant plant polysaccharide units by cooperating with cellobiose dehydrogenase enzyme (not necessarily as some organisms lack the CDH coding genes) resulting in production of oxidized and non-oxidized chains [64-67]. Glycoside hydrolase class 13 enzymes with alpha-amylase activity functions similar to AA-9 class enzymes which also act on polysaccharide units. Genes coding the AA-9 (LPMO) class enzymes were downregulated in low glucose-high lignin and also in only glucose (GSE14734) conditions, which might be due to its preference for lignocellulosic substrates [68]. The genes encoding these enzymes were also found to be down regulated in 40- hours samples, while these genes were highly expressed in 96-hours (GSE69461), which can be explained by expression of LPMO's during the initial days of fungal incubation [69].
Several studies have reported that occurrence of plant polymers such as cellulose, xylan in the growth medium of T. reesei induces the increased production of cellulolytic and hemicellulolytic enzymes [70-73]. The oligosaccharides such as sophorose [74, 75], β-cellobiono-1,5-lactone, xylobiose, D-galactose, D-xylose and lactose in the growth medium have also been reported to increase the genes encoding cellulolytic and hemicellulolytic enzymes [76-81]. It was always a fascinating fact that how these large insoluble polymers cellulose and xylan can induce the production of cellulases and hemicellulases, as the fungal cells usually do not incorporate these polymers [82]. Researchers investigated on this fact and postulated that soluble low molecular weight compounds derived from cellulose induce the hydrolytic enzyme production by the fungal cells. Another proposed mechanism is that, minimal levels of extracellular cellulases especially CEL7A and CEL6A produced by fungal cells act on cellulose liberating a soluble inducer which enter the cell and induces the production of hydrolytic enzymes [83, 84]. These facts can explain the higher expression of endoglucanases and exoglucanases in growth conditions mentioned above (Figure 4).
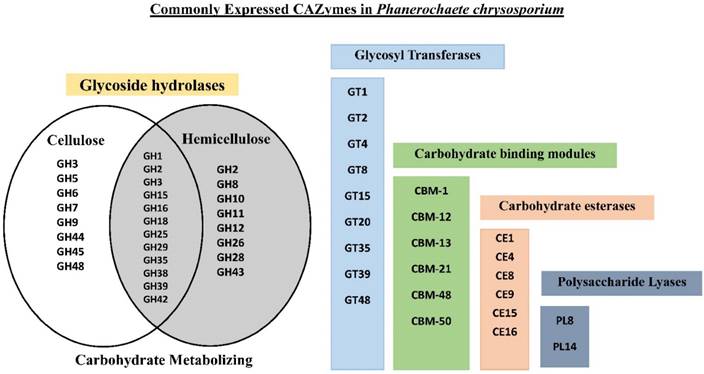
Venn diagram showing the cellulose degrading glycoside hydrolases (left), oligosaccharide metabolizing (center) hemicellulose degrading (right) and other CAZy enzyme classes.
Tentative network of genes coding for P. chrysosporium cellulose degrading enzymes and cellulose degradation mechanism; O (CAZymes involved in oligosaccharide degrading), C (cellulolytic CAZymes), CDH (cellobiose dehydrogenase encoding CAZymes) and LPMO (CAZymes coding for lytic polysaccharide monoxygenases).
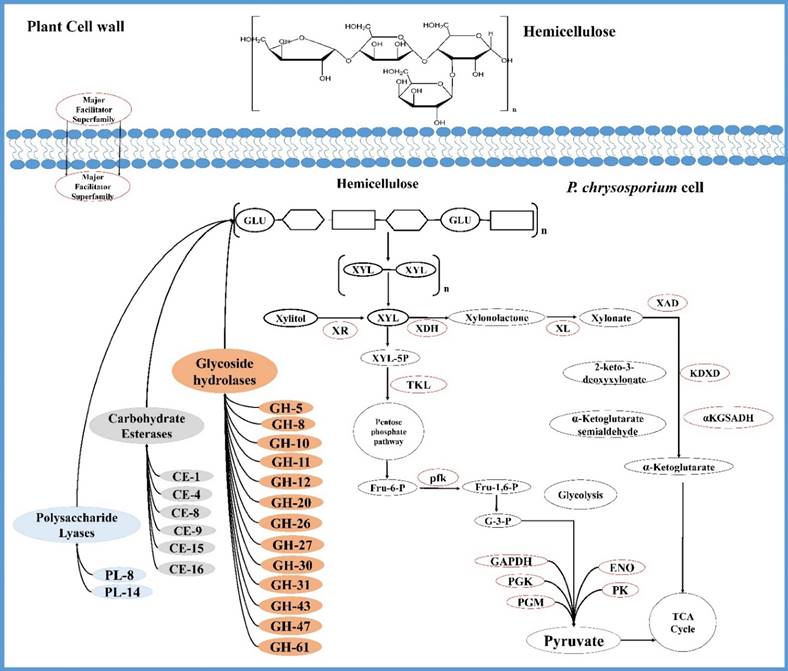
Most of the polysaccharide degrading microorganisms breakdown the hemicellulose units into monomeric sugars and acetic acid [85]. Most of the known hemicellulases are frequently classified based on their action on discrete substrates. Xylan forms the major carbohydrate constituent of hemicellulose and its degradation requires the parallel action of various hydrolytic enzymes [85]. Initially, xylan is degraded by endo-1,4-β-xylanase generating oligosaccharide chains and later xylan 1,4-β-xylosidase breaks the xylan oligosaccharides to xylose units [86]. Enzymes such as acetyl xylan esterases, α-L-arabinofuronosidases, α-4-O-methyl glucuronosidases and ferrulic, p-coumaric esterases are also efficiently required for the breakdown of wood xylan and mannans. along with the above mentioned major xylan degrading enzymes [85]. As mentioned by Pérez et al. (2002) degradation of highly occuring hemicellulosic polymer, O-acetyl-4-O-methylglucuronxylan takes place through synergistic action of endo-1,4-β-xylanases (which breaks down the whole polymer to oligosaccharide units) acetyl esterases (eliminates acetyl groups), α-glucuronidase or α-galactosidases removes galactose residues and finally β-mannosidase or β-glycosidase (cleaves the endomannose generated oligomer β-1,4-bonds) [85]. It was known that P. chrysosporium produces several endoxylanase encoding genes [87]. Glycoside hydrolases such as endoxylanases, β-xylosidases, β-galactosidases, β-mannosidases and other hemicellulases encoding genes such as GH-1, GH-2, GH-8, GH-10, GH-11, GH-26, GH-38, GH-43 and GH-47 were majorly found to be differentially expressed when P. chrysosporium was cultured in plant biomass containing growth mediums. We have observed that, process of hemicellulose degradation by P. chrysosporium occurs by combinatorial action of glycoside hydrolases and carbohydrate esterases (CE-1, 4, 8, 9, 15 and16) as observed in the gene expression profiles of GSE69008, GSE69461 datasets. Based on our metadata analysis we have found that P. chrysosporium secrete a wide range of hydrolytic enzymes involved in the degradation of lignin and hemicellulose followed by secretion of cellulolytic enzymes when cultured on complex natural plant biomass (Figure 5).
Whole genome and transcriptome studies of P. chrysosporium conducted in the recent years have revealed several significant facts on the molecular mechanisms employed by this white rot fungus during degradation of plant cell wall components. Our present study based on metadata analysis of P. chrysosporium gene expression data provides a list of common cellulolytic and hemicellulolytic enzymes expressed among various datasets under different growth conditions (customized medium and complex plant biomass containing medium). This study proves that gene expression of P. chrysosporium is strongly influenced by the growth substrates and the incubation periods. The gene expression profiles observed in GSE69008 and GSE69461 show that, when P. chrysosporium is cultured on plant biomass it initially secretes various lignin and hemicellulolytic enzymes followed by secretion of cellulolytic enzymes in the later growth stages. The future prospects of this work would be determining the precise functions of various significantly expressed hypothetical proteins, as most of them were classified as proteins with unknown functions. We believe that our present work will potentially enhance the present day understanding about P. chrysosporium and its lignocellulosic degrading patterns, and will also play a crucial role in developing overexpressed gene products of P. chrysosporium (Supplementary Material: Figure S3 and Table S1, S2) for targeting the cellulosic and hemicellulosic feedstocks.
Tentative network of genes coding for P. chrysosporium hemicellulose degrading enzymes and hemicellulose degradation mechanism.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by NSERC-RDF Fund to Wensheng Qin and Ontario Trillium Scholarship (OTS) to Ayyappa Kumar Sista Kameshwar.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Losordo Z, McBride J, Rooyen JV, Wenger K, Willies D, Froehlich A. et al. Cost competitive second-generation ethanol production from hemicellulose in a Brazilian sugarcane biorefinery. Biofuels, Bioproducts and Biorefining. 2016
2. Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW. et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. science. 2007;315:804-7
3. Zhao X, Zhang L, Liu D. Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels, Bioproducts and Biorefining. 2012;6:465-82
4. Lynd LR. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annual review of energy and the environment. 1996;21:403-65
5. Chandra RP, Bura R, Mabee W, Berlin dA, Pan X, Saddler J. Substrate pretreatment: The key to effective enzymatic hydrolysis of lignocellulosics? Biofuels - Springer. 2007:67-93
6. Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnology for biofuels. 2012;5:1
7. Scheller HV, Ulvskov P. Hemicelluloses. Plant Biology. 2010;61:263
8. Harholt J, Suttangkakul A, Scheller HV. Biosynthesis of pectin. Plant physiology. 2010;153:384-95
9. Keegstra K. Plant cell walls. Plant Physiology. 2010;154:483-6
10. Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53-78
11. Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink V. Novel enzymes for the degradation of cellulose. Biotechnology for biofuels. 2012;5:1
12. Gilbert HJ, Stålbrand H, Brumer H. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Current opinion in plant biology. 2008;11:338-48
13. Agger J, Viksø-Nielsen A, Meyer AS. Enzymatic xylose release from pretreated corn bran arabinoxylan: differential effects of deacetylation and deferuloylation on insoluble and soluble substrate fractions. Journal of Agricultural and food Chemistry. 2010;58:6141-8
14. Várnai A, Huikko L, Pere J, Siika-Aho M, Viikari L. Synergistic action of xylanase and mannanase improves the total hydrolysis of softwood. Bioresource technology. 2011;102:9096-104
15. Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J. et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nature biotechnology. 2004;22:695-700
16. Wymelenberg AV, Minges P, Sabat G, Martinez D, Aerts A, Salamov A. et al. Computational analysis of the Phanerochaete chrysosporium v2. 0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal Genetics and Biology. 2006;43:343-56
17. Abbas A, Koc H, Liu F, Tien M. Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Current genetics. 2005;47:49-56
18. Wymelenberg AV, Sabat G, Martinez D, Rajangam AS, Teeri TT, Gaskell J. et al. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. Journal of biotechnology. 2005;118:17-34
19. Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic acids research. 2014;42:D490-D5
20. Levasseur A, Piumi F, Coutinho PM, Rancurel C, Asther M, Delattre M. et al. FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal genetics and biology. 2008;45:638-45
21. Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I. et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic acids research. 2014;42:D26-D31
22. Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS. et al. FungiDB: an integrated functional genomics database for fungi. Nucleic acids research. 2011 gkr918
23. Wymelenberg AV, Gaskell J, Mozuch M, Sabat G, Ralph J, Skyba O. et al. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Applied and environmental microbiology. 2010;76:3599-610
24. Wymelenberg AV, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D. et al. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Applied and environmental microbiology. 2009;75:4058-68
25. Thuillier A, Chibani K, Belli G, Herrero E, Dumarçay S, Gérardin P. et al. Transcriptomic Responses of Phanerochaete chrysosporium to Oak Acetonic Extracts: Focus on a New Glutathione Transferase. Applied and environmental microbiology. 2014;80:6316-27
26. Wymelenberg AV, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J. et al. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Applied and environmental microbiology. 2011;77:4499-507
27. Gaskell J, Marty A, Mozuch M, Kersten PJ, BonDurant SS, Sabat G. et al. Influence of Populus genotype on gene expression by the wood decay fungus Phanerochaete chrysosporium. Applied and environmental microbiology. 2014;80:5828-35
28. Skyba O, Cullen D, Douglas CJ, Mansfield SD. Gene expression patterns of wood decay fungi Postia placenta and Phanerochaete chrysosporium are influenced by wood substrate composition during degradation. Applied and Environmental Microbiology. 2016 AEM. 00134-16
29. Korripally P, Hunt CG, Houtman CJ, Jones DC, Kitin PJ, Cullen D. et al. Regulation of Gene Expression during the Onset of Ligninolytic Oxidation by Phanerochaete chrysosporium on Spruce Wood. Applied and environmental microbiology. 2015;81:7802-12
30. Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R. et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research. 2013 gkt1183
31. Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP. et al. Fueling the future with fungal genomics. Mycology. 2011;2:192-209
32. de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453-4
33. Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246-8
34. Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007
35. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015 gkv007
36. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40
37. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research. 2012 gks042
38. Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321-32
39. Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic acids research. 2014;42:e91-e
40. Dashtban M, Schraft H, Qin W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int J Biol Sci. 2009;5:578-95
41. Coutinho PM, Stam M, Blanc E, Henrissat B. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends in plant science. 2003;8:563-5
42. Vuong TV, Wilson DB. Glycoside hydrolases: catalytic base/nucleophile diversity. Biotechnology and bioengineering. 2010;107:195-205
43. dos Santos Castro L, Antoniêto ACC, Pedersoli WR, Silva-Rocha R, Persinoti GF, Silva RN. Expression pattern of cellulolytic and xylanolytic genes regulated by transcriptional factors XYR1 and CRE1 are affected by carbon source in Trichoderma reesei. Gene Expression Patterns. 2014;14:88-95
44. Kubicek CP, Messner R, Gruber F, Mandels M, Kubicek-Pranz EM. Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose-inhibited beta-diglucoside permease. Journal of Biological Chemistry. 1993;268:19364-8
45. Ilmen M, Saloheimo A, Onnela M-L, Penttilä ME. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Applied and Environmental Microbiology. 1997;63:1298-306
46. Zeilinger S, Schmoll M, Pail M, Mach RL, Kubicek CP. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Molecular Genetics and Genomics. 2003;270:46-55
47. Ilmén M, Onnela M-L, Klemsdal S, Keränen S, Penttilä M. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Molecular and General Genetics MGG. 1996;253:303-14
48. Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnology advances. 2012;30:1575-88
49. Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28:799-808
50. Lairson L, Henrissat B, Davies G, Withers S. Glycosyltransferases: structures, functions, and mechanisms. Biochemistry. 2008;77:521
51. Linhardt R, Galliher P, Cooney C. Polysaccharide lyases. Applied biochemistry and biotechnology. 1987;12:135-76
52. Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiology and molecular biology reviews. 1998;62:1-34
53. Walmsley AR, Barrett MP, Bringaud F, Gould GW. Sugar transporters from bacteria, parasites and mammals: structure-activity relationships. Trends in biochemical sciences. 1998;23:476-81
54. Suzuki H, MacDonald J, Syed K, Salamov A, Hori C, Aerts A. et al. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC genomics. 2012;13:444
55. Shoseyov O, Shani Z, Levy I. Carbohydrate binding modules: biochemical properties and novel applications. Microbiology and Molecular Biology Reviews. 2006;70:283-95
56. Gilkes N, Warren R, Miller R, Kilburn DG. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. Journal of Biological Chemistry. 1988;263:10401-7
57. Tomme P, Tilbeurgh H, Pettersson G, Damme J, Vandekerckhove J, Knowles J. et al. Studies of the cellulolytic system of Trichoderma reesei QM 9414. European Journal of Biochemistry. 1988;170:575-81
58. Van Tilbeurgh H, Tomme P, Claeyssens M, Bhikhabhai R, Pettersson G. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei. FEBS letters. 1986;204:223-7
59. Wymelenberg AV, Sabat G, Mozuch M, Kersten PJ, Cullen D, Blanchette RA. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Applied and environmental microbiology. 2006;72:4871-7
60. Cohen-Kupiec R, Broglie KE, Friesem D, Broglie RM, Chet I. Molecular characterization of a novel β-1, 3-exoglucanase related to mycoparasitism of Trichoderma harzianum. Gene. 1999;226:147-54
61. Lodder AL, Lee TK, Ballester R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics. 1999;152:1487-99
62. Yoshida M, Igarashi K, Kawai R, Aida K, Samejima M. Differential transcription of β-glucosidase and cellobiose dehydrogenase genes in cellulose degradation by the basidiomycete Phanerochaete chrysosporium. FEMS microbiology letters. 2004;235:177-82
63. Eriksson KE. Enzyme mechanisms involved in cellulose hydrolysis by the rot fungus Sporotrichum pulverulentum. Biotechnology and Bioengineering. 1978;20:317-32
64. Quinlan RJ, Sweeney MD, Leggio LL, Otten H, Poulsen J-CN, Johansen KS. et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proceedings of the National Academy of Sciences. 2011;108:15079-84
65. Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VG, Igarashi K. et al. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PloS one. 2011;6:e27807
66. Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnology for biofuels. 2013;6:1
67. Bey M, Zhou S, Poidevin L, Henrissat B, Coutinho PM, Berrin J-G. et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Applied and environmental microbiology. 2013;79:488-96
68. Žifčáková L, Baldrian P. Fungal polysaccharide monooxygenases: new players in the decomposition of cellulose. Fungal ecology. 2012;5:481-9
69. Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S. et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnology for biofuels. 2015;8:1
70. Bisaria VS, Mishra S, Eveleigh DE. Regulatory aspects of cellulase biosynthesis and secretion. Critical reviews in biotechnology. 1989;9:61-103
71. Kubicek C. From cellulose to cellulase inducers: facts and fiction. Proceedings of the second TRICEL symposium on Trichoderma reesei cellulases and other hydrolytic enzymes: Foundation of Biotechnical and Industrial Fermentation Research, Espoo. 1993:181-8
72. Zeilinger S, Mach R. Xylanolytic enzymes of Trichoderma reesei: properties and regulation of expression. Curr Topics Cer Chem. 1998;1:27-35
73. Mach R, Zeilinger S. Regulation of gene expression in industrial fungi: Trichoderma. Applied microbiology and biotechnology. 2003;60:515-22
74. Hrmová M, Petráková E, Biely P. Induction of cellulose-and xylan-degrading enzyme systems in Aspergillus terreus by homo-and heterodisaccharides composed of glucose and xylose. Microbiology. 1991;137:541-7
75. Mandels M, Parrish FW, Reese ET. Sophorose as an inducer of cellulase in Trichoderma viride. Journal of Bacteriology. 1962;83:400-8
76. Aro N, Pakula T, Penttilä M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS microbiology reviews. 2005;29:719-39
77. Karaffa L, Fekete E, Gamauf C, Szentirmai A, Kubicek CP, Seiboth B. D-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology. 2006;152:1507-14
78. Kubicek C, Penttilä M. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. Trichoderma and Gliocladium. 1998;2:49-71
79. Morikawa Y, Ohashi T, Mantani O, Okada H. Cellulase induction by lactose in Trichoderma reesei PC-3-7. Applied microbiology and biotechnology. 1995;44:106-11
80. Stricker AR, Grosstessner-Hain K, Würleitner E, Mach RL. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryotic cell. 2006;5:2128-37
81. Stricker AR, Steiger MG, Mach RL. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS letters. 2007;581:3915-20
82. Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Current genomics. 2013;14:230-49
83. El-Gogary S, Leite A, Crivellaro O, Eveleigh D, El-Dorry H. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proceedings of the National Academy of Sciences. 1989;86:6138-41
84. Carle-Urioste JC, Escobar-Vera J, El-Gogary S, Henrique-Silva F, Torigoi E, Crivellaro O. et al. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. Journal of Biological Chemistry. 1997;272:10169-74
85. Pérez J, Munoz-Dorado J, de la Rubia T, Martinez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. International Microbiology. 2002;5:53-63
86. Jeffries TW. Biodegradation of lignin and hemicelluloses. Biochemistry of microbial degradation: Springer. 1994:233-77
87. Kirk TK, Cullen D. Enzymology and molecular genetics of wood degradation by white-rot fungi. Environmentally friendly technologies for the pulp and paper industry Wiley, New York. 1998:273-307
Author contact
![]() Corresponding author: Wensheng Qin, Email: wqinca, Tel: 807-343-8467
Corresponding author: Wensheng Qin, Email: wqinca, Tel: 807-343-8467

 Global reach, higher impact
Global reach, higher impact