10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(3):295-307. doi:10.7150/ijbs.16968 This issue Cite
Review
LncRNAs: From Basic Research to Medical Application
1. Cardiovascular Department, The Affiliated Hospital of Medical College, Ningbo University, No.247, Renmin Road, Jiangbei District, Ningbo, China;
2. Ningbo Institute of Medical Science, No.42-46, Yangshan Road, Jiangbei District, Ningbo, China.
Received 2016-7-24; Accepted 2016-11-2; Published 2017-2-23
Abstract
This review aimed to summarize the current research contents about long noncoding RNAs (lncRNAs) and some related lncRNAs as molecular biomarkers or therapy strategies in human cancer and cardiovascular diseases. Following the development of various kinds of sequencing technologies, lncRNAs have become one of the most unknown areas that need to be explored. First, the definition and classification of lncRNAs were constantly amended and supplemented because of their complexity and diversity. Second, several methods and strategies have been developed to study the characteristic of lncRNAs, including new species identifications, subcellular localization, gain or loss of function, molecular interaction, and bioinformatics analysis. Third, based on the present results from basic researches, the working mechanisms of lncRNAs were proved to be different forms of interactions involving DNAs, RNAs, and proteins. Fourth, lncRNA can play different important roles during the embryogenesis and organ differentiations. Finally, because of the tissue-specific expression of lncRNAs, they could be used as biomarkers or therapy targets and effectively applied in different kinds of diseases, such as human cancer and cardiovascular diseases.
Keywords: lncRNA, molecular biomarker, therapy strategy, cancer, cardiovascular diseases
Introduction
The insights of the genome have changed overwhelmingly over the past decades. It has been a long time before we realize our unawareness of microcosm. There is no doubt that the advent of sensitive, high-throughput sequencing (NGS) has given rise to an unheard-of ability to detect novel transcripts [1]. Those “nonsense and dark materials of genome” unravel their mystery, play more important roles, and participate in various biological processes. However, those discoveries don't mean the end, but seem to open a door to take us toward a more in-depth research on microcosm. Until now, except for messengers RNA (mRNA) and other functional RNAs, such as transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar (snoRNA), and micro RNA (miRNA), we know very little on how lncRNAs function, how many different types of lncRNAs exist, or even whether most of them carry biological significance [1]. This review focused on the basic research of lncRNAs and their medical applications in human cancer and cardiovascular diseases.
Definition and classification of lncRNAs
The present definition of lncRNA, that is, an RNA molecule with a size longer than 200 nucleotides that is not translated into a protein, may be arbitrary without considering the following two issues [2]. 1). About the molecular size: A cutoff of 200 nucleotides was more based on the principle of RNA binding acid to silica columns during RNA purification [3]. While a protein coding gene (PCG) is usually defined as a transcript that contains an open reading frame (ORF) longer than 100 amino acids [4], thus a lncRNA might contain a longer ORF but not synthesize polypeptides. Besides, polypeptides shorter than 100 amino acids can be functional in organisms and are not outgrowth of canonical proteins [5]. 2) About the functions: The same RNA can contain both PCG and non-coding functions [6]. Coding transcript can lose their ability to encode a protein, and noncoding transcripts can acquire a coding function [6]. Such above issues obscures a clear annotation of lncRNAs. The definition by Mercer et al is more moderate, who defined lncRNAs as RNA molecules that may function as either primary or spliced transcripts and could not be classified into known classes of small RNAs or structural RNAs [7]. Compared with small ncRNAs, such as miRNAs, which have been defined in more detail and comprehensively, the definition of lncRNA is still extensive with complex contexts. However, finally some difference has been made following an in-depth research of lncRNAs.
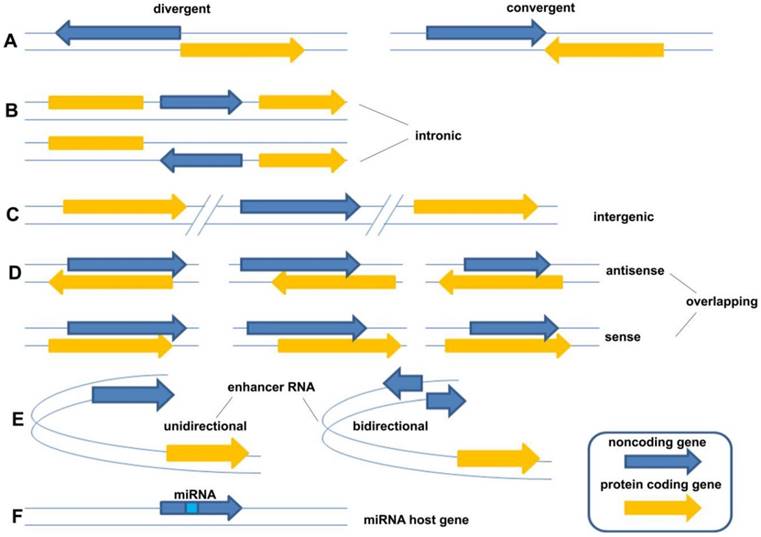
LncRNAs constitute various RNA molecules. It is difficult to classify them because of their broad spectrum of molecular splicing and cellular functions as a result of implementing different modes of action. The traditional classification is based on their locations of transcripts from genome, including five broad categories: (1) sense, (2) antisense, (3) bidirectional, (4) intronic, and (5) intergenic [8]. However, as new forms of lncRNAs were continually being found, the traditional classification of lncRNA could not cover the whole field and should be described in more detail. Sandre et al., who viewed it more widely, classified lncRNAs into eight categories: (A) divergent (pancRNA: They originate from the opposite strand of the same promoter region of protein coding gene as the adjacent) and convergent (They encoded on the opposite strands and facing each other); (B) intronic (They transcribed from an intron of another gene); (C) intergenic (They located distant from other genes, usually >10kb); (D) overlapping sense (They overlapped with other genes on the same strand) and overlapping antisense (They overlapped with other genes on the opposite strand); (E) enhancer RNA (They expressed as uni- or bidirectional transcripts); (F) miRNA host gene (Figure 1) [9]. Compared with the former classification, the latter described the relationship between lncRNA gene and their neighborhood gene in detail. However, they still belong to one criterion in essence. Following the development of new forms of lncRNAs, the classification needs to be continuously improved taking more criteria into account, such as their splicing modes or their final working mechanisms.
The classification of lncRNAs: (A) divergent (pancRNA: They originate from the opposite strand of the same promoter region of protein coding gene as the adjacent) and convergent (They encoded on the opposite strands and facing each other); (B) intronic (They transcribed from an intron of another gene); (C) intergenic (They located distant from other genes, usually >10kb); (D) overlapping sense (They overlapped with other genes on the same strand) and overlapping antisense (They overlapped with other genes on the opposite strand); (E) enhancer RNA (They expressed as uni- or bidirectional transcripts); (F) miRNA host gene [9].
Research methods and strategies of lncRNAs
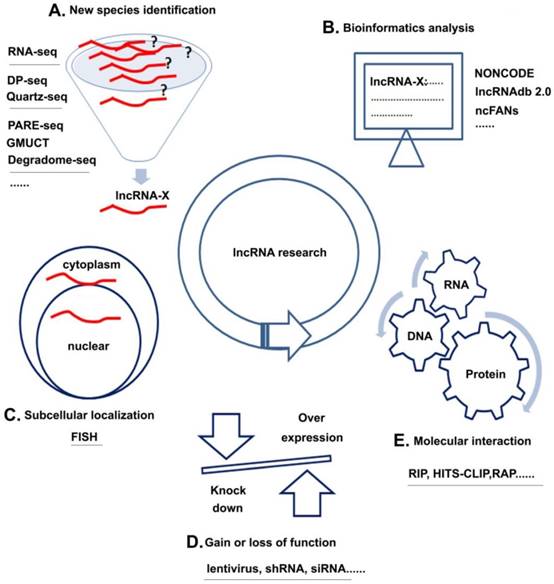
Unlike miRNAs, which have been extensively studied, the research of lncRNAs is still in its infancy. The following sections discuss four research strategies of lncRNAs with several common methods for each strategy (Figure 3): (1) new species identification, (2) subcellular localization, (3) molecular interaction, and (4) gain of loss of function and (5) bioinformatics analysis.
New species identification
Although some difficulties exist in terms of lncRNA discovery because of tissues specificity and low expression levels, many kinds of methods still exist for identifying new lncRNAs, which were well reviewed by Kashi et al. [10]. Each kind of technology was used to certain purpose. Among them, RNA-seq might be the most wide-spread method used not only for RNA expression detection but also for novel lncRNA discovery [10]. Sometimes, because of limited samples sizes for RNA-seq, single-cell transcript sequencing were needed, including Smart-Seq, which allows the detection of alternative transcripts isoforms and single-nucleotide polymorphisms [11]; DP-Seq, which allows amplification of RNA from sample sizes as small as 50pg [10]; and Quartz-Seq, which reduces the background noise [12]. Besides, some methods could be used to map transcripts that are in the process of being degraded, including PARE-Seq [13], GMUCT [14], and Degradome-Seq [15], by which decapped transcripts are adapter ligated and reverse transcribed [10].
Subcellular localization
Fluourescence in situ hybridization (FISH) is a method for visualization the subcellular localization of a certain lncRNA. For example, MALAT1 has been visualized by RNA-FISH, which indicated that it is rich in nuclear speckles of cells in interphase and is concentrated in mitotic interchromain granule clusters [16]. RNA-FISH was used to verify that MIR99HG (MONC) and MIR100HG were located in the nucleus [17]. Furthermore, RNA-FISH could also be combined with DNA-FISH to validate co-localization of a transcript with chromatin sequences [10].Alternatively, new recent technology of fluorescent in situ RNA sequencing (FISSEQ), which amplify cDNA in crosslinked cells and tissue samples [18], provide higher resolution and can identify a higher number of targets than RNA-FISH [10].
lncRNA research strategies and methods. A. New species identification: Using methods, such as RNA-seq et al could discover new kinds of lncRNAs; B. Subcellular localization: FISH could help to analyze the location of target lncRNAs; C. Gain or loss of function: over-expression or knock down the lncRNA by lentivirus, shRNA or siRNA could help to learn the basic function of lncRNAs in cell; D. Molecular interaction: Using methods like RIP et al could analyze the interactions among RNA, DNA and protein; E. Bioinformatics analysis: Various kinds of databases could help to learn about the interested lncRNAs.
Molecular interaction
On the basis of the working mechanisms of lncRNAs, molecular interactions occur among RNA, DNA, and proteins. The related technologies can also be classified into three categories: (1) RNA-protein interaction: RNA immunoprecipitation is used to analyze and purify RNAs associated with specific proteins by directing antibodies against the target protein [10]. High-throughput sequencing cross-linking immunoprecipitation (HITS-CLIP/ CLIP-Seq) is another technique to analyze the interaction between RNA and protein by cross-linking cell in vitro with UV light [10]. (2) RNA-DNA interaction: chromatin isolation by RNA purification (ChIRP) can analyze the relationship between RNA and chromatin by 20 nt-long biotinylated oligonucleotides which specifically recognize the target lncRNA [19]; RNA antisense purification (RAP) is another method which can be used to identify the genomic regions of chromatin that interact with RNAs using about 120nt long antisense RNA probes[20]; capture hybridization analysis of RNA targets (CHART) involves the purification of cross-linked protein, RNA, and DNA complexes by designing its probe on the region of potential open binding sites [10, 21]. (3) RNA and RNA interaction: rap-rna modification of the RAP method can provide a means to detect RNA-RNA interactions, even to distinguish the direct and indirect interaction between RNA transcripts by using different cross-linking methods [10]; Cross-linking, ligation and sequencing of hybrids (CLASH) is another technology for capturing direct RNA and RNA interaction by using UV cross-linking [22].
Gain or loss of function
Traditionally, if we want to learn about the function of a new molecule, we generally over-express or knock down the candidate target to investigate changes resulted by these means. LncRNAs could also follow the above methods. For example, using over-expression plasmid and siRNA (or shRNA) separately to up regulating and down regulating lncRNA, Cui et al., have verified that over-expression of HULC was able to accelerate lipogenesis in HepG2 and Huh7 cells, while HULC siRNA attenuate the lipogenesis in HepG2.2.15 cells [23]. For another, using the same methods, Nie et al., have found that knockdown of ANRIL expression could impair cell proliferation and induce cell apoptosis both in vitro and vivo [24]. In fact, in order to gain stable function of a gene, technology of lentivirus has been used for many years. Presently, although there has always been a dispute, exact gene editing techonologies, such as CRISPR/CAS9 and NgAgo have emerged. Compared to the traditionally methods, they might be expected to realize the more accurate gene editing than any time in the past.
Bioinformatics analysis
The lncRNA databases have grown rapidly following with the research of lncRNAs. The database of lncRNAdb (http://www.lncrnadb.org/) is a famous long non-coding RNA research database that provides full comments on functional lncRNAs [25]. The present version is lncRNAdb 2.0 [26]. Another well-known ncRNA research database NONCODE (http://www.noncode.org) also provides full comments on lncRNAs, including expression and functions predicted by their computer software (ncFANs) [27]. Now the version has been updated to NONCODE v4 [28]. LNCipedia (http://www.lncipedia.org) provides the sequence and full comments on structures of human lncRNAs[29-30]. LncRNA disease (http://cmbi.bjmu.edu.cn/lncrnadisease) provides full comments of reported disease-related lncRNAs[31]. NRED (http://jsm-research.imb.uq.edu.au/nred/) provides the lncRNA expression information of human and mouse based on chip data [32]. ChIPBase (http://deepbase.sysu.edu.cn/chipbase/) provides the map of lncRNA, comprehensive identification, and annotation of the transcription regulation [33]. It also integrated lncRNAs identified by high-throughput RNA-seq, their expression profile and even the transcription factor binding sites identified by ChIP-seq experiment [33]. fRNAdb (http://www.ncrna.org/) collects non-code transcripts with or without comments from H-invitation, NONCODE and RNAdb database and provides searching function of four kinds of ncRNAs, including miRNAs, short ncRNAs, mid-size ncRNAs and lncRNAs [34]. Starbase (http://starbase.sysu.edu.cn/mirLncRNA.php) has set up the most comprehensive experimental support by CLIP-Seq for the regulation network of miRNA and lncRNA, Protein (RNA-binding protein) and lncRNA (includes lncRNA, pseudogene, circular RNA), competing endogenous RNAs (ceRNA) regulatory networks and provides prediction tools for a lncRNA function [35]. In fact, the section of bioinformatics analysis should not be summarized as a separate section because the context of each of the aforementioned sections could be permeated with biological information. Necessary information can be searched before studying a new function of a known lncRNA, or more information can be supplemented after the research on a new lncRNA.
Working mechanisms of lncRNAs
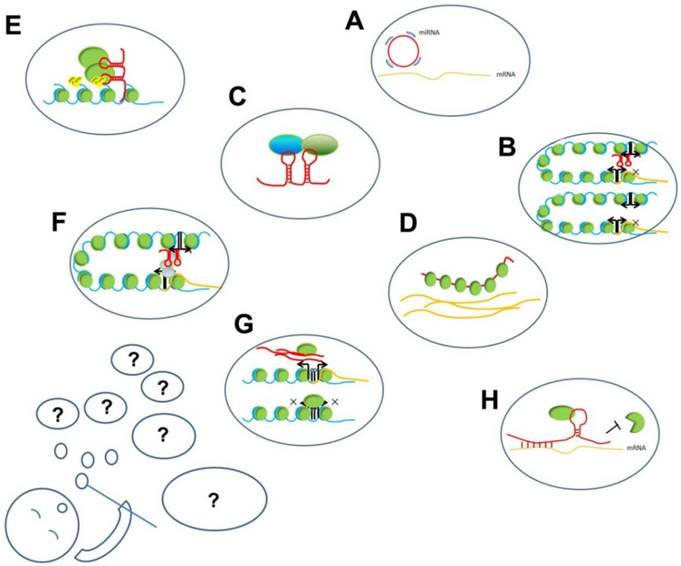
LncRNAs play important regulatory roles and participate in different levels of biological processes, such as chromatin remodeling, histone modification, and DNA methylation, and also serve as transcription factors or enhancers. The mechanisms underlying their functions can be summarized as interactions among RNA, DNA, and proteins (Figure 2). These interactions can be further summed up into two modes: (1) based on sequence hybridization and (2) based on secondary or tertiary structures.
The working mechanisms of lncRNAs. The present known working mechanisms of lncRNAs might be only a part of its all functions, just like several hubbles among all blowing group. A. ceRNA function of lncRNA: circRNA compete binding with miRNA to prevent miRNA binding with target mRNAs; B. Enhancer on/off: lncRNAs transcribed from an enhancer region interact with enhancer-promoter contact to inhibit the transcription of the protein-coding gene; C. lncRNAs acting as scaffold linking different proteins interaction; D. lncRNAs binding with proteins to prevent their actions; E. lncRNAs recruiting proteins, such as chromatin- modifying complexes to specific target sites in the genome; F. lncRNAs loop formation and transcription of the associated gene; G. lncRNA transcripts evicting proteins from chromatin, such as pancRNA prevent DNMT from methylating DNA in their promoter region, thereby ensuring mRNA transcription; H. Stabilizing of mRNA: lncRNA recruiting proteins to preventing degradation.
Interaction based on specific sequence hybridization
Some lncRNAs can mediate interactions by specific sequence hybridization with DNAs or other RNAs. For example, antisense intronic noncoding ras association domain family member 1 (ANRASSF1), which is transcribed in the antisense direction relative to the protein-coding mRNAs of the RAS association domain family member 1 (RASSF1) gene locus, could interact with genomic DNA, forming an RNA-DNA hybrid and regulating the expression of the sense gene at the pre-transcriptional level[36]. Phosphatase and tensin homolog pseudogene 1 antisense RNA (PTENP1-AS) may be an example of the RNA-RNA interaction. Because of the lack of a poly(A) tail, the stability and export to the cytoplasm of phosphatase and tensin homolog pseudogene 1 (PTENP1) is facilitated by interactions with PTENP1-AS, which has three variants: alpha, beta, and unspliced [37]. During this process, PTENP1-AS beta forms an RNA-RNA duplex with the PTENP1 transcript, altering its subcellular distribution and increasing its RNA stability [6], [37].
If the aforementioned examples of interaction between lncRNA and other RNAs are a universal phenomenon, the discovery of competing endogenous RNA (ceRNA) function of lncRNAs, which is of great biological significance, will not be surprising. LncRNAs can participate in the ceRNA network, by which lncRNAs cross talk with other RNAs by sharing the same miRNAs [38]. These exogenously expressed transcripts contain tandem repeats of miRNA response elements (MRE) sites that allow them to specifically bind a distinct miRNA or combination of miRNAs [39]. Once the miRNA(s) of interest is decoyed, it is unavailable to bind to its targets, leading to effective derepression of these transcripts [40], [41]. Such ceRNA function of lncRNAs has been well studied in human cancers. Metastasis-associated lung adenocarcinoma transcript 1(MALAT1) could sponge miR-145 to modulate the radio-sensitivity of high-risk human papillomavirus (HR-HPV+) cervical cancer [42]. Liu S. et al., reported that MALAT1 could competitively bind with miR-124 to up-regulate the RBG2 expression, and then promote HR-HPV (+) cervical cancer cell growth and invasion [43]. HOX transcript antisense RNA (HOTAIR) could cross talk with erb-b2 receptor tyrosine kinase 2 (HER2kny) via miR-331-3p in gastric cancer [44]. Li JT. et al., identified that HOTAIR could suppress the miR-568 to up-regulate the expression of S100 calcium binding protein A1 (S100A), and then facilitates the metastasis of breast cancer [45]. The B-Raf proto-oncogene, serine/threonine kinase (BRAF) pseudogene 1 is a newly reported lncRNA that functions as a ceRNA for BRAF, in part by sponging miR-30a, miR-182, and miR-876[46].
Interaction based on secondary and tertiary structures
LncRNAs can also form secondary and tertiary structures that enable more complex interactions with proteins. For example, cyclin-dependent kinase inhibitor 2B antisense RNA 1(ANRIL) was identified to recruit polycomb repressive complex-1 (PRC1) and polycomb repressive complex-2 (PRC2) to form heterochromatin surrounding the INK4b-ARF-INK4a locus, and to repress gene expression [47]. Similarly, imprinted maternally expressed transcript (H19) could interact with the protein complex heterogeneous nuclear ribonucleoprotein U/lysine acetyltransferase 2B/RNA polymerase II (hnRNP U/PCAF/RNAPol II), activating miR-200 family by increasing histone acetylation, and then altering the miR-200 pathway, thus contributing to mesenchymal-to-epithelial transition and suppression of tumor metastasis in hepatocellular carcinoma (HCC) [48]. Moreover, MALAT1 could promote cell proliferation, migration, and invasion via PRKA kinase anchor protein 9 (AKAP-9) in gastrointestinal cancer [49]. It also could combine with splicing factor proline/glutamine-rich (SFPQ) to release polypyrimidine tract binding protein 2 (PTBP2) from the SFPQ/PTBP2 complex to promote the growth and metastasis of colon cancer [50]. Besides, tumor protein p53 pathway corepressor 1 (TP53COR1/LincRNA-p21) recruits heterogeneous nuclear ribonucleoprotein K (hnRNP-K) to the promoter of p21, which is pivotal for the efficient binding of p53 to the p21 promoter to initialize the transcription of p21, such that lincRNA-p21 could activate the expression of p21 [51].
Although the interaction mechanisms were identified, in many cases, each interaction could not exist independently; they always worked together. Considering the aforementioned context as an example, the working mechanisms of lincRAN-p21 could be summarized as a mode of lincRNA-p21(lncRNA)-p53(protein) promoter of p21(DNA) interaction.
Functions of lncRNAs during the embryogenesis and organ differentiation
lncRNA transcripts were found at all examined stages during the early differentiation process by a single-cell sequencing study [52]. Furthermore, in the following various differentiation periods, they might also act as important functions. In this section, we would like to discuss several lncRNAs involved in the embryogenesis and organ differentiation.
Imprinted genes are monoallelically expressed according to the parent of origin and are critical for embryogenesis [53]. H19 is a well-known imprinted gene, which is exclusively expressed from the maternal allele and governs normal embryogenesis [54]. Maternally expressed 3 (meg3) is another imprinted gene, which has effects on neighboring genes in cis and results in perinatal lethality [55].
X-chromosome inactivation (XCI) is also an interesting phenomenon during embryogenesis. Xist was found as an XCI gene in knockout mice two decades ago [56-57]. Lacking Xist could cause female mice die during the first half of gestation, while male mice were unaffected [9]. However, knockout female mice with a single X-chromosome (XO) lacking Xist could survive [9]. Thus, it can be explained by the failure to adjust the X-chromosome gene dosage and causes embryonic lethality [9].
There are so many kinds of lncRNAs related to various organ differentiations that we could not cover them all. However, we would like to focus more about on lncRNAs related to cardiovascular differentiation.
Fendrr is an example of lncRNAs demonstrated to play an essential role in cardiovascular differentiation and embryo survival [9]. Grote et al. have verified that loss of Fendrr lead to impaired differentiation of tissues derived from lateral mesoderm, the heart and the body wall, and eventually result in embryonic death [58-59]. Embryos lacking Fendrr lead to upregulation of some transcription factors which controlled lateral plate or cardiac mesoderm differentiation, coupled with a drastic reduction in PRC2 occupancy along with decreased H3K27 trimethylation and/or an increase in H3K4 trimethylation at their promoters [59].
Braveheart (Bvht) has been verified as another cardiac-expressed lncRNA [60]. It epigenetically regulates cardiomyocyte differentiation [61]. It was cardiac-enriched approximately threefold than other tissues, but expressed similar with E14.5 embryos and adults; It was also not regulated late after hemodynamic stress [60]. In addition to its canonical role in regulation of MesP1, it may play “housekeeping” roles in adult hearts to stimulate and maintain cardiomyocyte fate [60].
Kcnq1ot1 induces transcriptional silencing by histone methylation of the overlapping Kcnq1/Kv7.1 potassium channel gene and other genes at the same genomic locus [62-64]. It is expressed at high levels in embryonic hearts, but reduced 5 times in adult hearts. Its counterparts mRNA, Kcnq1, which encodes the Kv7.1 slow delayed rectifying potassium channel increased during the same period. Kcnq1 is vital for normal cardiomyocyte repolarization which ends the action potential and cardiomyocyte contraction. Such a combination of opposite changes, in accordance with the need for enhanced pump function during the transition between developing embryonic and fully functioning adult hearts.
By using RNA-sequencing, Kurian et al. have identified 3 previously uncharacterized lncRNAs, TERMINATOR, ALIEN, and PUNISHER, which were specifically expressed in undifferentiated pluripotent stem cell, cardiovascular progenitors, and differentiated endothelial cell, respectively [65]. They provide a comprehensive transcriptomic roadmap that sheds new light on the molecular mechanisms underlying human embryonic development, mesodermal commitment, and cardiovascular specidication [65]. By the same method, Matkovich et al. identified 321 lncRNAs present in the heart and 117 of which exhibit a cardiac-enriched pattern of expression [60]
It is not difficult to understand that some lncRNAs are necessary regulators during embryogenesis and organ differentiations. No matter what kinds of regulation modes are, they were involved in different biogenesis, and would cause more or less impacts on embryogenesis and organ differentiations. Besides, because of the tissue specificity, their aberrant expressions might be finally related to different human diseases.
Medical applications of lncRNAs in human diseases
The best results of basic research are transforming the discoveries into diagnostic methods and therapeutic strategies for human diseases. Because of the tissue-specific characteristics of lncRNAs, they would be the next generation of biomarkers or targets for human diseases such as human cancer and cardiovascular diseases.
Biomarkers and therapy strategies for human cancers (Table 1)
lncRNAs, which isolated from tumor cells or circulating system, could provide readily-available, inexpensive and stable blood-borne diagnostics to more readily detect cancers and cancer subtypes[66]. Yang et al. analyzed eight kinds of lncRNAs in 240 patients with HCC and reported that highly upregulated in liver cancer (HULC) had a significant association with vascular invasion and was a positive factor for HCC overall survival and disease-free survival time [67]. They also reported that H19 was overexpressed in patients with hepatitis B [67]. Furthermore, they indicated that H19 and maternally expressed 3 (MEG3) were both considered to be risk factors for high alpha fetoprotein (AFP) level [67]. Tang J., et al have reported that compared with the traditional biomarkers of HCC (AFP), three lncRNAs (RP11-160H22.5, XLOC_014172 and LOC149086) might be the potential biomarker for the tumorigenesis prediction and XLOC_014172 and LOC149086 for metastasis prediction in the future [68]. Zhao et al., have reported that SPRY4 intronic transcript 1 (SPRY4-IT1) is an independent prognostic factor of overall survival in patients with urothelial carcinoma of the bladder (UCB) [69]. The work of Kristina P Sørensen.et al., firstly investigated the prognostic potential of lncRNA profiles for patients with breast cancer, which can distinguish metastatic patients from non-metastatic patients with sensitivity more than 90% and specificity of 64 to 65% [70].
Working mechanisms and medical applications of lncRNAs in human cancer
| LncRNA | Working mechanisms | Medical applications |
|---|---|---|
| ANRASSF1 | form an RNA/DNA hybrid and recruits PRC2 to the RASSF1A promoter[76] | Biomarker : breast cancer[77] |
| ANRIL | chromatin remodeling: CDKN2A/B [47], Kruppel-like factor 2 (KLF2) and p21 locus by interacting with PRC1 and PRC2 [24, 78] | Biomarker : acute lymphoblastic leukemia (ALL) [79], ovarian cancer[80], non-small cell lung cancer (NSCLC)[24] |
| H19 | 1. regulate IGF2 imprinting[81]; 2. miR-675 precursor[82-85]; 3. ceRNA function by sponging of let-7[86-88], miR-138[89], miR-200a [89]; 4. chromatin remodeling by binding MBD1[90]; 5. interact with Slug or EZH2 and regulate E-cadherin expression[91-92] | Biomarker: hepatocellular (HCC)[67] Cancer therapy: BC-819[72], BC-821, siRNA and anti-miR* for lung cancer[93]; Cancer therapy: double promoter toxin vector P4-DTA-P3-DTA for pancreatic cancer, ovarian cancer and HCC[94-95]; Cancer therapy: DTA-H19 for unresectable pancreatic cancer[66, 73] |
| MALAT1 (NEAT2) | 1. bind to unmethylated Pc2 promotes E2F1 SUMOylation, leading to activation of the growth-control gene program[96]; 2. ceRNA function by sponging of miR-145[42], miR-124[43], miR-205[97]; 3. directly interact with Sp1 and LTBP3 promoter to increase expression of LTBP3 gene[98]; 4. bind to SFPQ and releasing PTBP2 from the SFPQ/PTBP2 complex[50] | Biomarker: predicting prostate cancer risk[99] |
| HOTAIR | 1. chromatin remodeling chromatin remodeling HOXD gene locus[100-101]; 2. recruit and bind to PRC2[102-103] and epigenetically represses miR-34a[104]; 3. ceRNA function by sponging of miR-331-3p[44]; 4. associate with the immunoprecipitated Ago2 (Argonaute2) complex, and cleaved by the Ago2 complex in the presence of miR-141[105]. | Biomarkers: risk factors for high AFP level, HCC[106], breast cancer[107], colorectal cancer (CRC), gastric cancer (GC)[108] Cancer therapy strategy: Triple-negative breast cancer (TNBC)[109] |
| LincRNA-p21 (TP53COR1) | interact with hnRNP K leads to gene silencing and cell apoptosis[51] | NA |
| PTENP1 | 1. ceRNA function by binding with miR-21[110], miRNA sponge of miR-17[111],miR-19b[111],miR-20a[111]; 2. PTEN mRNA stability[6] | Biomarker: GC[111-112] Cancer therapy: SB-BV hybrid vector for HCC therapy[111] |
| PTENP1 asRNA (α,β,unspliced) | 1. the α isform suppress the expression of PTEN by localize on the PTEN promoter and interact with EZH2 and DNMT3a[37]; 2. the β isform involved as a miRNA sponge mediated by PTENP1[37] | NA |
| BRAFP1 | ceRNA[46] | NA |
| MEG3 | 1. interact with p53 by promoting p53 binding to GDF15 promoter[113]; 2. dissociate the transcription factor SOX2 from the BMP4 promoter[114]; 3. ceRNA function by sponging of miR-21[115]; | Biomarker: risk factors for high AFP level, CRC [116], Tongue squamous cell carcinoma (TSCC) [117], GC [118] |
| TERRA | promoted recruitment of lysine-specific demethylase 1 (LSD1) to unprotected telomeres and interacting with MRE11 (double strand break repair nuclease)[119] | NA |
| PVT1 | 1. ceRNA function by sponging of miR-200 family[120]; 2. interact with NOP2 to protects it from degradation[121]; 3. associate with enhancer of zeste homolog 2 (EZH2) to repress p15 and p16[122]; 4. recruit EZH2 to the large tumor suppressor kinase 2 (LATS2) promoter and represses LATS2 transcription[123] | Biomarker: HCC, GC[122, 124], pancreatic cancer[125], CRC[126] |
| HULC | 1. activate ACSL1 promoter[23] 2. inhibit the miR-9 promoter[23]; 3. ceRNA function by sponging of miR-372 [127]; | Biomarkers : HCC[128] , pancreatic cancer[129] |
| FER1L4 | ceRNA function by sponging of miR-106-5p[130-131] | Biomarkers: diagnosis of CRC[130], GC[132] |
| PCA3 | form PRUNE/PCA3 double-strand RNA to control PRUNE[133] | Biomarker : prostate cancer[134-135] |
| CCAT1 | 1. ceRNA function by sponging of let-7, miR-218-5p[136], miR-155[137], miR-490[138]; 2. interact with CTCF and modulate chromatin conformation at these loop regions[139] | NA |
| ATB | 1. ceRNA function by sponging of miR-200c to mediate EMT[140-141]; 2. promote organ colonization of disseminated tumor cells by binding IL-11 mRNA, autocrine induction of IL-11, and trigger STAT3 signaling [141]. | NA |
| HNF1A-AS1 | bind with DNMT1 to regulate cyclinD1[142] | NA |
| NKILA | interact with NF-KB/I-KB to form a stable complex , serving as NF-KB modulators to suppress metastasis[143] | NA |
| PANDAR | interact with NF-YA to pro-apoptotic genes[144] | Biomarker : hepatocellular carcinoma[145] |
| PCAT-1 | 1. interact with myc[146]; 2. repress BRCA2 3'-UTR imparts a high sensitivity to PARP1[147] | Biomarker: esophageal squamous carcinoma (ESCC)[148], CRC[149] |
| HOST2 | ceRNA function by sponging of let-7b[150] | NA |
| GAPLINC | ceRNA function by sponging of miR-211-3p to compete with CD44[151] | Biomarker: GC[151] |
| ROR | 1. ceRNA function by sponging of miR-145 [152-153]and miR-205[154]; 2. suppress p53 translation through direct interaction with the heterogeneous nuclear ribonucleoprotein I (hnRNP I)[155] | NA |
| FAL1 | interact with BMI1 and regulate a number of genes[156] | NA |
| TARID | interact with TCF21 promoter and GADD45A to directs de-methylation of TCF21[157] | NA |
| UCA | 1. activate STAT3 and repress miR-143 to activate mTOR to regulate HK2[158]; 2. interact with hnRNP1 to suppress p27 protein level[159] | NA |
| TUG1 | modulate HOXB7 to participating in AKT and MAPK pathways[160] | NA |
| RUNXOR | utilize its 3'-UTR terminal fragment to directly interact with RUNX1 promoter chromosomal translocation[161] | NA |
| SPRY4-IT1 | interact with ZNF-703[162] | Biomarkers: urothelial carcinoma of the bladder[69], GC[163], prostate cancer[164], ESCC[165] |
Because of the heterogeneity of human cancer, the personalized medicine for human cancer has been proposed for many years, and the precision medicine has become the most important field for cancer research and treatment. However, the etiologies of human cancer are so complex that the results need to be transformed by different levels of clinical trials. Several strategies of cancer therapy have been developed presently.
Treating cancer by interference of RNA expression has developed as a treatment modality because growth-promoting lncRNAs could be inactivated through antisense technologies [66]. LncRNAs could also function as mRNA or miRNA sponges to inhibit the growth-promoting, pro-tumorigenic signaling pathway so that synthetically-engineered lncRNAs may be employed through replacement therapy to inhibit tumor cells [71]. Dextran nanoparticles can deliver chemotherapy to the nucleus, which may be used to attach cytotoxic agents to lncRNAs [66]. The plasmid DTA-H19 was designed to express a diphtheria toxin subunit controlled by the H19 promoter [72], [73], which could be injected into tumor and lead to a H19-dependent activation of diphtheria toxin within the tumor [45]. Another emerging method is to increase the activity of tumor suppressor by neutralizing the inhibitory effects of lncRNAs through antisense oligo nucleotides [66]. These compounds, which are directly taken up by cells, can be injected subcutaneously without any delivery vehicle [66]. They can cross endosomal membranes and enter the nucleus to inhibit PRC2 interaction with lncRNAs[46]. These compounds could also cross the blood brain barrier without a lipid carrier [66].
Biomarkers and therapy strategies in human cardiovascular diseases (Table 2)
Although lncRNA in cardiovascular have not been researched as extensively and thoroughly as in human cancer, some research has indicated that lncRNA could be biomarkers for cardiovascular diseases. Kumarswamy et al., have validated mitochondrial lncRNA uc022bqs.1 (LIPCAR) as a novel biomarker of cardiac remodeling that predicts future death in patients with heart failure [74]. Myocardial infarction (MI) is another kind of important cardiovascular disease. Vausort et al., have verified about five kinds of lncRNAs as biomarkers for predicting the outcome of MI in about 414 patients, including hypoxia inducible factor 1A antisense RNA 2 (HIA1A-AS2), cyclin-dependent kinase inhibitor 2B antisense RNA 1 (ANRIL), potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1 (KCNQ1OT1), myocardial infarction-associated transcript (MIAT), and MALAT1) [75]. Their results are summarized as follows: (1) HIA1A-AS2, CNQ1OT1 and MALAT1 were higher in patients with MI than in healthy volunteers (P<0.01), while levels of ANRIL were lower in patients with MI (P=0.003); (2) compared with patients with non-ST-segment-elevation MI, patients with ST-segment-elevation MI had lower levels of ANRIL (P<0.001), KCNQ1OT1 (P<0.001), MIAT (P<0.001), and MALAT1 (P=0.005) ; (3) levels of ANRIL were related to age, diabetes mellitus, and hypertension; (4) HIA1A-AS2 elevated in patients presenting within 3 h of chest pain onset than in patients presenting later; (5) ANRIL, KCNQ1OT1, MIAT, and MALAT1 were significant univariable predictors of left ventricular dysfunction as assessed by an ejection fraction ≤40% at 4-month follow-up; and (6) ANRIL and KCNQ1OT1 improved the prediction of left ventricular dysfunction by a model, including demographic features, clinical parameters, and cardiac biomarkers based on the multivariable and reclassification analyses [75].
Conclusions
In summary, we have separately discussed several items of lncRNAs, including their definition and classification, their related research methods and strategies, their possible working mechanisms, their function during embryogenesis and organ differentiation, their significance in human cancer and cardiovascular diseases.
However, each section might have their limitations due to our unawareness of them. We could not make an exact definition for lncRNAs. And following with the discovery of new lncRNAs, we might be confronting with the difficulties from classifications. The present research methods could realize the interaction among RNA, DNA and protein, and provide important bioinformatics clues on basic research, we still need further techniques to reach a whole comprehension on lncRNA network mechanisms. We need not only learn about the real mechanisms of lncRNAs during embryogenesis and development, but also the novel changes in various kinds of diseases, and finally realize the destination of promoting human health by our knowledge.
Working mechanisms and medical applications of lncRNAs in human cardiovascular diseases
| LncRNA | Working mechanisms in cardiovascular diseases | Medical application |
|---|---|---|
| LIPCAR | NA | Biomarkers: cardiac remodeling predicts future death in patients with heart failure[74] |
| ANRIL | NA | Biomarkers: significant univariable predictors of left ventricular dysfunction[75] |
| KCNQ1OT1 | NA | Biomarkers: significant univariable predictors of left ventricular dysfunction[75] |
| MALAT1 | NA | Biomarkers: significant univariable predictors of left ventricular dysfunction[75] |
| MIAT | ceRNA: function by sponging of miR-150-5p[166] | Biomarkers: significant univariable predictors of left ventricular dysfunction[75] |
| MHRT | Sequesters Brg1 from its genomic DNA targets to prevent chromatin remodeling [167] | Biomarkers : myocardial infarction (MI)[168] |
| UCA1 | NA | Biomarkers: Acute myocardial infarction (AMI) [169] |
| LincRNA-p21 | Binding with MDM2 and release MDM2 repression of p53 [170] | NA |
| RNCR3 | acts as a ceRNA, and forms a feedback loop with Kruppel-like factor 2 and miR-185-5p to regulate cell function, atheroprotective role in atherosclerosis[171] | NA |
| ROR | promotes cardiac hypertrophy via interacting with miR-133[172] | NA |
Acknowledgements
This work was supported by Ningbo Municipal Natural Science Foundation (Grant Number: 2014A610219); Ningbo University Science Foundation (Grant Number: XYY15016); National Natural Science Foundation (Grant Number: 81602136).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651-69
2. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577-87
3. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484-8
4. Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176
5. Washietl S, Findeiss S, Muller SA, Kalkhof S, von Bergen M, Hofacker IL. et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011;17:578-94
6. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
8. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
9. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491-509
10. Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta. 2016;1859:3-15
11. Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777-82
12. Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T. et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31
13. German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P. et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941-6
14. Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H. et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854-66
15. Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758-62
16. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925-38
17. Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13:171
18. Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442-58
19. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667-78
20. Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973
21. Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK. et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465-9
22. Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci U S A. 2011;108:10010-5
23. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X. et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846-57
24. Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R. et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268-77
25. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146-51
26. Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB. et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168-73
27. Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y. et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112-5
28. Zhao Y, Yuan J, Chen R. NONCODEv4: Annotation of Noncoding RNAs with Emphasis on Long Noncoding RNAs. Methods Mol Biol. 2016;1402:243-54
29. Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K. et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246-51
30. Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J. et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363-4
31. Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X. et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983-6
32. Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122-6
33. Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177-87
34. Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y. et al. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145-8
35. Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202-9
36. Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16:3251-66
37. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D. et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440-6
38. Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601-12
39. Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30
40. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-6
41. Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043-50
42. Lu H, He Y, Lin L, Qi Z, Ma L, Li L. et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37:1683-91
43. Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016;37:633-40
44. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92
45. Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J. et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454
46. Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319-32
47. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662-74
48. Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS. et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577-86
49. Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY. et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166-74
50. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736-48
51. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K. et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777-90
52. Yan L, Yang M, Guo H, Yang L, Wu J, Li R. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131-9
53. Rancourt RC, Harris HR, Barault L, Michels KB. The prevalence of loss of imprinting of H19 and IGF2 at birth. FASEB J. 2013;27:3335-43
54. Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34-9
55. Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, Rice KA. et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643-52
56. Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131-7
57. Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156-66
58. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-14
59. Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579-85
60. Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW 2nd. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci U S A. 2014;111:12264-9
61. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML. et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570-83
62. Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493-9
63. Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P. et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713-28
64. Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232-46
65. Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB. et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278-90
66. Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027-38
67. Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers. 2015;2015:191029
68. Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505-15
69. Zhao XL, Zhao ZH, Xu WC, Hou JQ, Du XY. Increased expression of SPRY4-IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Int J Clin Exp Pathol. 2015;8:1954-60
70. Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M. et al. Long non-coding RNA expression profiles predict metastasis in lymph node-negative breast cancer independently of traditional prognostic markers. Breast Cancer Res. 2015;17:55
71. Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics. 2014;9:13-20
72. Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D. et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol. 2008;180:2379-83
73. Scaiewicz V, Sorin V, Fellig Y, Birman T, Mizrahi A, Galula J. et al. Use of H19 Gene Regulatory Sequences in DNA-Based Therapy for Pancreatic Cancer. J Oncol. 2010;2010:178174
74. Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A. et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569-75
75. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668-77
76. Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT. et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9:e1003705
77. Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A. et al. Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol. 2016;37:2933-40
78. Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP. et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:50
79. Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J. et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492-4
80. Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497-505
81. Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem. 2000;275:29562-9
82. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313-6
83. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-8
84. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-22
85. Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295
86. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101-12
87. Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z. et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163-9
88. Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S. et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076-84
89. Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513-25
90. Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A. 2013;110:20693-8
91. Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E. et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014;1843:1414-26
92. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-21
93. Hasenpusch G, Pfeifer C, Aneja MK, Wagner K, Reinhardt D, Gilon M. et al. Aerosolized BC-819 inhibits primary but not secondary lung cancer growth. PLoS One. 2011;6:e20760
94. Amit D, Hochberg A. Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int J Clin Exp Med. 2012;5:296-305
95. Amit D, Tamir S, Hochberg A. Development of targeted therapy for a broad spectrum of solid tumors mediated by a double promoter plasmid expressing diphtheria toxin under the control of IGF2-P4 and IGF2-P3 regulatory sequences. Int J Clin Exp Med. 2013;6:110-8
96. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773-88
97. Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL. et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322-31
98. Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J. et al. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem. 2014;289:29365-75
99. Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y. et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091-102
100. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-93
101. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-23
102. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320-6
103. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
104. Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH. et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802
105. Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V. et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550-65
106. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243-50
107. Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu Y. et al. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res Treat. 2015;152:199-208
108. Ma G, Wang Q, Lv C, Qiang F, Hua Q, Chu H. et al. The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol. 2015;141:2139-45
109. Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC. et al. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget. 2015;6:11150-61
110. Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W. et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13:3086-97
111. Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM. et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71-81
112. Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH. et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137:1128-35
113. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R. et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731-42
114. Zhuang W, Ge X, Yang S, Huang M, Chen P, Zhang X. et al. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells. 2015;33:1985-97
115. Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104-13
116. Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851-9
117. Jia LF, Wei SB, Gan YH, Guo Y, Gong K, Mitchelson K. et al. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer. 2014;135:2282-93
118. Sun M, Xia R, Jin F, Xu T, Liu Z, De W. et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065-73
119. Porro A, Feuerhahn S, Lingner J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 2014;6:765-76
120. Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83
121. Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C. et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278-90
122. Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM. et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82
123. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R. et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol Cancer Ther. 2016;15:1082-94
124. Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63:442-9
125. Xie Z, Chen X, Li J, Guo Y, Li H, Pan X. et al. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016
126. Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H. et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164-71
127. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y. et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366-83
128. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J. et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687-96
129. Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346
130. Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F. et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323-32
131. Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P. et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445
132. Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613-7
133. Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI. et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112:8403-8
134. Zhang A, Zhang J, Kaipainen A, Lucas JM, Yang H. Long non-coding RNA: A newly deciphered "code" in prostate cancer. Cancer Lett. 2016;375:323-30
135. Mouraviev V, Lee B, Patel V, Albala D, Johansen TE, Partin A. et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:14-20
136. Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W. et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583
137. Chen L, Wang W, Cao L, Li Z, Wang X. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Mol Cells. 2016;39:330-6
138. Zhou B, Wang Y, Jiang J, Jiang H, Song J, Han T. et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201-10
139. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-31
140. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652-63
141. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666-81
142. Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160-72
143. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370-81
144. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621-9
145. Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother. 2015;72:113-8
146. Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V. et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900-8
147. Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S. et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651-60
148. Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS. et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36:2501-7
149. Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H. et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588
150. Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T. et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841-52
151. Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y. et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890-902
152. Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330-8
153. Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127:1585-94
154. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y. et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287
155. Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D. et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340-50
156. Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J. et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344-57
157. Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D. et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604-14
158. Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951-5
159. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M. et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008
160. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243
161. Wang H, Li W, Guo R, Sun J, Cui J, Wang G. et al. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. 2014;135:2783-94
162. Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y. et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51
163. Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36:6751-8
164. Lee B, Mazar J, Aftab MN, Qi F, Shelley J, Li JL. et al. Long noncoding RNAs as putative biomarkers for prostate cancer detection. J Mol Diagn. 2014;16:615-26
165. Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li SQ. et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743-54
166. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ. et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116:1143-56
167. Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST. et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102-6
168. Zhang J, Gao C, Meng M, Tang H. Long Noncoding RNA MHRT Protects Cardiomyocytes against H2O2-Induced Apoptosis. Biomol Ther (Seoul). 2016;24:19-24
169. Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X. et al. Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction. Biomed Res Int. 2016;2016:8079372
170. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C. et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452-65
171. Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD. et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248
172. Jiang F, Zhou X, Huang J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS One. 2016;11:e0152767
Author contact
![]() Corresponding author: duyantao_2000com
Corresponding author: duyantao_2000com

 Global reach, higher impact
Global reach, higher impact