10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(2):157-166. doi:10.7150/ijbs.17440 This issue Cite
Research Paper
Regulatory Axis of miR-195/497 and HMGA1-Id3 Governs Muscle Cell Proliferation and Differentiation
1. Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences, Shenzhen University, Shenzhen, Guangdong, 518060, China.
2. Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, College of Optoelectronic Engineering, Shenzhen University, Shenzhen, Guangdong, 518060, China.
3. Department of Physiology, Shenzhen University Health Science Center, Shenzhen, Guangdong, 518000, China.
* These authors contributed equally to this work.
Received 2016-9-2; Accepted 2016-11-13; Published 2017-1-15
Abstract
Myocytes withdraw from the cell cycle to differentiate during muscle development. Given the capacity of microRNAs (miRNAs) to regulate gene expression during development, we screened for miRNAs that were associated with muscle development. S-Poly(T) Plus analysis of 273 miRNAs in porcine longissimus dorsi muscles revealed 14 miRNAs that were strongly upregulated with age of postnatal muscle development in vivo, including miR-195 and miR-497. These two miRNAs were also strongly upregulated at late differentiation stages of mouse skeletal myoblast C2C12 cells, and demethylation treatment induced significant upregulation of miR-195/497. Manipulation of miR-195/497 expression resulted in dramatic changes in the proliferation and differentiation of C2C12 cells. We identified high-mobility group AT-hook 1 (Hmga1) mRNA as a highly conserved target of miR-195/497 in C2C12 myoblasts. Overexpression of miR-195/497 or Hmga1 silencing in C2C12 cells promoted myogenic differentiation. Moreover, we showed that miR-195/497 repressed Hmga1, which in turn downregulated one of the HMGA1 downstream targets Id3, whose inhibitory effect on myogenic differentiation is well established. Our study revealed a subset of potential development-associated miRNAs and suggests a novel regulatory axis for myogenesis in which miR-195/497 promote myogenic differentiation by repressing the HMGA1-Id3 pathway.
Keywords: miR-195/497, Hmga1, myoblast, skeletal muscle, differentiation, proliferation.
1. Introduction
Pork is the most widely consumed meat in the world. In the field of biomedicine, pigs are anatomically and physiologically analogous to humans. Knowledge of muscle development in pigs is important to improve production and quality of pork and provides understanding of human muscle disease. Myogenesis is an important biological process in all stages of skeletal muscle growth [1, 2]. During myogenesis, myoblasts proliferate, withdraw from the cell cycle, and differentiate to form myotubes [3]. Myogenesis is a tightly regulated process that is promoted mainly by basic helix-loop-helix (bHLH) factors including MyoD, myogenic factor 5, myogenin, E-proteins, and myocyte enhancer factor 2 (MEF2) [4]. MicroRNAs (miRNAs) are involved in regulating various biological processes, including cell proliferation and differentiation, and the function of a subset of miRNAs during muscle development has been well established [5].
Clearly, a complete understanding of the roles of miRNAs in myogenesis is imperative for elucidating skeletal muscle growth. In this study, we profiled the expression patterns of 273 miRNAs in longissimus dorsi muscle of pigs during postnatal growth and identified a set of development-associated miRNAs. Of these, miR-195 and miR-497 (hereafter called miR-195/497) were strongly upregulated in an age-dependent manner. MiR-195/497 are known cancer repressors that govern the expression of many cell-cycle-associated genes [6-8]. Recently, miR-195/497-mediated myogenic proliferation and renewal have been characterized during muscle development [9, 10]. Here, we revealed the role of miR-195/497 in myotube formation during myogenesis.
The high-mobility group A1 gene (Hmga1) is an architectural transcription factor that participates in various nuclear processes that modulate numerous biological processes, including myogenic differentiation [11, 12]. In this study, manipulating the activity of miR-195/497 showed that they regulate the Hmga1 expression in muscle cells. We found that in myoblasts, HMGA1 induced the expression of Id3, one of the major repressors of cell differentiation.
2. Material and methods
2.1. Cell culture
Human embryonic kidney cell lines HEK293T and mouse skeletal myoblast C2C12 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM; Corning Cellgro, Manassas, VA, USA) containing 10% fetal bovine serum (FBS, Biowest, Nuaillè, France) in a humidified incubator with 5% CO2 at 37°C. Mouse C2C12 myoblasts were transferred to DMEM containing 2% horse serum (Gibco, Grand Island, NY, USA) to initiate differentiation and mimic normal myogenesis.
2.2. Animals and muscle collection
The castrated male hybrid pigs (Landrace × Large White × Duroc) were obtained from College of Animal Science of South China Agricultural University. Feeding conditions and experimental designs have been detailed in the study of Wu et al. [13]. Briefly, the pigs were housed in a controlled environment and were provided free access to food and water according to the NRC2012 (Nutrient requirements of swine, 2012). We collected the porcine longissimus dorsi muscles at postnatal days 35 (weaning), 63 (nursery), 98 (grow-finish), and 161 (reach market weight) with six pigs in each group. The C57BL/6 male mice were purchased from the Guangdong Medical Laboratory Animal Center. Mouse skeletal muscles were collected at postnatal days 2 (newborn), 14 (suckling), 28 (weaning), 42 (sexually mature) and 63 (adult). All the protocols were approved by the Animal Ethical and Welfare Committee of Shenzhen University (Approval No. AEWC-2014-001004).
2.3. Plasmid construction
Hmga1 3′UTR luciferase reporter: To construct the Hmga1 3'UTR luciferase reporter plasmid, the wild-type Hmga1 3′UTR sequence was amplified from mouse genomic DNA using primers Hmga1-UTR-Wt-F (5′-CCAGAATTCACTTGTCAGCCTTGAAGCATCGGAG-3′) and Hmga1-UTR-Wt-R (5′-CCACTGAGCATGGGGGAAGGAAATCAATGAGTG-3′). The PCR products were then subcloned into the downstream region of the Firefly luciferase reporter gene of the miRGlo vector (Promega, Madison, WI, USA). Hmga1-UTR mutants were generated by site-directed mutagenesis using primers Hmga1-UTR-Mut-F (5′-ATGTTCATTCTTAGATACCCACTATTGCTGCTGCTACCAGCGTCAGATGTT-3′) and Hmga1-UTR-Mut-R (5′-CAGGAGGATGAACATCTGACGCTGGTAGCAGCAGCAATAGTGGGTATCTAA-3′).
Myogenin promoter luciferase assay: The mouse myogenin promoter was subcloned into the vector pGL4 (Promega, Madison, WI, USA) at the XhoI-MluI sites. The myogenin promoter was amplified from mouse genomic DNA using primers MyoG-Pro-F (5'-CCACTCGAGGTCTCTAGCTGCATATGTAGCAGAAGATGG-3') and MyoG-Pro-R (5'-GGTACGCGTCAGGTCGCAAAAGGCTTGTTCCTGCCACTG-3').
Hmga1 overexpression plasmid: The sequences of wild-type Hmga1 were amplified from the cDNA from C2C12 cells using primers Hmga1-F (5′-CACCGAATTCAGTCGGGGCTATTTCTGGTGTTGGC-3′) and Hmga1-R (5′-CACCGGATCCGCTGGTGCCTGGTGCGGTAGTGTGC-3′). The PCR products were then subcloned into the pLVX-puro vector (Clontech, Mountain View, CA, USA).
Hmga1 shRNA plasmids. Plasmid vectors containing shRNA targeted to Hmga1 and nonspecific control (sh-NC) were constructed based on pLVX-hU6 vector. The target sequences of sh-Hmga1-1 and sh-Hmga1-2 were 5'-AAGTCACCACAGCTCCAGGGA-3' and 5'-GTGAAGTGCCAACTCCGAAGA-3'.
miRNA dTuD inhibitor plasmids: miRNA inhibitors against miR-195 or miR-497 were generated using the two-step PCR method as detailed in our previous study [14].
2.4. Lentivirus packaging and transduction
HEK293T cells were used to package lentiviral particles. Briefly, the cells were transfected using poly-ethylenimine (PEI) transfection reagent with the packing plasmids (17.1 µg) in a ratio of 3:1:3 as follows: psPAX2 encoding HIV Gag-Pol (Addgene 12260; Addgene, Cambridge, MA, USA), pVSVg encoding the VSV-G glycoprotein (Addgene 8454; Addgene, Cambridge, MA, USA), and a lentiviral vector (6.9 µg). At 48 h and 72 h after transfection, the cell-free supernatants were harvested and used for cell infection using 8 ng/ml of polybrene.
2.5. Fluorescence reporter assays
For the myogenin promoter reporter assay, C2C12 cells were transfected with myogenin promoter luciferase reporter plasmids (100 ng), miRNA mimics (100 µM), and in-house modified SV40 promoter-driven Renilla luciferase vector (pSV40-R.Luc; Promega, Madison, WI, USA) as an internal control. For the 3' UTR reporter assay, C2C12 cells were transfected with 3' UTR reporter plasmids (100 ng) and miRNA mimics (100 µM). The miRNA mimics (RiboBio, Guangzhou, Guangdong, China) were transfected into C2C12 cells using K2 transfection reagent (Biontex Laboratories GmbH, München, Germany) following the manufacturer's instructions. PEI transfection reagent (Polyplus, Illkirch, France) was used for the transfection of plasmids. The luciferase activity was measured using a Lumat3 LB9508 luminometer (Berthold Technologies, Bad Wildbad, Germany).
2.6. Ethynyl deoxyuridine (EdU) assay
The EdU incorporation assay was performed using an EdU assay kit (Ribobio, Guangzhou, Guangdong, China) following the manufacturer's instructions. EdU labeling was conducted for 1 h for the C2C12 cells stably expressing miRNA or miRNA inhibitor (dTuD-miR-195 and dTuD-miR-497).
2.7. RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted with RNAiso Reagent (TaKaRa, Dalian, China) following the manufacturer's instructions. The expression levels of the miRNAs were evaluated with our previously developed S-Poly(T) Plus method [15, 16] (Geneups, Shenzhen, Guangdong, China), using SNORNA202 and SNORNA234 as the porcine and mouse miRNA endogenous references, respectively. Briefly, RNA are polyadenylated and reverse-transcribed into cDNA. The RT primer consists of a universal reverse primer sequence, a universal Taqman probe sequence and 17 bases that are complementary to target miRNA with tailed poly(A). The PCR reactions were carried out in the StepOnePlus Real-Time PCR System (Applied Biosystems, Grand Island, NY, USA) with a miRNA-specific forward primer and a universal reverse primer. The primers used can be found in Supplementary Table 1.
2.8. Western blotting and cell immunostaining
Western blotting and antibodies: total proteins of the C2C12 cells were extracted with RIPA buffer supplemented with a protease inhibitor cocktail (Roche, Mannheim, Germany). Protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Beijing, USA). The primary antibodies used are as follows: β-tubulin (Cat #10094-1-AP, 1:5,000; Proteintech, Wuhan, Hubei, China), myogenin (Cat # ab1835, 1:2000; Abcam, Cambridge, MA, USA), HMGA1 (Cat #ab129153, 1:1,000; Abcam, Cambridge, MA, USA), and DNMT1 (Cat # 24206-1-AP; Proteintech, Wuhan, Hubei, China). Horseradish peroxidase (HRP)-conjugated secondary antibodies (1:10,000; Bio-Rad, Hercules, CA, USA) were also used.
Cell immunostaining: The C2C12 cells were cultured on glass coverslip in 24-well plates, then fixed with 4% formaldehyde in PBS buffer, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and blocked with 1% bovine serum albumin for 15 min. The primary antibody anti-myosin heavy chain (1:100; Abcam, Cambridge, MA, USA) was used. Indirect immunofluorescence was detected with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibodies (Cat #A-11001; Invitrogen, Carlsbad, CA, USA).
2.9. Demethylation
C2C12 myoblasts were cultured in 35-mm dishes and treated with 5-aza-2′-deoxycytidine (Sigma, St. Louis, MO, USA) at a final concentration of 2 μM for 48 h in growth medium (GM) or low horse serum differentiation medium (DM).
2.10. Statistical analysis
All results were expressed as the mean ± SD, and at least three independent individuals or replicates were used per treatment. Two-tailed Student's t-tests were used to determine the statistical significance using STATGRAPHICS (Centurion XVI.I) software (StatPoint Technologies, USA). Short time-series expression miner hierarchical cluster analysis (v1.3.9) software was used to analyze the short time series miRNA expression data from quantitative RT-PCR.
3. Results
3.1. miR-195/497 were upregulated in skeletal muscles during postnatal growth
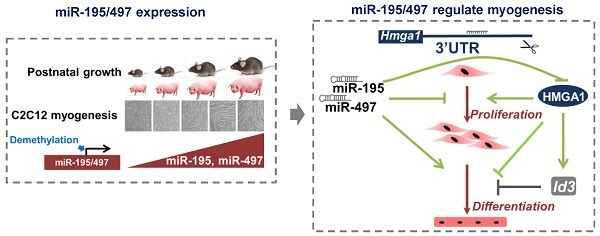
To explore muscle development-associated miRNAs, we employed S-Poly(T) Plus quantitative real-time PCR to analyze the temporal expression patterns of database-annotated pig miRNAs in longissimus dorsi muscle from 35 to 161 postnatal days. Of the 273 analyzed miRNAs, 42 were differentially expressed by more than 2.5-fold between 35 and 161 days (Fig.1A). After two rounds of screening, 9 miRNAs were downregulated during myogenesis (Fig.1B and C). 33 miNRAs were upregulated during myogenesis and 14 miRNAs showed a robust and progressive increase with age (Fig. 1D and E). MiR-195, miR-497 and miR-29c were the three most prominently upregulated miRNAs (Fig.1E). Because miR-195/497 are both members of the miR-15 family, we performed a simultaneous study of the roles of miR-195/497 in other pig skeletal muscles. Meanwhile, we found that miR-195/497 were upregulated in semitendinosus, semimembranosus, gluteus medius, and triceps brachii muscles during postnatal development (Fig. 1F and G). The upregulation of miR-195/497 was also found in mouse hindlimb muscle during postnatal development (Fig. 1H and I). These findings show that the upregulation of miR-195/497 is associated with postnatal development. Moreover, mouse skeletal myoblasts (C2C12 cells) proliferate in GM, withdraw from the cell cycle, and differentiate to myotubes when the culture is shifted to DM. As shown in Fig. 1J and K, miR-195/497 were induced in C2C12 cells by DM. Therefore, the upregulation of miR-195/497 is also associated with myogenic differentiation.
miR-195 and miR-497 are upregulated during postnatal muscle growth and myogenic differentiation. (A) Workflow used to identify muscle development-associated miRNAs. (B-E) 42 miRNAs (≥2.5) expression profile during postnatal growth in porcine longissimus dorsi muscles. (F, G) Expression patterns of miR-195 (F) and miR-497 (G) in different porcine muscles during postnatal growth. The error bars indicate the means ± standard deviations of the samples from six pigs. aP <0.05, bP <0.01, cP <0.001. (H, I) Expression patterns of miR-195 (H) and miR-497 (I) in mouse tibialis anterior muscle during postnatal growth. The error bars indicate the means ± standard deviations of the samples from eight mice. aP <0.05, bP <0.01, cP <0.001. (J, K) Upregulation of miR-195 (J) and miR-497 (K) during C2C12 myogenic differentiation. C2C12 cells were cultured in growth medium (GM) and then switched to differentiation medium (DM). DM1, 3, 5, and 7 indicate DM for 1, 3, 5, and 7 days, respectively. The error bars indicate the means ± standard deviations of four independent cell samples. Quantitative RT-PCR was performed to analyze the expression of the miRNAs using SNORNA202 and SNORNA234 as porcine and mouse endogenous references, respectively. aP <0.05, bP <0.01, cP <0.001.
3.2. miR-195/497 blocked myoblast proliferation but promoted myogenic differentiation
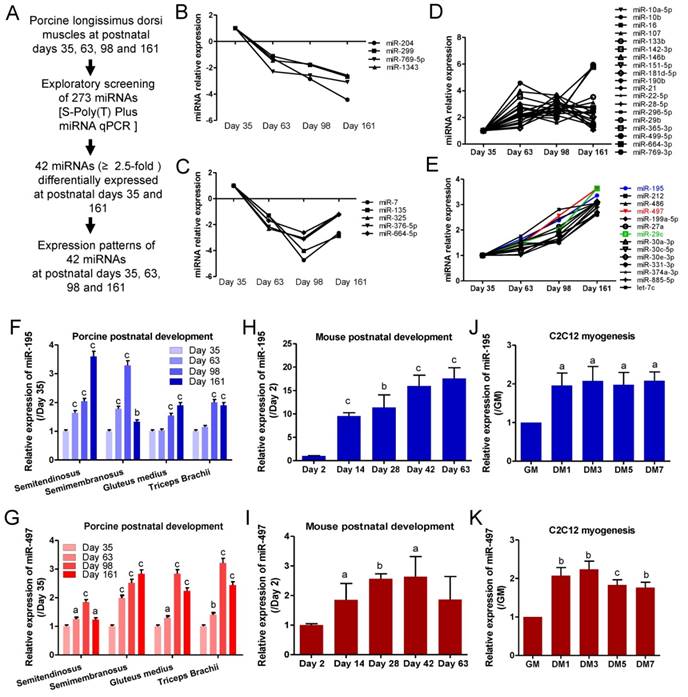
MiR-195/497 are known tumor-suppressive miRNAs and their anti-proliferative functions have been reported in many cancer cells, including breast cancer [6], hepatocellular carcinoma [7], and thyroid cancer cells [8]. miR-195/497 have been reported to regulate myoblast proliferation and self-renewal through regulating the cell cycle [9, 10]. Here, we confirmed the anti-proliferative activities of miR-195/497 in vitro by transfecting miRNA mimics and inhibitors of miR-195/497 in C2C12 myoblasts. Quantitative PCR confirmed the efficiency of miRNA mimics or inhibitors on miR-195/497 expression in myoblasts (Fig. 2A and B). The EdU incorporation assays showed that mimics of miR-195 or miR-497 reduced cell proliferation (0.63-fold, P = 9.1 E-05 and 0.41-fold, P = 6.3 E-05, respectively) compared with the controls (Fig. 2C, Supplementary Figure 1a). In contrast, synthetic inhibitors against miR-195 or miR-497 had the opposite effect (1.29-fold, P = 0.0051 and 1.37-fold, P = 0.0163, respectively; Fig. 2D, Supplementary Figure 1b).
We next assessed the function of miR-195/497 in myogenic differentiation, although Wei et al. [10] reported that miR-195/497 have no effect on myotube formation. Because myogenesis is regulated by a series of muscle-specific regulatory genes such as myogenin, we performed a myogenin promoter-driven luciferase assay. We found that the depletion of miR-195/497 repressed the luciferase activity (0.23-fold, P = 1.14E-10 and 0.46-fold, P =1.05E-05, respectively; Fig. 2E), indicating miR-195/497 had positive effects on myogenic differentiation in C2C12 myoblasts. This is further confirmed by western blot that the level of myogenin protein was lower in C2C12 cells transfected with the inhibitors of miR-195/497 (Fig. 2F). Moreover, immunostaining assays showed that inhibition and overexpression of miR-195/497 repressed and enhanced the formation of myotubes, respectively (Fig. 2G). Therefore, miR-195 and miR-497 both are required for myogenic gene expression and myotube formation on the skeletal muscle cells.
miR-195/497 repress myoblast proliferation and promote differentiation. (A) Relative miR-195/497 levels measured by quantitative PCR in C2C12 myoblasts transfected with miRNA mimics (miR-195 or miR-194), compared with mimic control. (B) Quantitative PCR showing decreased expression of miR-195/497 in C2C12 myoblasts transfected with miRNA inhibitors against miR-195 or miR-497 and their control. ***P <0.001. (C) Overexpression of miR-195/497 reduces the rate of myoblast proliferation. The cell proliferation analysis was performed by EdU incorporation of C2C12 transfected with miR-195/497 mimics and controls (NC). Quantification of EdU-positive cells from 10 random fields per sample. The cell number of the NC was set to 1.0. The error bars indicate the means ± standard deviations of three independent cell samples. ***P <0.001. (D) Inhibition of miR-195/497 enhances the rate of myoblast proliferation. Knockdown of miR-195/497 increases the rate of myoblast proliferation. The other details are as described in C. *P <0.05, **P <0.01. (E) miR-195/497 inhibitors reduce the transcriptional activity of myogenin. C2C12 cells transfected with the myogenin promoter luciferase reporter, the pSV40-R.Luc vector, and miR-195 inhibitor, miR-497 inhibitor, or inhibitor control (IN-miR-NC) were transferred to DM for 3 days. The luciferase activity of the NC was set to 1.0. The error bars indicate the means ± standard deviations of four independent cell samples. ***P <0.001. (F) Western blots demonstrate that inhibition of miR-195/497 reduces the abundance of myogenin in C2C12 cells. β-tubulin served as the loading control. (G) Immunostaining images of myogenin (green) showing that miR-195/497 inhibitors and mimics repressed and enhanced myotube formation at DM4 (DM for 4 days), respectively. Scale bar, 200 μm. (Right) Quantification of fused myonuclei is presented relative to the control (100%). The error bars indicate the means ± standard deviations of eight measurements. *P <0.05, ***P <0.001.
3.3. HMGA1 represses myogenic differentiation and is a common target of miR-195 and miR-497
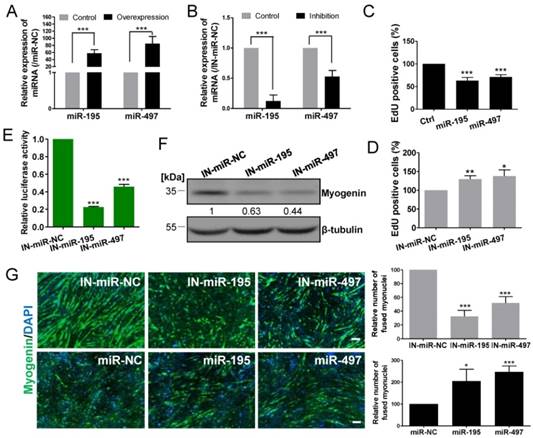
We investigated the possible mechanism by which miR-195/497 modulate myogenesis. Based on the miRWalk database [17], miR-195 and miR-497 had 45 common predicted targets (Fig. 3A), including Hmga1 whose expression is required for embryonic stem cell differentiation [18]. The 3' UTR of Hmga1 contains one putative miR-195 and miR-497 binding site (Fig. 3B), which was conserved among vertebrates (Fig. 3C). Consistent with the previous study [12], the abundance of the HMGA1 protein was reduced in C2C12 cells after the induction of the myogenic program (Fig. 3D), which is opposite to the upregulated expression of the miR-195/497. The pattern of myogenin abundance confirmed the state of differentiation (Fig. 3D). These findings prompted us to investigate whether miR-195/497 regulated the expression of HMGA1 during myogenesis.
miR-195/497 downregulate the expression of the HMGA1 protein by targeting the 3' UTR of Hmga1. (A) The number of targets of miR-195/497 predicted by miRWalk. (B) The predicted binding sites for miR-195 and miR-497 in the 3' UTR of Hmga1. (C) The predicted binding sites of miR-195/497 in the 3'UTR of Hmga1 is highly conserved among mammals. (D) The abundance of the HMGA1 protein is reduced during C2C12 myogenic differentiation. Western blot analysis of myogenin abundance confirmed the process of myogenic differentiation in C2C12 cells. GM, growth medium; DM1, 3, 5, and 7 indicate differentiation medium for 1, 3, 5, and 7 days, respectively. β-tubulin served as the loading control. (E-F) Western blots confirm the efficiency of overexpression (E) and silencing of Hmga1 (F) in C2C12 cells. (G) HMGA1 promotes cell proliferation. C2C12 cells were infected with lentiviral vectors expressing Hmga1 or silenced Hmga1 (sh-Hmga1) and maintained in GM. The error bars indicate the means ± standard deviations of three independent cell samples. (H) HMGA1 inhibits myogenic differentiation. C2C12 cells stably expressing Hmga1 or silenced Hmga1 (sh-Hmga1) were transferred into DM for 4 days. Immunostaining images of myogenin (green) showing that overexpression and silencing of Hmga1 repressed and enhanced myotube formation, respectively. Scale bar, 200 μm. ***P <0.001. (I) Western blots showing the overexpression of HMGA1 reduced the abundance of myogenin, whereas reduced abundance of HGMA1 resulted in a higher abundance of myogenin. The data are the means of three independent cell samples. *P <0.05. (J, K) Hmga1 3' UTR or 3' UTR Mut were co-transfected with miR-195 (J) or miR-497 (K) mimics or mimic control (miR-NC) into HEK293A cells. The data were normalized to the Renilla luciferase activity. The luciferase activity of C2C12 cells transfected with miR-NC was set to 1.0. The error bars indicate the means ± standard deviations of four independent cell samples. **P <0.01. (L) miR-195/497 repress HMGA1 protein expression in C2C12 cells. Western blot shows that the transfection of miR-195/497 mimics decreased the abundance of HMGA1 protein at DM4 (DM for 4 days). β-tubulin served as the loading control. The protein level in cells transfected with miR-NC was set to 1.0. The data indicate the means of three independent cell samples. **P <0.01, ***P <0.001.
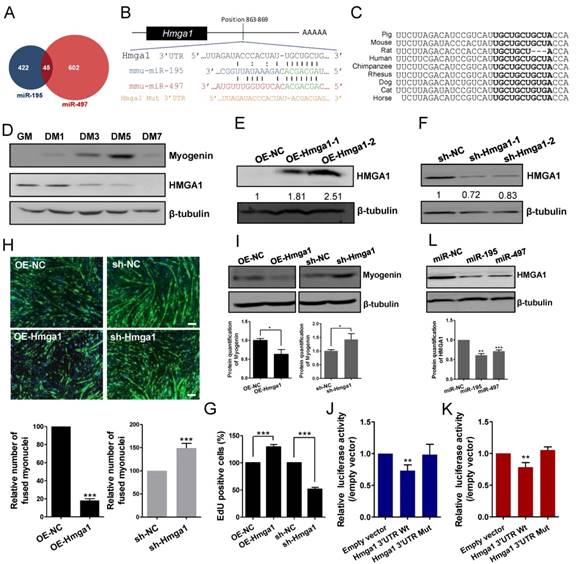
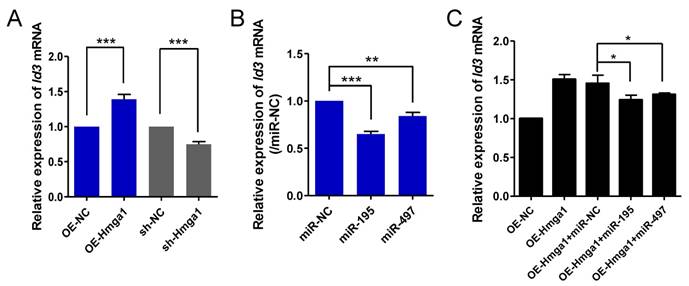
miR-195/497 reduce HMGA1-derived Id3 expression. (A) HMGA1 promotes the expression of Id3 mRNA. qRT-PCR of Id3 in C2C12 cells stably overexpressing (OE) Hmga1, silenced Hmga1 (sh-Hmga1), or empty vector (sh-NC). The expression levels of Id3 mRNA in the cells expressing NC were set to 1.0. The error bars indicate the means ± standard deviations of three independent cell samples. ***P <0.001. (B) miR-195/497 repress the expression of Id3 mRNA. qRT-PCR of Id3 in C2C12 cells transfected with miR-195 mimics, 497 mimics, or mimic control (miR-NC). **P <0.01 and ***P <0.001. (C) miR-195/497 reduce the HMGA1-drived Id3 expression. C2C12 cells stably overexpressing (OE) Hmga1 were transfected with miR-195/497 mimics or NC. *P <0.05.
HMGA1 proteins have been shown to regulate global chromatin composition and functions as repressors in myogenic differentiation through regulating the myogenic gene expression [12]. Little is known about its role in myogenic proliferation and myotube formation. We therefore generated C2C12 myoblasts stably expressing Hmga1 or silenced Hmga1 (sh-Hmga1). The efficient overexpression and silencing of Hmga1 were confirmed by western blot analysis (Fig.3E and F). We found that the overabundance of HMGA1 in C2C12 cells increased the cell proliferation rate (1.29-fold, P = 3.43E-07; Fig. 3G), blocked myotube formation (Fig. 3H). Consistent with the previous study [12], overexpression of HMGA1 reduced the myogenic genes such as myogenin (Fig. 3I). In contrast, the silencing of Hmga1 repressed the cell proliferation rate (Fig. 3G), promoted myotube formation and increased myogenin abundance (Fig. 3I). Therefore, HMGA1 and miR-195/497 have opposite functions in cell myogenesis.
The Hmga1 3' UTR luciferase reporter assays in C2C12 cells showed that the ectopic miR-195/497 significantly repressed the luciferase activity (0.73-fold, P = 7.01E-04 and 0.78-fold, P = 2.78E-05, respectively), whereas the mutations introduced into the 3' UTR binding site abolished the repression (Fig. 3J and K). Consistent with the 3' UTR luciferase assay, ectopic miR-195/497 reduced the abundance of HMGA1 in C2C12 cells (Fig. 3L). Together, miR-195/497 repressed Hmga1 expression by targeting its 3' UTR in the mouse myoblasts.
3.4. miR-195/497-HMGA1-Id3 regulatory axis in myogenesis
Ids proteins (Id1-4) are known to be a strong inhibitor of myogenic differentiation [19]. It has been shown that transcription of Id3 is directly controlled by HMGA1 in C2C12 cells [20]. We confirmed that HMGA1 positively regulated the expression of Id3 mRNA in myoblasts (Fig. 4A). It is possible that miR-195/497 modulate HMGA1-driven Id3 expression in C2C12 cells. First, we measured the Id3 expression level in C2C12 cells transfected with miR-195/497 mimics. We found that overexpression of miR-195/497 in myoblasts downregulated Id3 mRNA expression (0.64-fold, P = 1.30E-07, 0.84-fold, P = 0.0067, respectively; Fig. 4B). Then, we investigated whether miR-195/497 attenuated the HMGA1-driven Id3 mRNA expression. Following the transfection of cells overexpressing Hmga1 with miR-195 or miR-497 plasmids, we noted a degree of attenuation of Id3 mRNA (Fig. 4C), indicating that miR-195/497 reduced the HMGA1-driven increase in Id3 mRNA expression.
3.5. miR-195/497 expression was regulated by demethylation in C2C12 cells
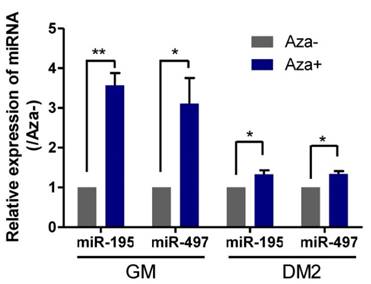
Because genome-wide demethylation promotes myogenic differentiation [21] and DNA methylation of CpG islands upstream of the genes encoding miR-195/497 has been demonstrated to be responsible for the downregulation of these miRNAs in human breast cancer cells [6], we investigated whether demethylation induced the expression of miR195/497 in C2C12 myoblasts. We measured the expression levels of miR-195/497 in C2C12 cells treated with the DNA-demethylating agent 5-aza-2'-deoxycytidine in GM and DM2 (DM for 2 days) and compared them with their expression levels in untreated C2C12 cells. Following demethylation, we found that miR-195/497 were induced by 5-aza-2'-deoxycytidine in both proliferating and differentiating cells (Fig. 5). Therefore, DNA demethylation may contribute to the increase of miR-195/497 expression levels in differentiating C2C12 cells.
Demethylation treatment in C2C12 cells induces the expression of miR-195/497. Demethylation treatment increases the expression of miR-195/497 in C2C12 cells. qRT-PCR of miR-195/497 in C2C12 cells with (Aza+) or without (Aza-) the DNA-demethylating agent 5-aza-2'-deoxycytidine in GM and DM2 (DM for 2 days). The expression levels of miR-195/497 in the cells without treatment (Aza-) were set to 1.0. The error bars indicate the means ± standard deviations of three independent cell samples per group. *P <0.05 and **P <0.01.
4. Discussion
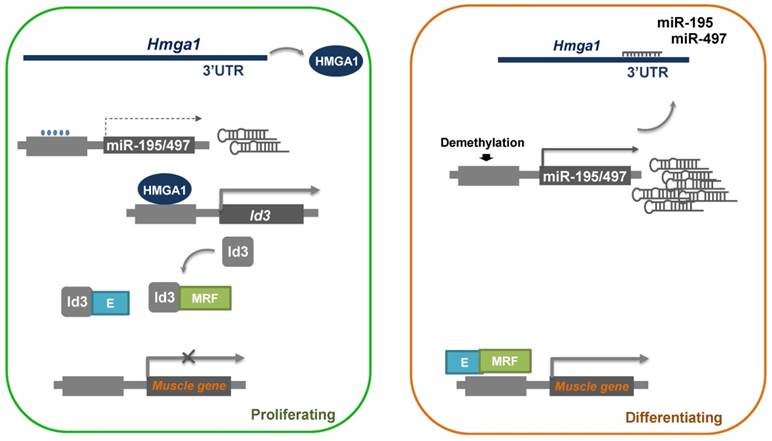
In this study, we demonstrated a role of miR-195/497 in myogenic differentiation. MiR-195/497 has been implicated in both muscle development and cell proliferation in two previous studies [10, 22]. Here miR-195/497 was found to inhibit the expression of a well-known differentiation repressor, Id3, by directly targeting Hmga1. We found that the overexpression of miR-195/497 or the attenuation of Hmga1 accelerated myotube formation and decreased the expression of Id3 mRNA, which indicated a novel role for miR-195/497 in regulating myogenic differentiation through a regulatory axis of HMGA1-Id3. It is well known that Ids proteins inhibit myogenic differentiation by preventing the heterodimerization of myogenic regulatory factors and E proteins [23]. Based on our results, we propose a model to account for the functions of miR-195/497 during myogenesis, as shown in Fig. 6.
Our miRNA screening detected 42 miRNAs as development-associated miRNAs, and about 30% of these miRNAs (miR-21, miR-27a, miR-29b/c, miR-30a/c/e, miR-133b, miR-135, miR-199a, miR-206, and miR-486) have been previously reported to play important roles in muscle development and differentiation [24-28]. For example, miR-21 was found to enhance the activity of PI3K/Akt/mTOR signaling by targeting the transforming growth factor TGFβI during porcine skeletal muscle development [24]. MiR-206 and miR-486 were shown to induce C2C12 myogenic differentiation by targeting paired box Pax7 [28]. Our results indicate the miR-195 and miR-497 can join the collection of muscle-restricted miRNAs with roles in regulating myogenesis. Whether the other 70% of the miRNAs modulate muscle development remains to be seen.
miR-195/497 have been reported to regulate myoblast proliferation and self-renewal through regulating the cell cycle [22]. Wei et al. [10] recently reported that forced expression of miR-195/497 in C2C12 cells blocked the G1-S phase transition. Skeletal muscle is in some ways similar to cardiac muscle. In the neonatal rat cardiomyocyte and myoblast cell line H9c2, miR-195 inhibited mitotic progression by blocking cells in the G2 phase [29]. Indeed, miR-195/497 were shown to engage a subset of cell cycle regulators such as checkpoint kinase 1 [29], cell division cycle 25a/b, G1/S-specific cyclin-D1/2 [22], and cyclin E1 [10]. In this study, we identified a novel target gene Hmga1, which encodes HMGA1, which has been reported to maintain proliferation in papillary carcinoma and gastric and colorectal cancer cells [30, 31]. We showed here for the first time that an increased abundance of HMGA1 increased the proliferative rate of myocytes (Fig. 3). The powerful induction of cell cycle withdrawal before terminal differentiation by miR-195/497 likely involves the actions of many target genes in myocytes.
MiR-195/497 has been reported to govern neural, adipogenic, and osteoblastic differentiation [32-34]. In this study, we employed several methods to determine the pro-differentiation role of miR-195/497 (Fig. 2). The opposite effect of HMGA1 further supports the idea that miR-195/497 positively modulate myogenic differentiation (Fig. 3); although, in a previous study, miR-195/497 were reported to have no significant effect on differentiation [10]. Thus, we have discovered a novel role of miR-195/497 in promoting myogenic differentiation during skeletal muscle development.
Deficiencies in muscle differentiation-induced (MDI) miRNAs such as miR-26 and miR-206 have been found to delay the normal kinetics of muscle regeneration after injury in mice [35, 36]. Indeed, accumulation of myocytes and the delay of novel myofiber formation are commonly observed in mice lacking MDI miRNAs [35, 36], indicating an accumulation of myocytes can block the transition from proliferation to differentiation. Although our results in vitro show the contributions of miR-195/497 to both proliferation and differentiation, further studies using detailed analysis of the mechanisms of miR-195/497 and their targets in mice lacking miRNA in the skeletal muscle injury model are warranted.
Proposed model of the miR-195/497-HMGA1-Id3 regulatory axis in myogenesis. In proliferating myoblasts, HMGA1 is abundant and drives the expression of Id3, which prevents the heterodimerization of MRF (myogenic regulatory factors) and E protein, which in turn represses the expression of muscle genes. In differentiating cells, miR-195/497 are highly expressed and repress the expression of Hmga1. As a result, HMGA1 is less abundant, and the complex of MRF and E protein forms and subsequently binds to the promoter of muscle genes, thereby inducing myogenic differentiation.
A previous study showed that miR-195/497 were upregulated in muscle stem cells during postnatal development and induction of miR-195/497 in stem cells resulted in cell quiescence [22]. Although, we showed that expression of miR-195/497 can be regulated by methylation. However, this effect may be indirect such as over-expression of MyoD or other transcription factors. Here we demonstrated that the upregulation of miR-195/497 in myocytes contributed to the progression of differentiation. Therefore, miR-195/497 have different functional roles in muscle stem cell and myoblasts. Although a full understanding of the mechanism of postnatal muscle development remains elusive, the results of this and previous studies show that miR-195/497 are critical in regulating the cell cycle in the transition between cell cycle arrest and proliferation and from proliferation to differentiation during muscle development.
Abbreviations
DM, differentiation medium; GM, growth medium; MDI, muscle differentiation-induced.
Supplementary Material
Additional File 1Supplementary Figures.
Supplementary Table 1.
Acknowledgements
We would like to thank Yueran Li, Yixuan Luo, Jiao Guo and Zixin Jin of ShenZhen University for providing technical assistance, and Qingyan Jiang and Dingyuan Feng of South China Agricultural University for providing us the porcine skeletal muscles.
Funding sources
This work was supported by the National Basic Research Program of China (973 Program) [2012CB124701]; the National Natural Science Foundation of China [81170047, 81370151, 81570046 and 31571199]; the Shenzhen Municipal Basic Research Program [JCYJ20150729104027220]; the Shenzhen Overseas High-Level Talents Innovation Program [YFZZ20111009]; and the Shenzhen High-tech Development Project [CXZZ20140828163951592].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421-35
2. White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:10-21
3. Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628-33
4. Perry RL, Rudnick MA. Molecular mechanisms regulating myogenic determination and differentiation. Frontiers in bioscience: a journal and virtual library. 2000;5:D750-67
5. Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Developmental biology. 2016;410:1-13
6. Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y. et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:1722-30
7. Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T. et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PloS one. 2013;8:e60155
8. Wang F, Jiang C, Sun Q, Yan F, Wang L, Fu Z. et al. miR-195 is a key regulator of Raf1 in thyroid cancer. OncoTargets and therapy. 2015;8:3021-8
9. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat Commun. 2014:5
10. Wei W, Zhang WY, Bai JB, Zhang HX, Zhao YY, Li XY. et al. The NF-kappaB-modulated microRNAs miR-195 and miR-497 inhibit myoblast proliferation by targeting Igf1r, Insr and cyclin genes. Journal of cell science. 2016;129:39-50
11. Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63-81
12. Brocher J, Vogel B, Hock R. HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation. BMC cell biology. 2010;11:64
13. Wu F, Zuo JJ, Yu QP, Zou SG, Tan HZ, Xiao J. et al. Effect of skeletal muscle fibers on porcine meat quality at different stages of growth. Genetics and molecular research: GMR. 2015;14:7873-82
14. Qiu H, Zhong J, Luo L, Kang K, Qu J, Peng W. et al. A PCR-based method to construct lentiviral vector expressing double Tough Decoy for miRNA inhibition. PloS one. 2015
15. Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y. et al. An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Scientific reports. 2015;5:15100
16. Kang K, Zhang X, Liu H, Wang Z, Zhong J, Huang Z. et al. A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PloS one. 2012;7:e48536
17. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. Journal of biomedical informatics. 2011;44:839-47
18. Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, Croce CM. et al. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:1496-8
19. Melnikova IN, Christy BA. Muscle cell differentiation is inhibited by the helix-loop-helix protein Id3. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1996;7:1067-79
20. Martinez Hoyos J, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C. et al. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer research. 2004;64:5728-35
21. Laker RC, Ryall JG. DNA Methylation in Skeletal Muscle Stem Cell Specification, Proliferation, and Differentiation. Stem cells international. 2016;2016:5725927
22. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nature communications. 2014;5:4597
23. Langlands K, Yin X, Anand G, Prochownik EV. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. The Journal of biological chemistry. 1997;272:19785-93
24. Bai L, Liang R, Yang Y, Hou X, Wang Z, Zhu S. et al. MicroRNA-21 Regulates PI3K/Akt/mTOR Signaling by Targeting TGFbetaI during Skeletal Muscle Development in Pigs. PloS one. 2015;10:e0119396
25. Agarwal P, Srivastava R, Srivastava AK, Ali S, Datta M. miR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochimica et biophysica acta. 2013;1832:1294-303
26. Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA. et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell death and differentiation. 2013;20:1194-208
27. Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song CC. et al. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. International journal of molecular sciences. 2014;15:296-308
28. Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Molecular and cellular biology. 2011;31:203-14
29. Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ. et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circulation research. 2011;109:670-9
30. Akaboshi S, Watanabe S, Hino Y, Sekita Y, Xi Y, Araki K. et al. HMGA1 is induced by Wnt/beta-catenin pathway and maintains cell proliferation in gastric cancer. The American journal of pathology. 2009;175:1675-85
31. Xing J, Cao G, Fu C. HMGA1 interacts with beta-catenin to positively regulate Wnt/beta-catenin signaling in colorectal cancer cells. Pathology oncology research: POR. 2014;20:847-51
32. Liu C, Teng ZQ, McQuate AL, Jobe EM, Christ CC, von Hoyningen-Huene SJ. et al. An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PloS one. 2013;8:e51436
33. Yun UJ, Song NJ, Yang DK, Kwon SM, Kim K, Kim S. et al. miR-195a inhibits adipocyte differentiation by targeting the preadipogenic determinator Zfp423. Journal of cellular biochemistry. 2015;116:2589-97
34. Almeida MI, Silva AM, Vasconcelos DM, Almeida CR, Caires H, Pinto MT. et al. miR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget. 2016;7:7-22
35. Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA. et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. The Journal of clinical investigation. 2012;122:2054-65
36. Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes & development. 2012;26:2180-91
Author contact
![]() Corresponding author: Deming Gou Tel: +86 0755 26527848 Fax: +86 0755 26527848 Email: dmgouedu.cn. Present Address: Nanhai Ave 3688, Shenzhen, Guangdong, P. R. China, 518060.
Corresponding author: Deming Gou Tel: +86 0755 26527848 Fax: +86 0755 26527848 Email: dmgouedu.cn. Present Address: Nanhai Ave 3688, Shenzhen, Guangdong, P. R. China, 518060.

 Global reach, higher impact
Global reach, higher impact