10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(2):245-253. doi:10.7150/ijbs.16818 This issue Cite
Research Paper
Melatonin Inhibits Glioblastoma Stem-like cells through Suppression of EZH2-NOTCH1 Signaling Axis
1. Department of Neurosurgery, Shandong Provincial Hospital Affiliated to Shandong University, Jinan 250021, Shandong, China;
2. Department of Neurosurgery, Qilu Hospital of Shandong University, Jinan 250012, Shandong, China;
3. Department of Histology and Embryology, Shandong University School of Medicine, Jinan, 250012, Shandong, China.
* These authors contributed equally to the study.
Received 2016-7-11; Accepted 2016-12-27; Published 2017-2-6
Abstract
Glioblastoma stem-like cells (GSCs) play essential roles in glioma growth, radio- and chemo-resistance, and recurrence. Elimination of GSCs has therefore become a key strategy and challenge in glioblastoma therapy. Here, we show that melatonin, an indolamine derived from I-tryptophan, significantly inhibited viability and self-renewal ability of GSCs accompanied by a decrease of stem cell markers. We have identified EZH2-NOTCH1 signaling as the key signal pathway that regulated the effects of melatonin in the GSCs. Instead of transcriptionally silencing gene expression by generating a methylated epigenetic mark at histone 3 at lysine 27 (H3K27), EZH2 regulates NOTCH1 expression by directly binding to the NOTCH1 promoter. Moreover, correlation between the expressions of EZH2 and NOTCH intracellular domain 1 (NICD1) was observed in the clinical tumor samples, evidently supporting the existence of EZH2-NOTCH1 interaction in the gliomas and GSCs. Collectively, we demonstrated that melatonin, a potential tumor inhibitor, performs its function partly by suppressing GSC properties through EZH2-NOTCH1 signaling axis.
Keywords: Melatonin, Glioblastoma stem-like cells, viability, self-renewal, EZH2, NOTCH1
Introduction
Cancer stem cells (CSCs) are subpopulations of cancer cells which have capacity to propagate tumors in vitro and in vivo, and have characteristics of normal stem cells. It's reported that glioblastomas multiforme (GBMs) also have a subpopulation of stem-like cells which are responsible for glioma proliferation, chemo- and radiotherapy resistance, and tumor recurrence [1-4]. Therapies that do not eradicate the GSC compartment are likely to achieve little success.
Melatonin (N-acetyl-5-methoxytryptamine) is an indolamine derived from I-tryptophan which is secreted by the pineal gland. It is well documented that melatonin is involved in the regulation of chrono-biological rhythms and endocrine function[5]. Subsequently, many other functions of melatonin have been reported such as immune-enhancing, anti-inflammatory and antioxidant properties [6-8]. In recent years, increasing evidence have demonstrated that melatonin has inhibitory effects on many cancer types such as breast, colon and gastric cancer[9-11]. It is also reported that melatonin inhibited glioma cell proliferation both in vitro and in vivo, in relation to its inhibitory role in key intracellular effectors such as PKC, Akt, and NF-kB[12]. However, the effects and mechanism in which melatonin can inhibit GSCs is far from clarified. Less clear are the extent and nature of the regulatory network involved and how these might affect melatonin function.
EZH2 is one of the core components of polycomb group (PcG) proteins. It functions as a lysine methyl-transferase, especially to catalyze tri-methylation of histone 3 at lysine 27 (H3K27me3). In a wide range of cancers, including glioblastomas, elevated expression of EZH2 is well recognized and its expression is strongly linked to tumor malignancy, invasiveness and poor prognosis[13, 14]. Recent studies also suggested that up-regulation of EZH2 has been identified as a marker for GSCs, possibly reflecting its role in the maintenance of stem cells[15]. EZH2 signaling regulates proliferation and self-renewal of GSCs and protects them from radiation-induced cell death[3, 16]. EZH2 modulators, on the other hand, have been shown to influence the development of GSC niche and tumor growth[17], suggesting EZH2 as a potential therapeutic target to eradicate glioblastomas.
NOTCH receptors and ligands are single-pass transmembrane proteins that play an important role in cell fate decisions during embryonic and postnatal development[18-20]. NOTCH receptors are overexpressed in many kinds of cancers such as pancreatic, cervical, breast, colon and lung cancer[20]. Previous study showed that NOTCH1 was decreased EZH2 was suppressed in Pancreatic Tumor[21].
In the current study, we provided the first evidence showing that melatonin can inhibit viability and self-renewal of GSCs, which is correlated with a decrease in the expression of PcG protein EZH2. Furthermore, we have identified NOTCH1 as the key signal molecule that regulates the EZH2-mediated effects of melatonin in the GSCs. Taken together, our findings suggest that melatonin, as a blocker of EZH2-NOTCH1 signaling axis, may be used as a potential therapeutic agent to prevent glioma progression.
Materials and methods
Cell Lines, GSC culture and Tissue Samples
Glioblastoma cell lines (U251 and T98G) were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) at 37℃ with 5% CO2. Tumor-spheres were obtained from the U251 and T98G cells cultured in DMEM/F12 medium (HyClone, Logan, UT, USA) supplemented with 2% B27 (Invitrogen, Carlsbad, CA, USA), 25ng/ml human recombinant basic fibroblast growth factor (bFGF; R&D systems, Minneapolis, MN, USA), 25ng/ml epidermal growth factor (EGF; R&D systems, Minneapolis, MN, USA) and 1% penicillin/streptomycin (Gibco, Birmingham, MI, USA). Half of the sphere-forming medium was replaced every other day. 67 GBM specimens and 12 normal human brain specimens were obtained from the Department of Neurosurgery at Provincial Hospital affiliated to Shandong University. The study was approved by Shandong University Ethics Committee and all of the patients provided written informed consent.
Quantitative Real-time PCR (qRT-PCR)
Trizol reagent (Gibco, Birmingham, MI, USA) was used for RNA extracting. Real-time ABI 7300 Fast Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) and SYBR Green PCR kit (Applied TaKaRa, Japan) were used for Real-time PCR [22]. The primer sequences are shown in Table 1.
Cell viability assay
Cell counting kit-8 (Dojindo, Japan) was used to monitor the proliferative viability of the cells. Briefly, 3000 cells were plated onto 96-well culture plates in a final volume of 100μL. After 48 hr, 10μL of CCK-8 solution was added into each well and incubated at 37℃ for 2 hr. The optical density of every sample was measured in a spectrophotometer at 450 nm. Experiments were repeated 6 times.
Oligonucleotide primers used for real-time PCR.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| EZH2 | 5'-TTGTTGGCGGAAGCGTGTAAAATC-3' | 5'-TCCCTAGTCCCGCGCAATGAGC -3' |
| NOTCH1 | 5'-AAGCTGCATCCAGAGGCAAAC-3' | 5'-TGGCATACACACTCCGAGAACAC-3' |
| NOTCH2 | 5'-GTTACAGCAGCCCTTGCCTGA-3' | 5'-CCATGGATACAAGGGTTACTTGCAC-3' |
| NOTCH3 | 5'-ATCGGCTCGGTAGTAATGCTG-3' | 5'-ACAACGCTCCCAGGTAGTCA-3' |
| NOTCH4 | 5'-TGCGAGGAAGATACGGAGTG-3' | 5'-GGACGGAGTAAGGCAAGGAG-3' |
| CCND1 | 5'-GGGCCACTTGCATGTTCGT-3' | 5'-CAGGTTCCACTTGAGCTTGTTCAC-3' |
| CCNE1 | 5'-CCGGTATATGGCGACACAAGA-3' | 5'-CAAACTGGTGCAACTTTGGAG-3' |
| CCNE2 | 5'-TGGGAACTTTGTCCTGTAACAATVA-3' | 5'-CACAAGGCAGCAGCAGTVAGTA-3' |
| CTNNB1 | 5'-GAGTGCTGAAGGTGCTATCTGTCT-3' | 5'-GTTCTGAACAAGACGTTGACTTGGA-3' |
| DVL2 | 5'-GACATGAACTTTGAGAACATGAGC-3' | 5'-CACTTGGCCACAGTCAGCAC-3' |
| HES1 | 5'-GGACATTCTGGAAATGACAGTGA-3' | 5'-AGCACACTTGGGTCTGTGCTC-3' |
| CD133 | 5'-AGTGGCATCGTGCAAACCTG-3' | 5'-CTCCGAATCCATTCGACGATA-3' |
| SOX2 | 5'-GTGAGCGCCCTGCAGTACAA-3' | 5'-GCGAGTAGGACATGCTGTAGGTG-3' |
| GAPDH | 5'-GCACCGTCAAGGCTGAGAAC-3' | 5'-TGGTGAAGACGCCAGTGGA-3' |
Immunohistochemistry (IHC)
Representative formalin-fixed and paraffin-embedded blocks were selected and cut into 5μm thick slices. IHC staining was performed using the standard avidin-biotin complex method. The primary antibodies used in the study were as follows: anti-EZH2 (Cell Signaling and Technology, Boston, MA, USA), anti-NOTCH1 (NOTCH intracellular domain 1, NICD1) (Cell Signaling and Technology, Boston, MA, USA). The staining scores were determined as previous method[23]. 0-4 scores were considered low expression an 6-16 scores were considered high expression.
Western Blotting
Wstern blotting was conducted as described[22]. The following primary antibodies were used: anti-CD133(Abgent, USA), anti-SOX2(Abgent, USA), anti-EZH2 (Cell Signaling and Technology, Boston, MA, USA), anti-NOTCH1 (NOTCH intracellular domain 1, NICD1) (Cell Signaling and Technology, Boston, MA, USA), anti-HES1 (Abcam, Cambridge, MA, USA) and anti-β-ACTIN (Zhongshan Golden Bridge Biotechnology, China).
Luciferase Reporter Assay
Luciferase reporter assay was conducted as previous method [22].
Chromatin Immunoprecipitation (CHIP)
The GSCU251 were cross-linked with 1 % formaldehyde, quenched by adding 125 mM glycine and lysed in lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl, pH 8.1, 1mM PMSF). The chromatin was subsequently sonicated to generate DNA fragments between 200-600 bps. Cleared lysates were then diluted and incubated with the antibodies overnight at 4℃. The following antibodies were used to perform precipitation: anti-H3K27me3 (Cell Signaling and Technology, Boston, MA, USA), anti-EZH2 (Cell Signaling and Technology, Boston, MA, USA) and anti-SUZ12(suppressor of zeste 12) (Abcam, Cambridge, MA, USA). An isotype matched IgG was used as a negative control. Cross-linking of DNA fragments was reversed by pronase and subsequently incubated at 42℃ for 2 hr and 68℃ for 8 hr. The NOTCH1 promoter DNA in the immunoprecipitates was detected by qRT-PCR. The following primers were used: F:5'-TAGGTCCCTCCCAGCCTTT-3'; R:5'-GCTGATTTATTTCTCCACCACGA-3'.
Plasmids and RNA interference
Knockdown by small hairpin RNA (shRNA) was performed using GV248 vector (GeneChem, Shanghai, China) following the manufacturer's instructions. The following sequences were used: EZH2, 5'-GAAATCTTAAACCAAGAAT-3'; scramble, 5'-TTCTCCGAACGTGTCACGT-3'. Overexpression and reconstitution of EZH2 were achieved using GV230 vector (GeneChem, Shanghai, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect the plasmids into the cells.
Statistical Analysis
Quantitative data were expressed as mean ± standard deviation (SD). Significance was tested by one-way ANOVA or two-tailed t-test in various groups. Differences in proportions were evaluated by the Mann-Whitney U test. Spearman rank correlation analysis was used to analyze the correlation between EZH2 and NOTCH intracellular domain 1 (NICD1) expression. P<0.05 was considered to be statistically significant.
Results
Melatonin Inhibited Proliferation and Self-renewal Ability of GSCs
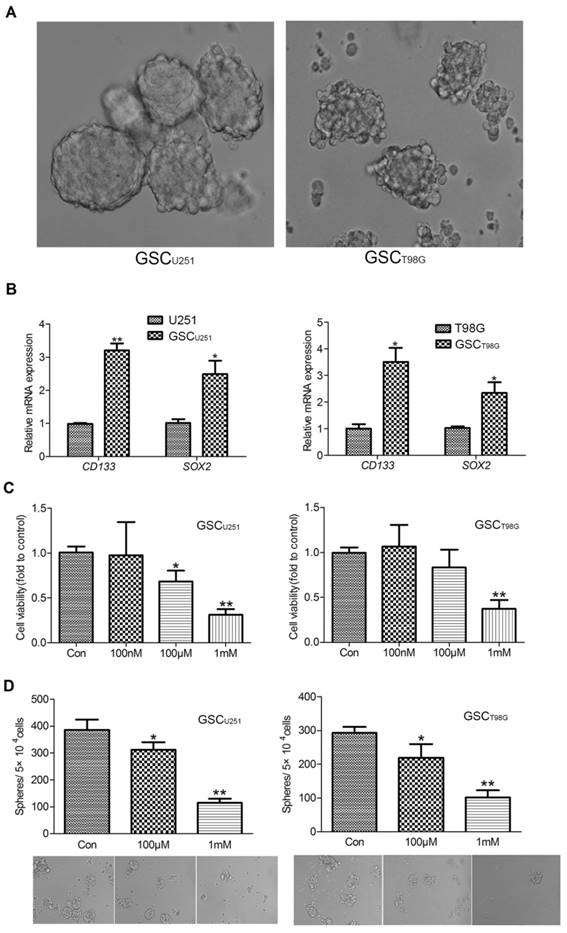
U251 and T98G glioblastoma cell lines were cultured in the stem cell enrichment medium (serum-free, supplemented with B27, bFGF and EGF) for more than 3 weeks until the diameter of the tumor-spheres reached to about 200μm (Figure 1A). We subsequently assessed the expression levels of CD133 and SOX2, two commonly-used GSC markers, in the glioma cell lines in comparison to the tumor-spheres derived from the respective cell line. The results showed that mRNA expressions of CD133 and SOX2 were significantly higher in the tumor-sphere cells (Figure 1B). These cells were then called the GSCU251 and GSCT98G based on their origins and used in the following experiments.
To investigate the effect of melatonin on the viability of GSCs, CCK-8 assay was used to evaluate cell number of the GSCU251 and GSCT98G treated with different concentrations of melatonin (0.1-1000μM). As shown in Figure 1C, melatonin inhibited viability of the GSCs in a dose-dependent manner (Figure 1C). Specifically, both 100μM and 1mM melatonin exerted notable growth-inhibitory effects on the GSCU251 and GSCT98G, even though the effect of 100μM on the GSCT98G is rather moderate.
As mentioned previously, GSCs are characterized by their self-renewal ability. To test the effect of melatonin on GSC self-renewal, sphere-forming assay was performed. About 5×104 dissociated GSCs were planted in each sphere-forming medium supplemented with 100μM or 1mM melatonin. After 10 days, number of the tumor-spheres was counted. As the result depicted in Figure 1D, melatonin significantly decreased formation of the tumor-spheres (Figure 1D). In addition, both mRNA and protein expression levels of the GSC markers, CD133 and SOX2 were also decreased (Figures 2A and 2B), supporting a role of melatonin in the regulation of stem cell properties of GSCs.
Melatonin Inhibited Viability and Self-renewal Ability of GSCs. (A) The representative images of the GSCU251 and GSCT98G (400×). (B) mRNA expression levels of CD133 and SOX2 were determined by qRT-PCR in the GSCU251 and GSCT98G and the original cell lines. Data were presented as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01vs. control. (C) The GSCU251 and GSCT98G were treated with 0, 100nM, 100μM, 1mM melatonin for 48 hr. Cell viability was determined by CCK-8 assay. Data were presented as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01 vs. results of the control w/o melatonin. (D) Morphological changes were shown in the GSCU251 and GSCT98G treated with 0, 100μM, 1mM melatonin for 10 days (200×). Tumor-sphere numbers were calculated. Data were presented as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01 vs. results of the control w/o melatonin.
EZH2 Was Involved in the Inhibitory Role of Melatonin in Regulation of GSC Properties. mRNA (A) and protein (B) levels of CD133, SOX2 and EZH2 were shown in the GSCU251 and GSCT98G treated with 1mM melatonin for 48 hr. Each bar represents mean ± SD from three independent experiments. *P < 0.05, **P < 0.01 vs. control. (C) The GSCU251 transfected with GV230-EZH2 (EZH2) or GV230 alone (Con) were treated with 0, 1mM melatonin for 48 hr. Cell viability was determined by CCK-8 assay. Data were presented as mean ± SD from three independent experiments. *P < 0.05, **P < 0.01. (D) The changes of morphology and tumor-sphere numbers were shown in the indicated cells (200×). Each bar represents mean ± SD from three independent experiments. **P < 0.01. (E) mRNA levels of CD133 were determined by qRT-PCR in the indicated cells. The result represents mean ± SD from three independent experiments. *P < 0.05, **P < 0.01.
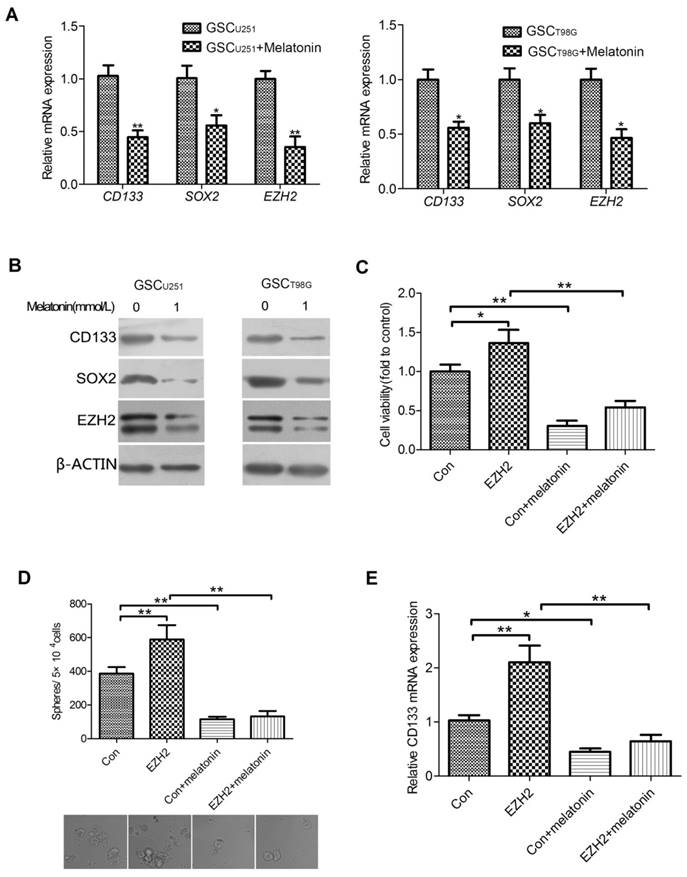
EZH2 Was Involved in the Inhibitory Role of Melatonin in Regulation of GSC Properties
It is well documented that EZH2 plays vital roles in the maintenance of GSCs[17]. Thus, it was hypothesized that the involvement of melatonin in regulation of GSC properties may be linked to EZH2. To test this hypothesis, we first investigated the effect of melatonin on EZH2 expression in the GSCU251 and GSCT98G. The GSCs were treated with 1mM melatonin for 48 hr. qRT-PCR analysis indicated that the mRNA expression of EZH2 was significantly decreased in the melatonin-treated GSCs as compared with those without melatonin (Figure 2A). Correspondingly, EZH2 protein level was markedly reduced (Figure 2B).
To determine whether down-regulation of EZH2 can explain, at least in part, the observed inhibitory effect of melatonin on GSCs, we knocked down EZH2 in the GSCU251 by introducing a GV248 vector of EZH2 shRNA. The results showed that depletion of EZH2 inhibited viability as well as self-renewal of the GSCU251 (Figures S1A and S1B). Meanwhile, CD133, the important GSC marker was also down-regulated (Figure S1C), all in consistency with the changes of GSCs treated with the effective concentrations of melatonin. On the other hand, when EZH2 was up-regulated in the GSCU251, the proliferative and self-renewal abilities of these cells were elevated (Figures 2C and 2D, lane2). A similar result was manifested with the mRNA level of CD133 (Figure 2E, lane2). More importantly, 1mM melatonin lowered GSC viability or self-renewal to a comparable level in both control and EZH2-overexpressed cells (Figures 2C and 2D, lane3,4). Likewise, the mRNA expression of CD133 were significantly reduced (Figure 2E, lane3,4). Collectively, these data suggested that EZH2 was not only required for the maintenance of GSCs, but also involved in the inhibitory role of melatonin in regulation of GSC properties.
NOTCH1 Was an EZH2 Target Gene Involved in the Effects of Melatonin
Global gene expression profile changes were analyzed in the GSCU251 knockdown of EZH2. Interestingly, NOTCH1 was one of the most significantly down-regulated genes which correlated with EZH2 depletion as evidenced by a 2.7-fold decrease in transcripts (unpublished data). As it is well established that NOTCH signaling plays a critical role in controlling GSC survival and multi-potency[24, 25], we speculated that NOTCH1 is involved in the EZH2-mediated effects of melatonin in the GSCs. To test this idea, qRT-PCR was performed in the GSCU251 and GSCT98G to assess the mRNA expressions of NOTCH family and NOTCH signaling pathway genes. The results showed that melatonin most significantly reduced mRNA expression of NOTCH1 in both of the GSCs compared with other NOTCH receptors. NOTCH1 signaling pathway components, such as CCND1, CCNE2 and HES1, were also deregulated (Figure 3A). Consistently, immunoblot analysis showed that the expressions of NOTCH intracellular domain 1 (NICD1) and HES1 were remarkably decreased when EZH2 was inhibited by melatonin (Figure 3B).
To further address whether NOTCH1 signaling is indeed regulated by EZH2 in the GSCs, dual-luciferase reporter assay was performed. As the result shown in Figure 3C, a notable reduction in the luciferase activity of NOTCH1 promoter region was coupled with knockdown of endogenous EZH2, whereas up-regulation of EZH2 in the GSCU251 increased NOTCH1 transcriptional activity (Figure 3C). A comparable result was observed in the GSCT98G (data not shown), confirming a positive role of EZH2 in NOTCH1 trans-activation. Moreover, immunoblot detected significant inhibition of NICD1 and HES1 protein levels in the GSCs depleted of EZH2. Over-expression of EZH2, on the other hand, resulted in an opposite effect, suggesting a functional control of EZH2 over NOTCH1 activity (Figure 3D).
We next asked the specific mechanism of EZH2 in NOTCH1 trans-activation. Chromatin immunoprecipitation(ChIP) assay was conducted in the GSCU251 by using the antibodies detecting EZH2, tri-methylation of histone 3 at lysine 27 (H3K27me3) or suppressor of zeste 12 (SUZ12), another key component of Polycomb Repressive Complex 2 (PRC2). Interestingly, significant enrichment for endogenous EZH2 protein was detected at the NOTCH1 promoter, which, however, can be blocked with the addition of 1mM melatonin. In contrast, no detection of H3K27me3 or SUZ12 occurred at the promoter region of NOTCH1 (Figure 3E). These data indicated that EZH2 may function through directly binding to the NOTCH1 promoter in the GSCs and this effect is independent of H3K27me3 or PRC2.
Clinical Relevance Existed between EZH2 and NOTCH1 Activation
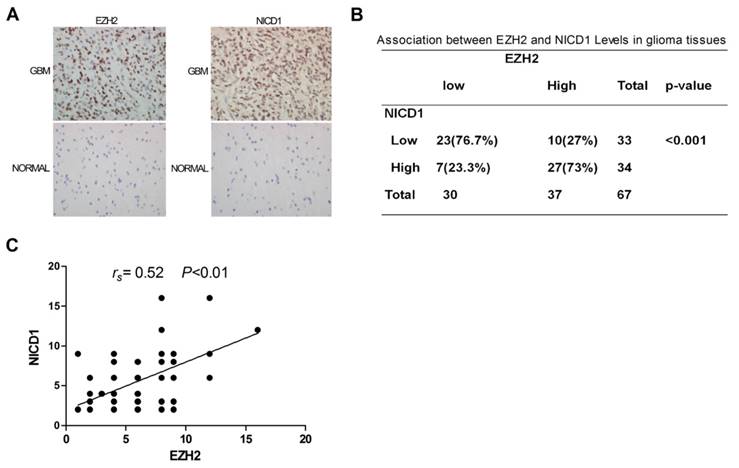
To probe for evidence of clinical relevance between EZH2 and NOTCH1 activation, 67 GBM specimens and 12 normal human brain tissues were analyzed for the expressions of EZH2 and NOTCH intracellular domain 1 (NICD1) by immuno-histochemistry (IHC). The results showed that remarkable up-regulation of these two proteins was noted in 37 (EZH2) or 34 (NICD1) of 67 GBM specimens compared to normal brain specimens (P<0.01; Figure 4A), among which 73% of the EZH2high specimens were also NICD1high ( P< 0.001; Figure 4B). Additionally, there was significant positive correlation between expressions of EZH2 and NICD1, evidently supporting NOTCH1 a potential target of EZH2 in human GBMs (Figure 4C).
NOTCH1 Was an EZH2 Target Gene Involved in the Effects of Melatonin. (A) The GSCU251 and GSCT98G were treated with 1mM melatonin for 48 hr. mRNA expression levels of NOTCH family and NOTCH signaling pathway genes were determined by qRT-PCR. Data were presented as mean ± SD from three independent experiments. *P < 0.05 vs. control. (B) Protein levels of EZH2, NICD1 and HES1 were analyzed by immunoblot in the GSCU251 and GSCT98G treated with 1mM melatonin for 48 hr. β-ACTIN served as a loading control. (C) The GSCU251 were transfected with pGL2-NOTCH1; a vector-encoding EZH2-shRNA (shEZH2) or scrambled-shRNA (shSC), or a vector-encoding wild-type EZH2 (EZH2) or its comparative control (Con); and pSV-Renilla. Values in graphs represent mean of FLuc:RLuc activity ± SD performed in triplicate. *P < 0.05 vs. control. (D) Protein levels of NOTCH intracellular domain 1(NICD1) and HES1 were examined by immunoblot in the EZH2 depleted (shEZH2) or over-expressed (EZH2) GSCs. β-ACTIN was used as a loading control. (E) The GSCU251 were treated with 0, 1mM melatonin for 48 hr. ChIP analysis was performed by using antibodies against EZH2, tri-methylation of histone 3 at lysine 27 (H3K27me3) or suppressor of zeste 12 (SUZ12) with primers targeted to the promoter region of NOTCH1. Isotype matched IgG was used as a negative control. Data are presented as mean ± SD from three independent experiments. *P < 0.05, **P<0.01 vs. control.

Clinical Relevance Existed between EZH2 and NOTCH1 Activation. (A) IHC was performed to detect EZH2 and NOTCH intracellular domain 1(NICD1) levels in the GBM specimens and samples of normal brain specimens (400×). (B) Distribution of EZH2 and NICD1 expression in 67 GBM specimens. (P < 0.001). (C) Correlation between expression of EZH2 and NICD1 in 67 GBM specimens (P < 0.01).
Discussion
In view of the potent tumor-propagation capacity of GSCs, GSCs-targeted therapies have attracted increasing attention in recent years[26-28]. Here, we show that melatonin inhibited GSCs viability and self-renewal accompanied by a decrease in the expression of stem cell markers. Furthermore, we found a reduction in the expression levels of EZH2 and NOTCH1 signaling pathway components, therefore adding a new perspective to interpret the anti-tumor effects of melatonin.
EZH2 is an important epigenetic regulator that is implicated in the maintenance of tumor stem cells. It was reported that overexpression of EZH2 is frequently observed in malignant gliomas and GSCs[15]. Inhibition of EZH2 disrupted the morphological conversion and impaired tumorigecity of the GSCs in NOD-SCID mice[17]. In this study, we demonstrated that EZH2 expression is correlated with GSC propagation, self-renewal as well as GSC marker expression, providing further direct evidence for the function of EZH2 in modulating the “stemness” of the GSCs. More strikingly, melatonin lowered GSC viability or self-renewal to a comparable level in both control and EZH2-overexpressed cells. A similar result was manifested with the expression level of CD133, suggesting that the effects exerted by melatonin in the GSCs was, at least in part, dependent on EZH2 expression.
To probe for the downstream target of EZH2 in the melatonin-mediated effects, SAGE database analysis of potential EZH2 target genes were examined. NOTCH1 was one of the most significantly down-regulated genes. Coincidentally, melatonin remarkably reduced NOTCH1 in the GSCs as well as other NOTCH1 signaling pathway components (e.g. CCND1, CCNE2 and HES1), which are regulated by NOTCH1. The active NOTCH1 protein segment, NOTCH intracellular domain 1 (NICD1) was also deregulated, suggesting that NOTCH1 might mediate the effects of EZH2 upon treatment of melatonin.
We next asked the specific mechanism of EZH2 in NOTCH1 trans-activation. Substantial studies have indicated that EZH2 usually functions via transcriptional repression through its histone methylation activity on H3K27. For instance, biological interconversion between GSCs and differentiated non-GSCs is associated with the gain or loss of EZH2/PRC2-mediated tri-methylation of histone 3 at lysine 27 (H3K27me3) on pluripotent or development-associated genes (e.g. NANOG, WNT1, BMP5)[29]. Given the high divergent effects of EZH2 depending on context and tumor cell-type specificities, more studies began to reveal a H3K27me3-independent role of EZH2 in the GSCs. Suvà et al. reported that C-MYC was a downstream target of EZH2 by direct binding to C-MYC promoter via SUZ12 [17]. Kim et al. stated that AKT-mediated phosphorylation at S21 in EZH2 facilitated STAT3 methylation, resulting in increased tyrosine phosphorylation and activation of STAT3 in the GSCs[3, 30]. In our study, we found that EZH2 regulated NOTCH1 expression by directly interacting with the NOTCH1 promoter. This result is consistent with the H3K27me3-independent effects of EZH2 in the above studies and suggested a specific mechanism for melatonin in the anti-GSCs effects. Furthermore, significant correlation between the expressions of EZH2 and NOTCH intracellular domain 1 (NICD1) was observed in the tumor samples from GBM patients, evidently supporting the existence of EZH2-NOTCH1 signaling pathway in the malignant gliomas.
In conclusion, this study demonstrated that melatonin, a tumor inhibitor, performs its function partly by regulating GSC properties through EZH2-NOTCH1 signaling axis. As for the clinical application of melatonin in GBM, the concentration applied in vitro is much higher than the concentration given to patients. So the effective concentration could be achieved through local drug delivery, which need further study in future.
Supplementary Material
Figure S1.
Abbreviations
GSC: Glioblastoma stem-like cell; GBMs: glioblastomas multiforme; PcG: polycomb group; H3K27me3: tri-methylation of histone 3 at lysine 27; bFGF: basic fibroblast growth factor; EGF: epidermal growth factor; CCK-8: Cell counting kit-8; IHC: immunohistochemistry; CHIP: chromatin immunoprecipitation; shRNA: small hairpin RNA; NICD1: NOTCH intracellular domain 1; SUZ12: suppressor of zeste 12; PRC2: Polycomb Repressive Complex 2.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81471517), Natural Science Foundation of Shandong Province (ZR2013HQ060 and ZR2014HM008), Science and Technology Development Project of Shandong Province, China (2014GGB14439 and 2014GGB14257).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hemmati HD, Nakano I, Lazareff JA. et al. Cancerous stem cells can arise from pediatric brain tumors. P Natl Acad Sci USA. 2003;100:15178-83
2. Bao S, Wu Q, McLendon RE. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756-60
3. Kim E, Kim M, Woo DH. et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839-52
4. Chen J, Li Y, Yu TS. et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522-6
5. Lewy AJ. Melatonin and human chronobiology. Cold Spring Harb Sym. 2007;72:623-36
6. Carrillo-Vico A, Lardone PJ, Alvarez-Sanchez N. et al. Melatonin: buffering the immune system. Int J Mol Sci. 2013;14:8638-83
7. Mauriz JL, Collado PS, Veneroso C. et al. A review of the molecular aspects of melatonin's anti-inflammatory actions: recent insights and new perspectives. J Pineal Res. 2013;54:1-14
8. Manchester LC, Coto-Montes A, Boga JA. et al. Melatonin: an ancient molecule that makes oxygen metabolically tolerable. J Pineal Res. 2015;59:403-19
9. Hill SM, Belancio VP, Dauchy RT. et al. Melatonin: an inhibitor of breast cancer. Endocr-Relat Cancer. 2015;22:R183-204
10. Motilva V, Garcia-Maurino S, Talero E. et al. New paradigms in chronic intestinal inflammation and colon cancer: role of melatonin. J Pineal Res. 2011;51:44-60
11. Xin Z, Jiang S, Jiang P. et al. Melatonin as a treatment for gastrointestinal cancer: a review. J Pineal Res. 2015;58:375-87
12. Martin V, Herrera F, Carrera-Gonzalez P. et al. Intracellular signaling pathways involved in the cell growth inhibition of glioma cells by melatonin. Cancer Res. 2006;66:1081-8
13. Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor-a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2:349-50
14. Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613-8
15. Bian EB, Li J, He XJ. et al. Epigenetic modification in gliomas: role of the histone methyltransferase EZH2. Expert Opin Ther Tar. 2014;18:1197-206
16. Kim SH, Joshi K, Ezhilarasan R. et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Rep. 2015;4:226-38
17. Suva ML, Riggi N, Janiszewska M. et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211-8
18. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770-6
19. Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598-608
20. Shao S, Zhao X, Zhang X. et al. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer. 2015;14:28
21. Bao B, Ali S, Banerjee S. et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335-45
22. Liu Q, Wang L, Wang Z. et al. GRIM-19 opposes reprogramming of glioblastoma cell metabolism via HIF1alpha destabilization. Carcinogenesis. 2013;34:1728-36
23. Fu Y, Chen J, Pang B. et al. EZH2-induced H3K27me3 is associated with epigenetic repression of the ARHI tumor-suppressor gene in ovarian cancer. Cell Biochem Biophys. 2015;71:105-12
24. Gursel DB, Berry N, Boockvar JA. The contribution of Notch signaling to glioblastoma via activation of cancer stem cell self-renewal: the role of the endothelial network. Neurosurgery. 2012;70:N19-21
25. Zhu TS, Costello MA, Talsma CE. et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061-72
26. Nakano I. Stem cell signature in glioblastoma: therapeutic development for a moving target. J Neurosurg. 2015;122:324-30
27. Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem Pharmacol. 2010;80:654-65
28. Safa AR, Saadatzadeh MR, Cohen-Gadol AA. et al. Emerging targets for glioblastoma stem cell therapy. J Biomed Res. 2015 [Epub ahead of print]
29. Natsume A, Ito M, Katsushima K. et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013;73:4559-70
30. Fouse SD, Costello JF. Cancer Stem Cells Activate STAT3 the EZ Way. Cancer Cell. 2013;23:711-3
Author contact
![]() Corresponding authors: Dr. Qian Liu, Department of Histology and Embryology, Shandong University School of Medicine, 44# Wenhua Xi Road, Jinan, 250012, Shandong, China. Tel/Fax: 86-531-88382047, E-mail: qian_liu1980com. Dr. Qi Pang, Department of Neurosurgery, Shandong Provincial Hospital Affiliated to Shandong University, 324# Jingwu Road, Jinan, 250021, Shandong, China. Tel/Fax: 86-531-87938911, E-mail: pangqiedu.cn
Corresponding authors: Dr. Qian Liu, Department of Histology and Embryology, Shandong University School of Medicine, 44# Wenhua Xi Road, Jinan, 250012, Shandong, China. Tel/Fax: 86-531-88382047, E-mail: qian_liu1980com. Dr. Qi Pang, Department of Neurosurgery, Shandong Provincial Hospital Affiliated to Shandong University, 324# Jingwu Road, Jinan, 250021, Shandong, China. Tel/Fax: 86-531-87938911, E-mail: pangqiedu.cn

 Global reach, higher impact
Global reach, higher impact