10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(2):254-264. doi:10.7150/ijbs.17053 This issue Cite
Research Paper
Sirtuin 3 is required for osteogenic differentiation through maintenance of PGC-1ɑ-SOD2-mediated regulation of mitochondrial function
1. Department of Orthopedic, Tangdu Hospital, Fourth Military Medical University, #569 Xinsi Road, Xi'an 710038, China;
2. Department of Orthopedic, BaoJi Centre Hospital, #8 Jiangtan Road, Bao ji 721008, China.
Received 2016-8-1; Accepted 2016-10-19; Published 2017-2-12
Abstract
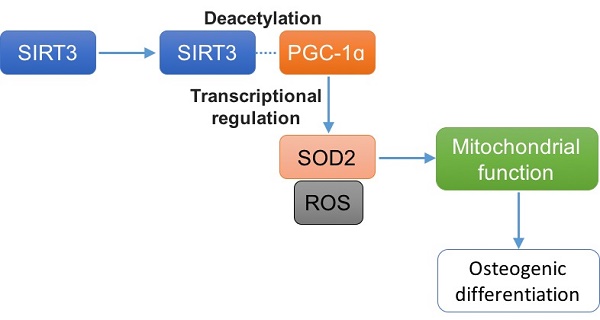
Osteogenic differentiation is crucial for the maintenance of bone homeostasis. Sirtuin 3 (SIRT3), a member of sirtuins family, functions as a critical deacetylase that regulates many key proteins. In the current study, we aimed to clarify the role of SIRT3 in osteogenic differentiation and the possible mechanisms, using mouse pre-osteoblastic MC3T3-E1 cells. Expression of SIRT3 was substantially increased in differentiated MC3T3-E1 cells. Knock down of SIRT3 significantly decreased alkaline phosphatase (ALP) staining, and mRNA expression of runt-related transcription factor 2 (Runx2) and collagen type I ɑ 1 (Col1ɑ1), and osteocalcin in differentiated MC3T3-E1 cells. Overexpression of wild type but not mutant SIRT3 could reverse SIRT3 knockdown-resulted decrease of ALP staining. Complex I, II, III, IV, and V activities, oxygen consumption and mitochondrial membrane potential were significantly decreased by SIRT3 knockdown. Moreover, SIRT3 knockdown reduced mitochondrial density, increased mitochondrial size and decreased the expression of NRF1 and TFAM. Knock down of SIRT3 decreased mRNA and protein expression of SOD2 and increased ROS level. Overexpression of SOD2 significantly suppressed SIRT3 knockdown-induced decrease of mitochondrial function and osteogenic differentiation. SIRT3 knockdown resulted in a significant decrease of PGC-1ɑ protein expression but not mRNA expression. Overexpression of wild type but not mutant SIRT3 could reverse SIRT3 knockdown-resulted decrease of PGC-1ɑ protein expression. Moreover, we detected a direct interaction between SIRT3 and PGC-1ɑ and SIRT3 knockdown reduced SIRT3 and PGC-1ɑ interaction, resulting in a reduction of PGC-1ɑ protein stability and PGC-1ɑ-binding in the promoters of SOD2. Overexpression of PGC-1ɑ blocked SIRT3 knockdown-induced decrease of SOD2 expression, increase of ROS level, and decrease of mitochondrial function and biogenesis, leading to improvement of osteogenesis. Overall, the data provide a better understanding of the role of SIRT3 in osteogenic differentiation.
Keywords: sirtuin 3, osteogenic differentiation, peroxisome proliferator-activated receptor-γ co-activator-1ɑ, superoxide dismutase 2, mitochondrial function
Introduction
Osteoporosis is characterized by bone metabolic disorder, including decrease of bone mineral density, increase of bone loss and higher risk of fractures, which is believed to affect nearly 50 % post-menopausal women over the age of 50 [1]. In general, bone structure and function is controlled by the balance between bone resorption and formation. Thus, the dynamic interaction between osteoblast-induced bone formation and osteoclast-induced bone resorption is critical for homeostasis of bone metabolism. In response to various genetic or environmental factors, bone resorption could be activated and bone formation could be inhibited, leading to bone metabolic disorder such as osteoporosis [2, 3]. Therefore, osteogenic differentiation, a key process of osteoblast-induced bone formation, is crucial for the maintenance of bone function and regulation of osteogenic differentiation is an effective method for treating diseases characterized by reduction of bone formation, including osteoporosis [4].
The sirtuins (SIRTs) family, which are the mammalian homologues of the Sir2α in yeast, is a group of evolutionarily highly-conserved nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases that regulate a battery of cellular events including growth, DNA duplication and repair, mitochondrial biogenesis, oxidative stress, apoptosis, and autophagy, and are indeed involved in various pathological conditions, including metabolic disorder, cancer, neurodegeneration, and aging, etc. [5-9]. To date, it is known that SIRTs contains 7 members, named SIRT1-7. Among them, SIRT3-5 play important roles in maintanence of mitochondrial function through regulating a variety of proteins that are responsible for mitochondrial metabolism [10]. We paid attention to SIRT3, which has been reported to deacetylate many key proteins regulating mitochondrial biogenesis, reactive oxygen species (ROS) generation, elimination and metabolism in mitochondria [11-13]. Moreover, it is found that SIRT3 could affect mitochondrial respiratory chain and thus regulate ATP-production [14, 15]. Recently, Huh et al. found that SIRT3 played a role in maintaining bone homeostasis [16]. They found that osteoclast differentiation was increased by silence of SIRT3, and that bone mass was decreased due to an increased number of osteoclasts in SIRT3-defficient mice [16]. However, the role of SIRT3 in osteogenic differentiation is not known.
The current study was designed to investigate the role of SIRT3 in osteogenic differentiation and explore the possible mechanisms, using mouse pre-osteoblastic MC3T3-E1 cells. We found that knock down of SIRT3 inhibited osteogenic differentiation via decreasing the stability of peroxisome proliferator-activated receptor-γ co-activator-1ɑ (PGC-1ɑ) and downregulation of superoxide dismutase 2 (SOD2) expression, and subsequently increasing ROS level and decreasing mitochondrial biogenesis and mitochondrial function.
Materials and methods
Cell culture
Mouse MC3T3-E1 cells were obtained from ATCC (Manassas, USA). Cells were cultured in ɑ-MEM (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone) at 37°C in an incubator containing 5% CO2 atmosphere. For the induction of osteogenic differentiation, MC3T3-E1 cells were cultured in osteogenic medium (a-MEM supplemented with 10% FBS, 20 mM β-glycerophosphate, 100 mg/mL ascorbic acid, and 10 nM dexamethasone (Sigma-Aldrich, USA)) for 12 days. The efficiency of differentiation was evaluated by staining with alkaline phosphatase (ALP).
Lentivirus and plasmid transfection
MC3T3-E1 cells were transfected with lentivirus carrying shSIRT3 (GeneChem, China) and purified with puromycin to establish cell lines in which SIRT3 expression was stably downregulated. For SOD2 or PGC-1ɑ overexpression, SOD2 or PGC-1ɑ was purified and amplified from mouse genomic DNA, and then cloned into a pCMV vector (Beyotime, China). For the transient overexpression of SIRT3, the pcDNA3.1-hSIRT3-WT-Flag plasmid (wild type SIRT3, Addgene) and pcDNA3.1-hSIRT3-H248Y-HA plasmid (SIRT3 HY, SIRT3 loss of deacetylase activity, Addgene) were obtained. pcDNA3.1-hSIRT3-WT-Flag (500 ng), pcDNA3.1-hSIRT3-H248Y-HA (500 ng), pCMV-SOD2 (500 ng) or pCMV-PGC-1ɑ (500 ng) plasmids and their empty vectors were transfected into cells using Lipofectamine 2000 (4 μl) (Invitrogen, USA) according to the manufacturer's instructions. Transfection of plasmids was conducted 2 days before the induction of osteogenic differentiation.
Real-time PCR
Total RNA was extracted from cells using a RNA isolation kit (TaKaRa, China). cDNA was synthesized using 500ng mRNA by a PrimeScriptTM RT reagent Kit (TaKaRa, China). qPCR was performed in a 20 μl reaction system using SYBR® Premix Ex Taq (TaKaRa, China). The Bio-Rad CFX96 Real-time PCR system (Bio-Rad, USA) was used to record and analyze the signal. The expression of target genes was normalized to β-actin, a house-keeping gene. The PCR reaction conditions were as follows: initial denaturation at 95 °C for 10 min followed by 30 cycles at 95 °C for 1 min, annealing at 53 °C for 1 min, extension at 72 °C for 1 min, and final extension at 72 °C for 5 min. The comparative cycle threshold (CT) (2-ΔΔCT) method was used to quantify the relative amount of RNA. The primers used in the study were shown in Table 1.
Western blot
We use RIPA buffer (Thermo Scientific, USA) to extract protein from cells according to the manufacture's protocols. After determination of protein concentration using BCA assay (Thermo Scientific, USA), the supernatants were mixed with loading buffer. In brief, the mixture containing 20 μg protein were used for separation on 10% SDS-PAGE, and then electro-transferred onto NC membranes (Millipore, USA). The membranes were then blocked in 5% non-fat milk for 1 h at room temperature and then incubated with primary antibodies (β-actin (Santa Cruz): 1: 500; SOD2 (Cell Signaling Technology): 1: 1000; SIRT3 (Cell Signaling Technology): 1: 1000; PGC-1ɑ (Abcam): 1:1000) overnight at 4 °C. After that, the membranes were incubated with HRP-conjugated goat anti-rabbit secondary antibody (1:5000. Thermo Scientific, USA) for 30 min at 37 °C. The bands were detected using an enhanced chemiluminescence reagent kit (Thermo Scientific, USA).
Primers sequences
| Gene | Forward | Reverse |
|---|---|---|
| β-actin | 5' AGGCCAACCGTGAAAAGATG 3' | 5' TGGCGTGAGGGAGAGCATAG 3' |
| SOD2 | 5' CTCAGGTCGCTCTTCAGC 3' | 5' AGCCTCCAGCAACTCTCC 3' |
| NRF-1 | 5' TCCATCTATCCGAAAGAG 3' | 5' GCCAAACACCTTGAAGAC 3' |
| PGC-1α | 5' AACAATGAGCCTGCGAAC 3' | 5' CCTCGTTGTCAGTGGTCA 3' |
| Runx2 | 5' TTCTCCAACCCACGAATGCAC 3' | 5' CAGGTACGTGTGGTAGTGAGT 3' |
| OCN | 5' CTGACAAAGCCTTCATGTCCAA 3' | 5' GCGCCGGAGTCTGTTCACTA 3' |
| Col1a1 | 5' GGGGCAAGACAGTCATCGAA 3' | 5' GAGGGAACCAGATTGGGGTG 3' |
| SIRT3 | 5' TTTCTTTCACAACCCCAAGC 3' | 5' AGGGATCCCAGATGCTCTCT 3' |
Evaluation of mitochondrial function
Transmission electron microscopy (TEM) was used to observe mitochondrial ultrastructure as previously described [17]. Cells were analyzed stereologically and mitochondrial density and size were calculated. Complex I, II, III, IV, and V activities were determined using the methods reported by Elschami et al [17]. Oxygen consumption activity was evaluated by the determination of complex II driven respiration as previously described [17]. For the determination of mitochondrial membrane potential, cells were incubated with rhodamine 123 (Rho123, 10μM) for 20 min at 37 °C. Subsequently, images were captured under confocal microscope (Olympus, Japan).
ROS determination
ROS level was detected by dihydroethidium (DHE), a superoxide sensitive probe. In brief, cells were incubated with 10 μM DHE at 37℃ for 30 min in dark environment. Subsequently, images were captured under confocal microscope (Olympus, Japan).
Chromatin Immunoprecipitations
Chromatin Immunoprecipitations (CHIP) was conducted to evaluate the transcriptional activity of PGC-1ɑ in the regulation of SOD2 using commercial kit (Thermo Scientific, USA). Cells were cross-linked using formaldehyde (1%) for 10 min, harvested, lysed in SDS lysis buffer, and then sheared by sonication. Sonicated cell lysate was immunoprecipitated using anti-PGC-1ɑ antibody (Cell Signaling Technology, USA) or normal mouse IgG (Abcam). DNA was extracted and used as templates for real-time PCR to detect the content of SOD2 promoters and the results were normalized to the respective input signals and expressed as folds of control.
Co-immunoprecipitation
Protein/protein interaction was evaluated by co-immunoprecipitation (Co-IP) according to manufacturer's protocols (Thermo Scientific, USA). Briefly, cells were lysed, centrifuged and used for immunoprecipitation. A small fraction of the supernatants was used for Input analysis, and the remainder were incubated with rabbit anti-SIRT3 antibody to couple the complex. After that, the extracts (immunoprecipitates) were then subjected to 10% SDS-PAGE. Western blot was then performed to detect SIRT3 and PGC-1ɑ expression in immunoprecipitates.
Statistical analysis
Data were obtained from at least three independent experiments and expressed as mean ± SEM. Results were analyzed using Graph Pad Prism. Statistical analysis was performed by one-way analysis of variance followed by a SNK-q test for multiple comparisons. Student's t-test was performed for comparison between two groups. P < 0.05 was considered to be statistically significant.
Results
SIRT3 is required for the osteogenic differentiation in MC3T3-E1 cells
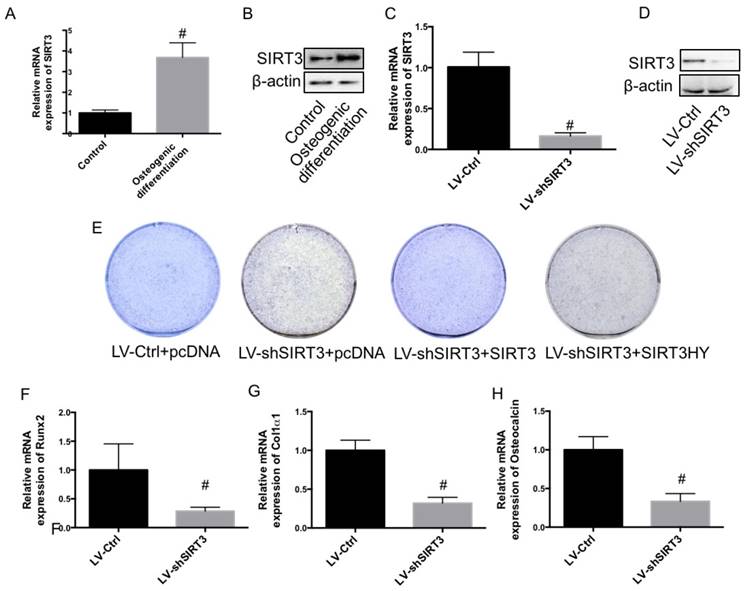
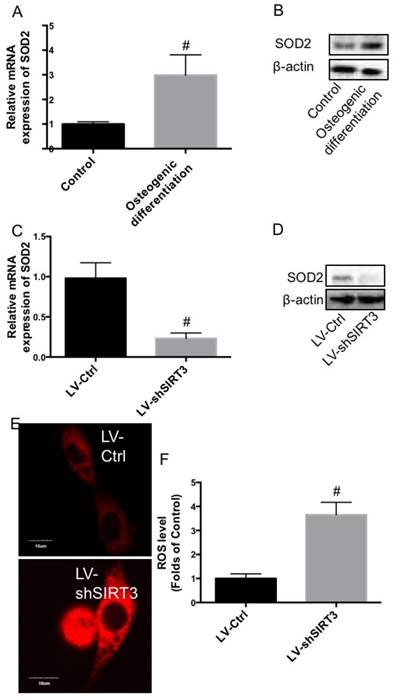
In the current study, using mouse pre-osteoblastic MC3T3-E1 cells, we investigated the role of SIRT3 in osteogenic differentiation. Compared with undifferentiated MC3T3-E1 cells, both mRNA and protein expression of SIRT3 was significantly increased in differentiated MC3T3-E1 cells (Fig. 1A and B), indicating a possible role of SIRT3 in osteogenic differentiation. MC3T3-E1 were transfected with lentivirus carrying shSIRT3 to stably knock down the expression of SIRT3. We showed that mRNA and protein expression of SIRT3 was significantly inhibited by knock down of SIRT3 (Fig. 1C and D). The results showed that knock down of SIRT3 significantly inhibited the osteogenic differentiation, as shown as decreased alkaline phosphatase (ALP) staining (Fig. 1E). Re-expression of SIRT3 in shSIRT3-transfected cells significantly reversed the decrease of ALP staining (Fig. 1E). However, Re-expression of SIRT3 loss of deacetylase activity could not alter the ALP staining (Fig. 1E), indicating that deacetylase activity of SIRT3 was required for SIRT3-exhibited regulation of osteogenic differentiation. Moreover, mRNA expression of key regulators of osteogenic differentiation, including runt-related transcription factor 2 (Runx2) and collagen type I ɑ 1 (Col1ɑ1), and osteocalcin (OCN), were markedly suppressed by SIRT3 knockdown, compared with control cells (Fig. 1E, F and G). The results indicated that SIRT3 expression and its deacetylase activity are required for osteogenic differentiation in MC3T3-E1 cells.
Knock down of SIRT3 decreased mitochondrial function and biogenesis in differentiated MC3T3-E1 cells
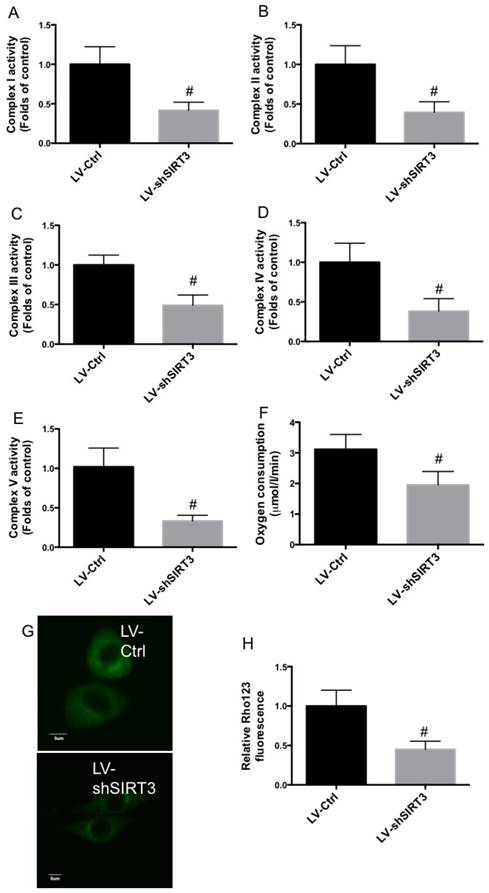
To explore the possible mechanism underlying the role of SIRT3 in the regulation of osteogenic differentiation, mitochondrial function and biogenesis was evaluated. As shown in Fig. 2A-E, Complex I, II, III, IV, and V activities were significantly decreased by SIRT3 knockdown, compared with that of control. In addition, knock down of SIRT3 notably inhibited oxygen consumption rate in differentiated MC3T3-E1 cells (Fig. 2F). Moreover, downregulation of SIRT3 markedly reduced Rho123 staining in differentiated MC3T3-E1 cells, indicating the reduction of mitochondrial membrane potential (Fig. 2G and H). The results indicated that downregulation of SIRT3 significantly decreased mitochondrial function.
Effect of knockdown of SIRT3 on osteogenic differentiation. MC3T3-E1 cells were induced to osteogenic differentiation, and SIRT3 expression was determined in pre-osteogenic MC3T3-E1 cells and differentiated MC3T3-E1 cells using real-time PCR (A) and western blot (B). MC3T3-E1 cells were transfected with lentivirus carrying shSIRT3 and SIRT3 expression was stably knocked down. mRNA (C) and protein (D) expression of SIRT3 was measured using real-time PCR and western blot, respectively. LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were transfected with SIRT3 or SIRT3HY plasmids and then induced to osteogenic differentiation. Osteogenic differentiation was measured using ALP staining (E). mRNA expression of Runx2 (F), Col1α1 (G), and OCN (H) in differentiated MC3T3-E1 cells transfected with LV-Ctrl and LV-shSIRT3 were determined by real-time PCR. #P<0.05, compared with control.
Effect of knockdown of SIRT3 on mitochondrial function. LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were induced to osteogenic differentiation, and Complex I (A), II (B), III (C), IV (D), and V (E) activities were determined and results were shown as folds of control. Oxygen consumption activity was evaluated by the determination of complex II driven respiration and results were expressed as µmol/l/min (F). Mitochondrial membrane potential was measured using Rho123 staining (G and H). Representative images were shown (G). Relative Rho123 fluorescence was calculated (H). #P<0.05, compared with control.
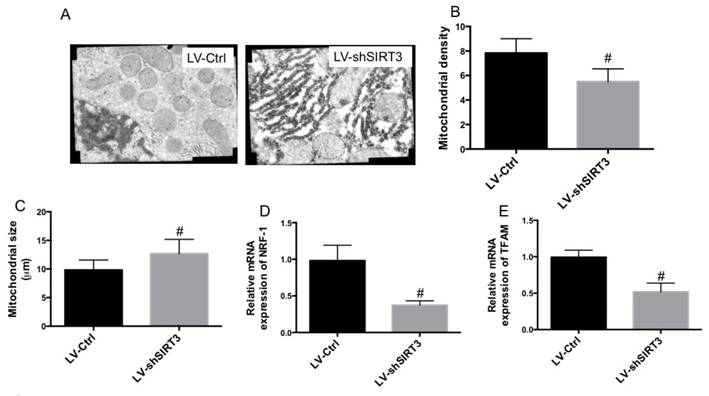
TEM was used to observe mitochondrial ultrastructure. In Fig. 3A, we found swollen mitochondria and broken mitochondrial cristae in MC3T3-E1 cells with knock down of SIRT3. The results also showed that downregulation of SIRT3 markedly reduced mitochondrial density and increased mitochondrial size (Fig. 3B and C). Moreover, the mRNA expression of key regulators of mitochondrial biogenesis, transcription factor nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (TFAM), was determined. The results showed that downregulation of SIRT3 markedly reduced the mRNA expression of both NRF1 and TFAM (Fig. 3D and E). The results indicated that downregulation of SIRT3 significantly decreased mitochondrial biogenesis. The results demonstrated that reduction of mitochondrial function and biogenesis may be involved in SIRT3 knockdown-mediated inhibition of osteogenic differentiation.
Downregulaton of SOD2 was involved in SIRT3 knockdown-mediated inhibition of osteogenic differentiation
ROS homeostasis is closely associated with mitochondrial function and biogenesis. In the next step, we further examined the effect of knock down of SIRT3 on the expression of SOD2, an important mitochondrial antioxidant. In Fig. 4A and B, we showed that compared with undifferentiated MC3T3-E1 cells, both mRNA and protein expression of SOD2 was significantly increased in differentiated MC3T3-E1 cells, showing a similar changing pattern with SIRT3. Downregulation of SIRT3 notably reduced the mRNA and protein expression of SOD2 (Fig. 4C and D). Moreover, knock down of SIRT3 markedly increased DHE staining, indicating the increase of ROS level (Fig. 4E and F). The data indicated that knock down of SIRT3 disturbed ROS homeostasis through downregulation of SOD2.
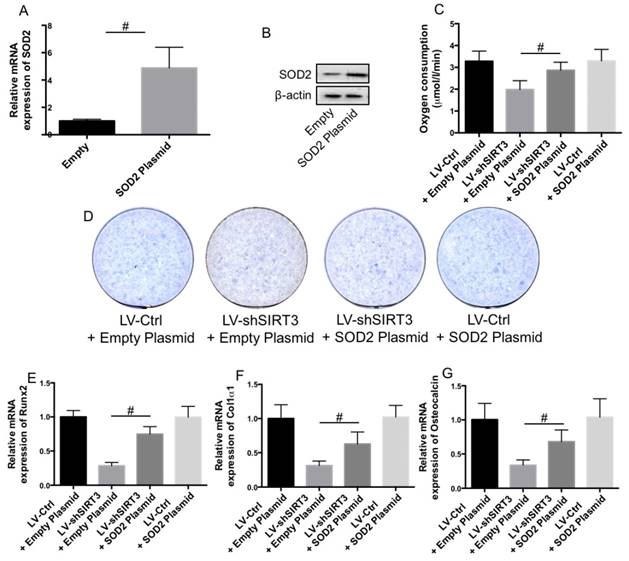
In order to evaluate whether downregulation of SOD2 was involved in SIRT3 knockdown-induced reduction of mitochondrial function and biogenesis and inhibition of osteogenic differentiation, LV-Ctrl and LV-SIRT3 MC3T3-E1 cells were transfected with plasmids carrying SOD2 or empty plasmids. SOD2 expression was significantly increased by plasmid transfection (Fig. 5A and B). Overexpression of SOD2 notably suppressed SIRT3 knockdown-induced reduction of oxygen consumption activity (Fig. 5C). Furthermore, Overexpression of SOD2 significantly blocked the reduction of ALP staining (Fig. 5D) and mRNA expression of Runx2 (Fig. 5E), Col1ɑ1 (Fig. 5F), and OCN (Fig. 5G), indicating that downregulation of SOD2 was involved in SIRT3 knockdown-induced reduction of mitochondrial function and biogenesis and inhibition of osteogenic differentiation.
Effect of knockdown of SIRT3 on mitochondrial ultrastructure and biogenesis. LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were induced to osteogenic differentiation. TEM was used to observe mitochondrial ultrastructure (A). Mitochondrial density (B) and size (C) were calculated. mRNA expression of NRF-1 (D) and TFAM (E) was examined using real-time PCR and results were shown as folds of control. #P<0.05, compared with control.
Reduction of PGC-1ɑ stability was involved in SIRT3 knockdown-mediated disturbance of ROS homeostasis
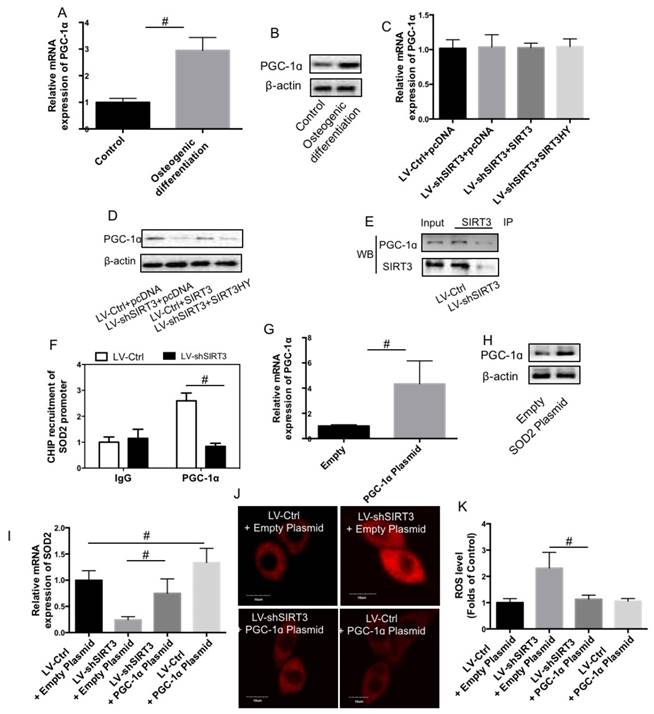
In order to examine the mechanism of SIRT3 knockdown-induced reduction of SOD2 expression, PGC-1ɑ expression was determined. As shown in Fig. 6A and B, compared with undifferentiated MC3T3-E1 cells, both mRNA and protein expression of PGC-1ɑ was significantly increased in differentiated MC3T3-E1 cells, showing a similar changing pattern with SIRT3. LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were transfected with SIRT3 or SIRT3HY plasmids and then induced to osteogenic differentiation. mRNA expression of PGC-1ɑ was not significantly affected by knock down of SIRT3 and transfection of wild type and mutant SIRT3 (Fig. 6C). However, knock down of SIRT3 remarkably decreased the protein level of PGC-1ɑ (Fig. 6D), indicating that downregulation of SIRT3 affected the stability of PGC-1ɑ. Transfection of wild type but not mutant SIRT3 could reverse the decrease of PGC-1ɑ protein expression induced by LV-shSIRT3-mediated knock down of SIRT3 (Fig. 6D), indicating that deacetylase activity of SIRT3 is required for PGC-1ɑ protein stability. Moreover, we showed that there was an evident protein-protein interaction between SIRT3 and PGC-1ɑ in differentiated MC3T3-E1 cells (Fig. 6E). Downregulation of SIRT3 significantly inhibited the interaction between SIRT3 and PGC-1ɑ (Fig. 6E), which may result in the reduction of PGC-1ɑ in differentiated MC3T3-E1 cells. Furthermore, we detected direct binding of PGC-1ɑ in the promoters of SOD2, which was markedly inhibited by downregulation of SIRT3 (Fig. 6F). LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were transfected with PGC-1ɑ plasmids and the results confirmed the transfection efficiency (Fig. 6G and H). Overexpression of PGC-1ɑ using plasmids suppressed SIRT3 knockdown-induced reduction of SOD2 mRNA expression (Fig. 6I) and increase of ROS level (Fig. 6J and K). The results indicated that reduction of PGC-1ɑ stability was involved in SIRT3 knockdown-mediated disturbance of ROS homeostasis through downregulation of SOD2 expression.
Downregulation of PGC-1ɑ was involved in SIRT3 knockdown-mediated reduction of mitochondrial function and biogenesis and osteogenic differentiation
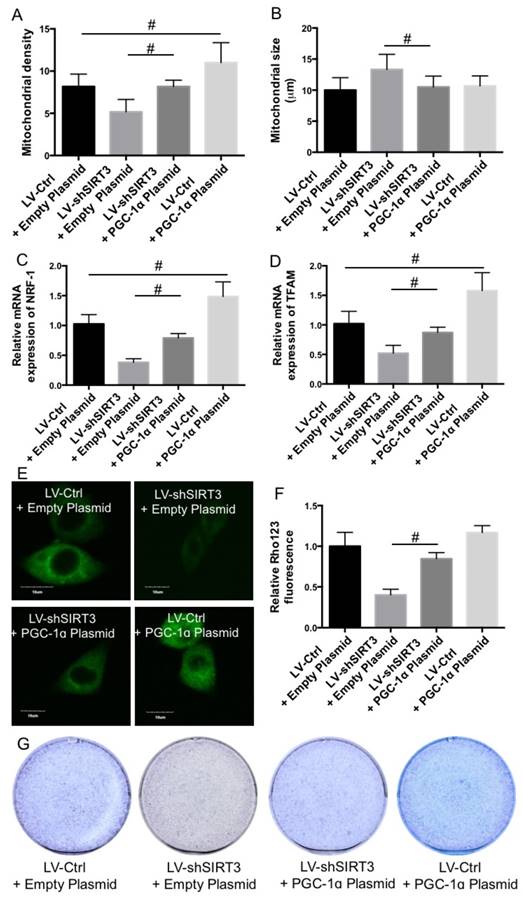
In the next step, we examined the role of downregulation of PGC-1ɑ in SIRT3 knockdown-mediated reduction of mitochondrial function and biogenesis and osteogenic differentiation. Overexpression of PGC-1ɑ suppressed SIRT3 knockdown-induced reduction of mitochondrial density (Fig. 7A) and increase of mitochondrial size (Fig. 7B). In addition, knock down of SIRT3-mediated reduction of NRF1 and TFAM expression was suppressed by overexpression of PGC-1ɑ (Fig. 7C and D). Overexpression of PGC-1ɑ suppressed SIRT3 knockdown-induced reduction of mitochondrial membrane potential, as reflected by increase of Rho123 fluorescence compared with that LV-SIRT3 MC3T3-E1 cells (Fig. 7E and F). Furthermore, overexpression of PGC-1ɑ markedly suppressed SIRT3 knockdown-induced reduction of ALP staining, indicating the improvement of osteogenic differentiation (Fig. 7G). The results indicated that downregulation of PGC-1ɑ was involved in SIRT3 knockdown-mediated reduction of mitochondrial function and biogenesis and osteogenic differentiation.
Effect of knockdown of SIRT3 on SOD2 expression and ROS level. MC3T3-E1 cells were induced to osteogenic differentiation, and SOD2 expression was determined in pre-osteogenic MC3T3-E1 cells and differentiated MC3T3-E1 cells using real-time PCR (A) and western blot (B). LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were induced to osteogenic differentiation. mRNA (C) and protein (D) expression of SOD2 was measured using real-time PCR and western blot, respectively. ROS level was determined by DHE staining (E and F). Representative images were shown (E). Relative DHE fluorescence was calculated (F). #P<0.05, compared with control.
Discussion
Previous study has shown that SIRT3 plays an important role in maintenance of bone homeostasis through regulation of osteoclast differentiation [16]. However, whether SIRT3 affects osteogenic differentiation is not known. In the current study, we evaluated the role of SIRT3 in osteogenic differentiation using MC3T3-E1 cells. We found that knock down of SIRT3 decreased ALP staining and the expression of key regulators, including Runx2, Col1ɑ1, and osteocalcin, in differentiated MC3T3-E1 cells. Re-expression of wild type but not mutant SIRT3 in shSIRT3-transfected cells significantly reversed the decrease of ALP staining. We concluded that SIRT3 and the deacetylase activity was required for osteogenic differentiation in MC3T3-E1 cells.
Maintenance of normal mitochondrial function and the homeostasis of biogenesis are critical for osteogenic differentiation [18-21]. Mitochondrial dysfunction is considered to be a fundamental pathological processes in the dysregulation of osteogenic differentiation [18, 19], and normal mitochondrial function is essential for physiological osteogenic differentiation [20]. Moreover, enhanced mitochondrial biogenesis was observed during osteogenesis and mitochondrial biogenesis is required for osteogenic differentiation [21]. The important role of SIRT3 functioning as a major mitochondrial deacetylase has been well documented [14, 22, 23]. There is a significant decrease of Complex I activity and reduction of oxygen consumption in Sirt3-/- animals [14, 23]. Moreover, previous study has found that SIRT3 functioned to regulate mitochondrial function and thermogenesis in brown adipocytes [22]. Consistent with these results, our findings showed that knock down of SIRT3 decreased Complex I-V activities, oxygen consumption activity, and mitochondrial membrane potential, induced changes of ultrastructure of mitochondria, and reduced mitochondrial density and biogenesis, indicating that SIRT3 was required for maintenance of mitochondrial function and biogenesis during osteogenic differentiation.
It is reported that SIRT3 could affect ROS homeostasis [8, 23-25], which is pivotal for keeping normal mitochondrial function and biogenesis [26-28]. Increased ROS generation in SIRT3 knockout mice results in oxidative stress in skeletal muscle, activates JNK and impairs insulin signaling [23]. The antitumor sensitizing effect of SIRT3 silencing involves an increment in ROS production [24]. SOD2 acts as an important sensor that mediates the effects of SIRT3 on ROS state [29, 30]. Previous finding have reported the deacetylation of SOD2 by SIRT3 in capillary endothelial cells [29] and hepatocytes [30]. In the present study, we found that downregulation of SIRT3 decreased both mRNA and protein expression of SOD2 and increased ROS level. It suggested that SIRT3 regulated SOD2 expression and acitivity in a cell-dependent manner, either through deacetylation or regulation of expression. Overexpression of SOD2 suppressed downregulation of SIRT3-mediated mitochondrial dysfunction and osteogenic differentiation. The findings demonstrated that downregulation of SOD2 was involved in SIRT3 knockdown-mediated reduction of mitochondrial function, mitochondrial biogenesis, and osteogenic differentiation.
Role of downregulation of SOD2 in the effect of SIRT3 knockdown on osteogenic differentiation. LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were transfected with SOD2 or empty plasmids and then induced to osteogenic differentiation. mRNA (A) and protein (B) expression of SOD2 in cells transfected with empty vector and SOD2 plasmid was measured using real-time PCR and western blot, respectively. Oxygen consumption activity was evaluated by the determination of complex II driven respiration and results were expressed as µmol/l/min (C). Osteogenic differentiation was measured using ALP staining (D). mRNA expression of Runx2 (E), Col1α1 (F), and OCN (G) in differentiated cells were determined by real-time PCR. #P<0.05, comparison between the two groups.
Role of PGC-1ɑ in the effect of SIRT3 knockdown on SOD2 expression and ROS level. MC3T3-E1 cells were induced to osteogenic differentiation, and SIRT3 expression was determined in pre-osteogenic MC3T3-E1 cells and differentiated MC3T3-E1 cells using real-time PCR (A) and western blot (B). LV-Ctrl and LV-shSIRT3 MC3T3-E1 cells were transfected with SIRT3 or SIRT3HY plasmids and then induced to osteogenic differentiation. mRNA (C) and protein (D) expression of PGC-1ɑ was measured using real-time PCR and western blot, respectively. LV-Ctrl and LV-SIRT3 MC3T3-E1 cells were induced to osteogenic differentiation. Co-IP was conducted to determine the interaction between PGC-1ɑ and SIRT3 (E). CHIP assay was conducted to the binding of PGC-1ɑ in the promoters of SOD2 (F). LV-Ctrl and LV-SIRT3 MC3T3-E1 cells were transfected with PGC-1ɑ or empty plasmids and then induced to osteogenic differentiation. mRNA expression of SOD2 (G) in differentiated cells were determined by real-time PCR.ROS level was determined by DHE staining (H and I). Representative images were shown (H). Relative DHE fluorescence was calculated (I). #P<0.05, comparison between the two groups.
Since the mRNA expression of SOD2 was affected by SIRT3 knockdown, we explored the mechanism of SIRT3-exhibited transcriptional regulation of SOD2. It is known that PGC-1α plays a role in the regulation of redox state, mitochondrial function [31, 32] and the transcription of SOD2 [33]. It has been reported that SIRT1-PGC-1α signaling prevents lipotoxicity-related apoptosis and oxidative stress in the kidney [34]. Moreover, SIRT3 could stimulate CREB phosphorylation, which may activate PGC-1α promoter directly [22]. These literatures suggest a potential interaction between SIRT3 and PGC-1α. In the current study, we found that knock down of SIRT3 remarkably decreased the protein level of PGC-1ɑ. Transfection of wild type but not mutant SIRT3 could reverse the decrease of PGC-1ɑ protein expression induced by knock down of SIRT3, indicating that deacetylase activity of SIRT3 is required for PGC-1ɑ protein stability. Overexpression of PGC-1α suppressed SIRT3 knockdown-induced reduction of SOD2 expression, increase of ROS level, decrease of mitochondrial density, increase of mitochondrial size, and reduction of mitochondrial membrane potential, leading to improvement of final osteogenic differentiation. The regulation of PGC-1α by SIRT3 deacetylase attributed to a protein/protein interactive regulation. Knock down of SIRT3 decreased PGC-1α protein stability, resulting in reduction of SOD2, NRF1 and TFAM expression, and increase of ROS level and decrease of mitochondrial function and biogenesis, eading to abnormal osteogenic differentiation.
In summary, in the current study, we found that SIRT3 was required for osteogenic differentiation through regulation of mitochondrial function and biogenesis via PGC-1α-SOD2 signaling. Further research into the molecular mechanism of SIRT3-PGC-1α interaction during osteogenic differentiation was needed. Overall, the data may contribute to a better understanding of the role of SIRT3 in osteogenic differentiation.
Role of PGC-1ɑ in the effect of SIRT3 knockdown on osteogenic differentiation. LV-Ctrl and LV-SIRT3 MC3T3-E1 cells were transfected with PGC-1ɑ or empty plasmids and then induced to osteogenic differentiation. Mitochondrial density (A) and size (B) were calculated. mRNA expression of NRF-1 (C) and TFAM (D) was examined using real-time PCR and results were shown as folds of control. Mitochondrial membrane potential was measured using Rho123 staining (E and F). Representative images were shown (E). Relative Rho123 fluorescence was calculated (F). Osteogenic differentiation was measured using ALP staining (G). #P<0.05, comparison between the two groups.
Abbreviations
SIRT3, sirtuin 3; ROS, reactive oxygen species; PGC-1ɑ, peroxisome proliferator-activated receptor-γ co-activator-1ɑ; SOD2, superoxide dismutase 2; CHIP, Chromatin Immunoprecipitations; DHE, dihydroethidium; Co-IP, Co-immunoprecipitation; ALP, alkaline phosphatase; Runx2, runt-related transcription factor 2; Col1ɑ1, collagen type I ɑ 1; OCN, osteocalcin; NRF1, transcription factor nuclear respiratory factor 1; TFAM, mitochondrial transcription factor A.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Briot K, Cortet B, Thomas T, Audran M, Blain H, Breuil V. et al. 2012 update of French guidelines for the pharmacological treatment of postmenopausal osteoporosis. Joint, bone, spine: revue du rhumatisme. 2012;79:304-13
2. Canalis E, Giustina A, Bilezikian JP. Mechanisms of anabolic therapies for osteoporosis. The New England journal of medicine. 2007;357:905-16
3. Huuskonen J, Vaisanen SB, Kroger H, Jurvelin C, Bouchard C, Alhava E. et al. Determinants of bone mineral density in middle aged men: a population-based study. Osteoporosis international: a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA. 2000;11:702-8
4. Zhu C, Zheng XF, Yang YH, Li B, Wang YR, Jiang SD. et al. LGR4 acts as a key receptor for R-spondin 2 to promote osteogenesis through Wnt signaling pathway. Cellular signalling. 2016;28:989-1000
5. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795-800
6. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587-91
7. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nature reviews Molecular cell biology. 2012;13:225-38
8. Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C. et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802-12
9. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121-5
10. Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends in biochemical sciences. 2010;35:669-75
11. Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R. et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and cellular biology. 2007;27:8807-14
12. Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. The Biochemical journal. 2012;444:1-10
13. Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B. et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular cell. 2011;44:177-90
14. Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A. et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447-52
15. Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP. et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PloS one. 2011;6:e23295
16. Huh JE, Shin JH, Jang ES, Park SJ, Park DR, Ko R. et al. Sirtuin 3 (SIRT3) maintains bone homeostasis by regulating AMPK-PGC-1beta axis in mice. Scientific reports. 2016;6:22511
17. Elschami M, Scherr M, Philippens B, Gerardy-Schahn R. Reduction of STAT3 expression induces mitochondrial dysfunction and autophagy in cardiac HL-1 cells. European journal of cell biology. 2013;92:21-9
18. Fang J, Yamaza H, Uchiumi T, Hoshino Y, Masuda K, Hirofuji Y. et al. Dihydroorotate dehydrogenase depletion hampers mitochondrial function and osteogenic differentiation in osteoblasts. European journal of oral sciences. 2016;124:241-5
19. Tsai PH, Chien Y, Chuang JH, Chou SJ, Chien CH, Lai YH. et al. Dysregulation of Mitochondrial Functions and Osteogenic Differentiation in Cisd2-Deficient Murine Induced Pluripotent Stem Cells. Stem cells and development. 2015;24:2561-76
20. Kwon IK, Lee SC, Hwang YS, Heo JS. Mitochondrial function contributes to oxysterol-induced osteogenic differentiation in mouse embryonic stem cells. Biochimica et biophysica acta. 2015;1853:561-72
21. Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem cells (Dayton, Ohio). 2008;26:960-8
22. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. The Journal of biological chemistry. 2005;280:13560-7
23. Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D. et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14608-13
24. Torrens-Mas M, Pons DG, Sastre-Serra J, Oliver J, Roca P. SIRT3 Silencing Sensitizes Breast Cancer Cells to Cytotoxic Treatments through an Increment in ROS Production. Journal of cellular biochemistry. 2016
25. Singh SP, Schragenheim J, Cao J, Abraham NG, Bellner L. PGC-1 alpha Regulates HO-1 Expression, Mitochondrial Dynamics and Biogenesis: Role of Epoxyeicosatrienoic Acid. Prostaglandins & other lipid mediators. 2016
26. Baldelli S, Aquilano K, Ciriolo MR. PGC-1alpha buffers ROS-mediated removal of mitochondria during myogenesis. Cell death & disease. 2014;5:e1515
27. Jing L, He MT, Chang Y, Mehta SL, He QP, Zhang JZ. et al. Coenzyme Q10 protects astrocytes from ROS-induced damage through inhibition of mitochondria-mediated cell death pathway. International journal of biological sciences. 2015;11:59-66
28. Olsen RK, Cornelius N, Gregersen N. Redox signalling and mitochondrial stress responses; lessons from inborn errors of metabolism. Journal of inherited metabolic disease. 2015;38:703-19
29. Gao J, Zheng Z, Gu Q, Chen X, Liu X, Xu X. Deacetylation of MnSOD by PARP-regulated SIRT3 protects retinal capillary endothelial cells from hyperglycemia-induced damage. Biochemical and biophysical research communications. 2016;472:425-31
30. Pi H, Xu S, Reiter RJ, Guo P, Zhang L, Li Y. et al. SIRT3-SOD2-mROS-dependent autophagy in cadmium-induced hepatotoxicity and salvage by melatonin. Autophagy. 2015;11:1037-51
31. Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V. et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115-24
32. St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S. et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397-408
33. Zhai X, Qiao H, Guan W, Li Z, Cheng Y, Jia X. et al. Curcumin regulates peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression by AMPK pathway in hepatic stellate cells in vitro. European journal of pharmacology. 2015;746:56-62
34. Kim MY, Lim JH, Youn HH, Hong YA, Yang KS, Park HS. et al. Resveratrol prevents renal lipotoxicity and inhibits mesangial cell glucotoxicity in a manner dependent on the AMPK-SIRT1-PGC1alpha axis in db/db mice. Diabetologia. 2013;56:204-17
Author contact
![]() Corresponding author: Honghui Sun. Department of Orthopedic, Tangdu Hospital, Fourth Military Medical University, #569 Xinsi Road, Xi'an 710038, China. Email: honghui_sun2015com.
Corresponding author: Honghui Sun. Department of Orthopedic, Tangdu Hospital, Fourth Military Medical University, #569 Xinsi Road, Xi'an 710038, China. Email: honghui_sun2015com.

 Global reach, higher impact
Global reach, higher impact