10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(3):349-357. doi:10.7150/ijbs.16635 This issue Cite
Research Paper
Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c
1. MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100076, P.R. China;
2. State Key Laboratory of Medical Molecular Biology, Department of Biochemistry and Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, P.R. China;
3. Department of Microbiology, Xinxiang Medical University, Xinxiang, Henan 453003, P.R. China.
Received 2016-6-28; Accepted 2017-1-14; Published 2017-2-25
Abstract
Sterol regulatory element binding proteins (SREBPs) are master regulators of hepatic lipid homeostasis. Aberrant expression of SREBPs frequently leads to lipid metabolism dysregulation. Long non-coding RNAs (lncRNAs) have been identified with diverse biological functions, but the effects of lncRNAs on lipid metabolism are rarely reported. Here, we identified a novel human specific lncRNA, lncHR1, as a negative regulator of SREBP-1c expression. Overexpression of lncHR1 inhibited expression of SREBP-1c and fatty acid synthase (FAS) and then repressed oleic acid-induced hepatic cell triglyceride (TG) and lipid droplet (LD) accumulation. In vivo, the data of established transgenic animals showed that mice with lncHR1 expression had less hepatic expression of SREBP-1c, FAS, Acetyl-CoA carboxylase α (ACCα), and less hepatic and plasma TG after being fed a high-fat diet. Therefore, we report a novel lncRNA which can decrease lipid metabolism by repressing SREBP-1c gene expression.
Keywords: Long non-coding RNA, SREBP-1c, triglyceride, lipid metabolism.
Introduction
The liver is central to lipogenesis, gluconeogenesis and cholesterol metabolism [1]. So it is understandable that lipid and lipoprotein abnormalities are frequently as a consequence of hepatitis virus infection, looked like the results of alcoholic and nonalcoholic liver disease. In the process, host positive and negative regulatory mechanisms are typically activated to re-establish lipid homeostasis [2, 3]. While lipogenic enzymes involved in energy storage via synthesis of fatty acid (FA) [4] and TG, are coordinately regulated at the transcriptional level during different lipid metabolic states [5]. Recent in vivo studies suggest that SREBP-1c is central to dietary regulation of most hepatic lipogenic genes.
SREBP-1c is an SREBP family member with transcriptional activity for TG synthesis [6]. SREBP-1c is involved in encoding enzymes catalyzing various steps in the FA and TG synthesis pathway, such as fatty acid synthase (FAS) [7] and acetyl-CoA carboxylase (ACCα) [8]. SREBP-1c facilitates FA synthesis and incorporation into TG [9] and abnormally high SREBP-1c causes hepatic TG accumulation and other lipid disorders [10].
Long non-coding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides that lack coding potential [11]. Most annotated lncRNAs are transcribed from RNA polymerase II and are then capped, spliced, and poly-adenylated [12]. lncRNAs regulate gene expression by diverse mechanisms [13]. LncRNAs are often expressed in a species- or tissue-specific manner making these molecules attractive therapeutic targets and suggesting unique functions for lncRNAs to treat diseases, in particular human cancers [14]. Increasing evidence suggests that lncRNAs contribute to oncogenic and tumor suppressor roles during tumorigenesis [15]. LncRNA expression is frequently dysregulated in cancer, and specific lncRNAs are correlated with cancer recurrence, metastasis and poor prognosis across diverse cancers [16]. Studies indicate [4] that in HCC, HULC may be involved in tumorigenesis or tumor progression. Recent evidence also suggests that lncRNAs are abnormally expressed in liver cancer [17, 18]. HULC is the first identified lncRNA specifically over-expressed in HCC [4] and it can accelerate hepatic cell growth by down-regulating p18 and facilitate the deregulation of lipid metabolism [19]. However, the molecular mechanisms underlying how lncRNAs affect lipid metabolism are unclear.
In this study, we identified a novel lncRNA molecule (lncHR1) that inhibited the SREBP-1c protein level and decreased TG synthesis in vitro. LncHR1 transgenic mice have less SREBP-1c level, TG and lipoproteins accumulation after being fed a high-fat diet (HFD). Thus, our data offer a practical and efficient platform for studying lncRNAs function in lipid metabolism.
Materials and methods
Cells and reagents
The Huh7 human hepatoma cell line was from Apath, Inc. (Brooklyn, NY, USA) and a 2-3+ cell line stably expressing the HCV genotype 1b replicon genome was provided by Dr Stanley Lemon (University of Texas Medical Branch, Galveston, TX). 293T, HepG2, Hep2, HeLa, MCF7, K562, RD, THP-1, TZMBL, and PANC-1 cells were obtained from ATCC and were cultured as described previously [20]. A549 (from ATCC) cells were cultured in Fischer's-12 media (Gibco, Grand Island, NY, USA). Antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA) for SREBP-1c, FAS and ACCα, Sigma (St. Louis, MO, USA) for β-actin, and Beyotime (Shanghai, China) for Histone 3. Horseradish peroxidase (HRP)-conjugated secondary antibodies were from Jackson Immuno Research (West Grove, PA, USA).
Human lncRNA microarray and Data Analysis
Huh7 cells were innoculated with HCV (JFH1) and cultured for 72 h. PBS served as a negative control. Total RNA in TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was extracted according to the manufacturer's instructions. Differentially expressed lncRNAs were screened using an Agilent Human long non-coding RNA 4*180K microarray chip. A hybridization map was analyzed and extracted using Agilent Feature Extraction (v10.7) software. Data were normalized using the Agilent GeneSpring software (Agilent Technologies). Hierarchical clustering was performed with Gene Cluster 3.0 and Eisen's Tree View (Stanford University, Palo Alto, CA). Differentially expressed genes were identified through the random variance model, and p values were calculated using a Student's paired t-test. Significance threshold for up-or down-regulated genes were assessed as fold change >2.0 or <2.0 and P < 0.05 was considered significant.
Plasmid construction
LncRNAs were cloned into BglII and XhoI sites of a pMIR-Empty-DFT vector. An overexpressing vector of lncHR1 with tissue nonspecific promoter was linearized with DNA restriction enzyme ClaI for establishing transgenic mice. shRNA targeting lncRNAs were cloned into BglII and XhoI sites of the pQSUP-R plasmid vector. Four paires of shRNA sequences are listed in Table S1.
Transfection and Luciferase Assays
Cells were cultured and transfected with related plasmid using LipoD293 (SignaGen, Rockville, MD, USA). Luciferase reporter activity was measured at 48 h after DNA transfection as described previously [21].
Western-blot
Western blot was performed to quantify protein according to standard protocols published in the literature [20].
Fluorescence in situ hybridization
Huh7 cells were mounted on glass slides, fixed in 4% paraformaldehyde and then permeabilized with 0.4% TritonX-100. Treated cells were pre-hybridized with RNA Hybridization Buffer. The denatured special probes (Table S1) were hybridized with cells. The next day, slides were washed, blocked, incubated with FITC conjugated anti-Digoxin and then counterstained with DAPI-antifade Solution. At last, the samples were visualized by Leica TCS-SL confocal microscope.
RT-PCR and quantitative real-time PCR
Total RNA from cells cultures and mouse livers were extracted using TRIzol reagent (Invitrogen). RT-PCR for lncHR1 was performed with a PrimeScript One Step RT-PCR kit Ver.2 from TAKARA (Kyoto, Japan). The qRT-PCR for lncHR1 were performed with a QuantiFast SYBR green RT-PCR kit (Qiagen, DüsseLDsorf, Germany) and reaction conditions were carried out according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). β-actin expression was an endogenous control to normalize the data. All primers appear in Table S1.
Animal experiment
C57BL/6J lncHR1 transgenic mice were established with a linearized overexpression vector for lncHR1 by the institute of Laboratory Animal Science, Chinese Academy of Medical Sciences. Primers identifying F0 transgenic mice using RT-PCR appear in Table S1. F1 male (4-weeks-of-age) transgenic mice were fed a high-fat diet (HFD) (45% fat) and had free access to water for 9 weeks. Weight was recorded weekly and mice were sacrificed during the last week. Serum were collected and stored at -80°C. Livers were harvested, weighed and immediately frozen in liquid nitrogen for further analysis. Serum LDL and VLDL were measured with a LDL and VLDL Assay Kit (Apply gen). All animal experiments were conducted under protocols approved by the Institute.
TG quantification
Intracellular and serum TG, and hepatic TG were measured with a TG Assay Kit (Apply gen), according to the manufacturer's instructions. TG values are expressed as mM (for serum) or µM TG/µg hepatic protein.
Oil red O staining
Cells were fixed and permeabilized for immunofluorescent assay. Pretreated cells were stained with freshly diluted oil red O dissolved in 60% isopropanol. Stained cells were washed with 50% ethylalcohol. Nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI). At least 5 random fields were observed under a Leica TCS-SL confocal microscope. The intensity was quantified with Image J software (NIH).
Statistical analysis
Bar graphs depict means ± SD of at least three independent experiments. A Student's test was used to analyze differences between means for two independent samples, and p<0.05 were considered statistically significant.
Results
Identification of a HCV-induced lncRNA implicated in SREBP-1c regulation
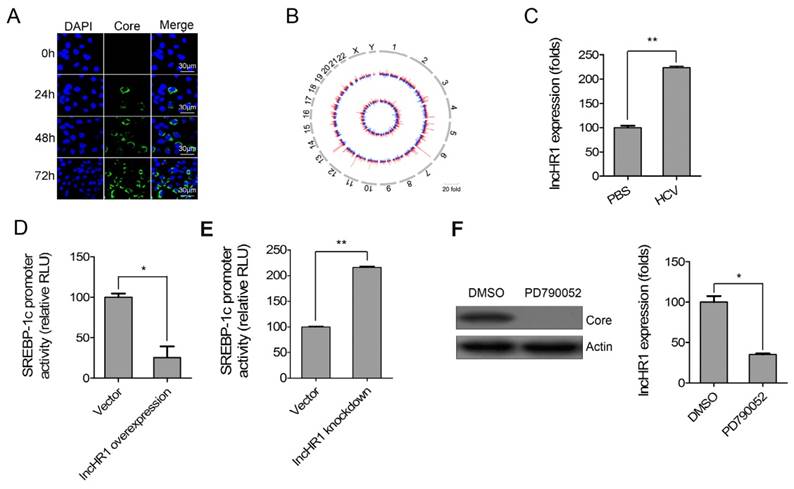
HCV infection enhances SREBP expression and lipid accumulation [22]. To systematically identify lncRNAs related to HCV infection and potentially involved in the regulation of hepatic lipid homeostasis, lncRNA microarray profiling and SREBP-1c promoter reporter gene were used. First, Huh7 cells were inoculated with HCV (JFH-1 strain) and infection was confirmed after staining with anti-HCV core antibody (Fig 1A). The circos plot indicated differentially expressed lncRNAs and mRNAs on each chromosome (outer track: lncRNAs; inner track: mRNA genes) (Fig 1B). In response to viral infection, differentially expressed lncRNAs (fold change > 2.0) were identified (Table S2). Five random differentially expressed lncRNAs were selected and their relative expression was confirmed with qRT-PCR (Fig S1). The results support our microarray data.
20 of the most up-regulated lncRNAs were cloned into an overexpression vector and then the SREBP-1c promoter reporter gene was co-transfected with each of the overexpression lncRNAs respectively in Huh7 cells. Interestingly, an lncRNA had a significant inhibitory effects on the SREBP-1c promoter and this was termed as lncHR1 (HCV regulated 1) in this report. We noted endogenous lncHR1 was increased more than 2-fold in Huh7 cells infected with HCV (Fig 1C). LncHR1 overexpression in Huh7 cells suppressed SREBP-1c promoter activity (Fig 1D). In addition, four short hairpin RNAs (1-4) of lncHR1 knockdown revealed that shRNA-1 could suppress lncHR1 nearly 80%, shRNA-3 and shRNA-4 had nearly 60% of low efficiency and shRNA-2 only 20% of low efficiency in Huh7 cells (Fig S2). Using shRNA-1 for subsequent knowdown experiments to assess the effect on SREBP-1c, the result showed that knockdown of endogenous lncHR1 enhanced SREBP-1c promoter activity (Fig 1E). When compared with parental HCV replicon 2-3+ cells, cells treated with the NS5A inhibitor PD790052 had much less lncHR1 expression (Fig 1F). Thus, lncHR1 is an HCV- related lncRNA and likely protects host cells from dysfunctional lipid synthesis.
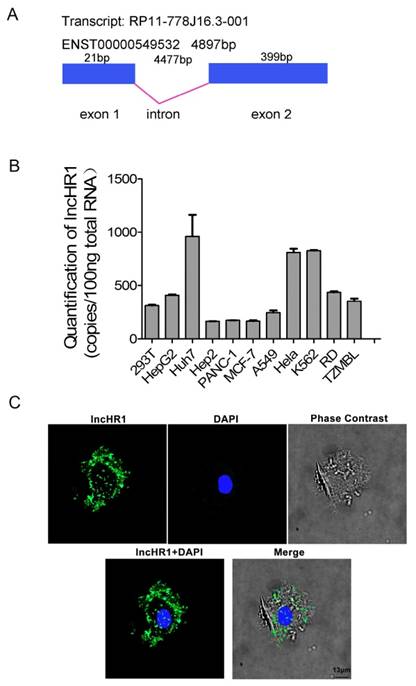
LncHR1 (ENST00000549532) is only one transcript from gene ENSG00000257400, which is contains 420 bp and the structure of lncHR1 gene was plotted (Fig 2A). Two potential proteins, 36 amino acids and 39 amino acids were predicted with ORF Finder (NCBI) (Fig S3A and B) but none matched known proteins or polypeptides according to BLAST. Abundance of lncHR1 was confirmed with absolute qRT-PCR in 11 cell lines (Fig 2B). RNA in situ hybridization in Huh7 cells was used to fluorescently label lncHR1. The results showed lncHR1 was concentrated in the nucleus and cytoplasm (blue for nucleus and green for lncHR1) (Fig 2C).
HCV-related lncRNA regulates SREBP-1c promoter activity. (A) Immunofluorescent assay confirmed HCV infection of Huh7 cells according to antibody staining against viral core protein. DAPI staining (blue) indicates nuclei. Scale bars, 30 μm. (B) Representative circos plot demonstrates differentially expressed lncRNAs (outer track) and mRNAs (inner track) in chromosomes. Red indicates up-regulated genes; blue is down-regulated genes. (C) LncHR1 was quantified in Huh7 cells in response to HCV infection. PBS treatment was used as a negative control. SREBP-1c promoter reporter gene was co-transfected with either lncHR1 overexpression (D) or knockdown (E) constructs in Huh7 cells, and luciferase reporter expression was measured and normalized. (F) HCV infection in 2-3+ replicon cells was cured with the NS5A inhibitor PD790052 (left), and lncHR1 expression was measured with qRT-PCR (right). Data are means ± SD. *p < 0.05, **p < 0.01.
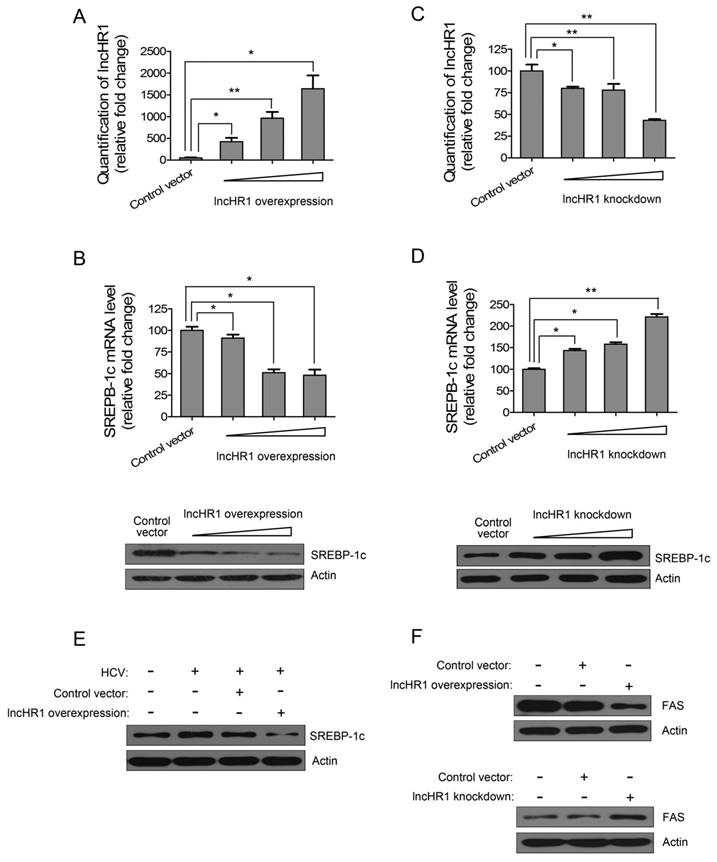
LncHR1 negatively regulates endogenous SREBP-1c and FAS
Hepatic TG synthesis is regulated by SREBP-1c and FAS. To confirm the effect of lncHR1 on endogenous SREBP-1c, Huh7 cells were transfected with an lncHR1 overexpression construct for 48 h and an empty vector was a negative control. We noted lncHR1 expression increased in a dose dependent manner (Fig 3A). In contrast, mRNA of endogenous SREBP-1c gradually decreased (Fig 3B up). Western blot data indicated that SREBP-1c protein was reduced in dose dependent manner (Fig 3B down). In addition, shRNA of lncHR1 knockdown could suppress lncHR1 more than 70% in Huh7 cells (Fig 3C). Using this system to assess the effect of lncHR1 depletion on SREBP-1c, we noted that compared with the GFP shRNA control vector, lncHR1 knockdown elevated both endogenous SREBP-1c mRNA and protein (Fig 3D). Thus, lncHR1 is a negative regulator of SREBP-1c.
Overexpression of lncHR1 abolished HCV-stimulated SREBP-1c induction (Fig 3E), indicating that lncHR1 up-regulation during HCV infection may be protective against excessive lipid synthesis. And our data showed that lncHR1 regulated negatively FAS protein level (Fig 3F). Thus, the effect of lncHR1 on FAS mRNA expression is similar to FAS protein level (Fig S4). So, lncHR1 suppressed SREBP-1c and FAS.
LncHR1 inhibits oleic acid-induced hepatic cell TG accumulation
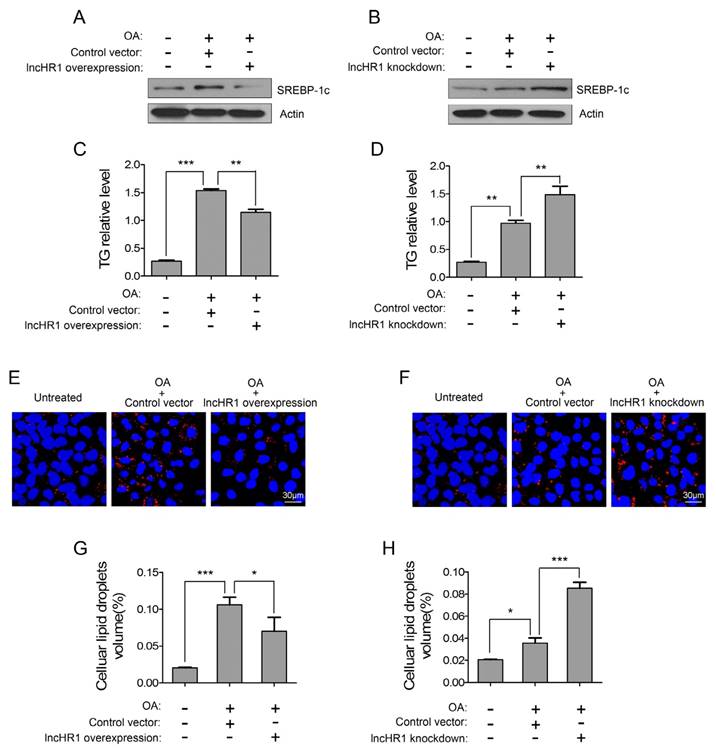
SREBP-1c is a key factor to active lipogenic gene expression and facilitate TG accumulation, but the regulation of hepatic TG by lncRNA has not been reported. Therefore, we assessed a potential functional role of lncHR1 for oleic acid (OA)-induced TG accumulation in cultured cells. OA treatment for 24 h increased SREBP-1c protein and intracellular TG in Huh7 cells, but simultaneously overexpressed lncHR1 abolished OA-induced SREBP-1c level (Fig 4A) and TG accumulation (Fig 4C). Accumulated TG is normally stored in cytosolic lipid droplet (LD). We observed that compared with untreated controls, OA treatment significantly increased LD; however overexpressed lncHR1 reduced LD to those control (Fig 4E). Consistently, the volume of intracellular LD was also reduced by lncHR1 (Fig 4G). In contrast, knockdown of lncHR1 produced the opposite effects. Therefore lncHR1 knockdown increased OA-induced SREBP-1c level (Fig 4B), intracellular TG (Fig 4D), and cytosolic LD volume (Fig 4F and H). These data suggest that lncHR1 can suppress TG accumulation and may be a protective molecule against hepatic steatosis.
LncHR1 has lipid-lowering effects in HFD-induced transgenic mice
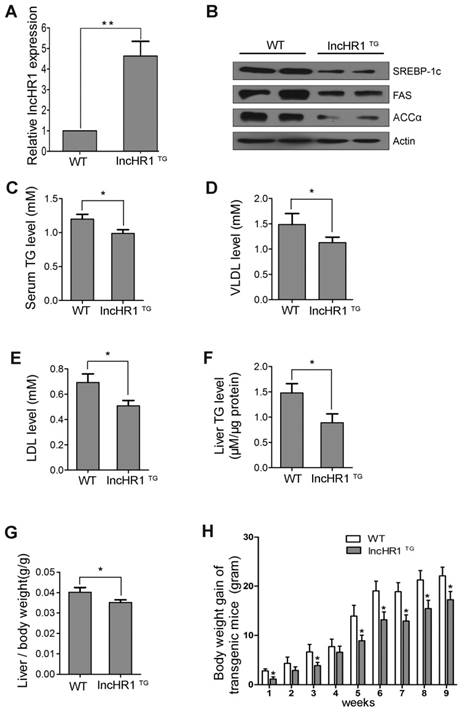
Sequence alignments indicate 42.0% homology with the human SREBP-1c promoter sequence and the mouse SREBP-1c promoter, including LXRE sites [23]. A knockout animal model is not available due to the lack of homologous lncHR1 gene in mice. Thus, we established transgenic mice expressing lncHR1 ubiquitously. Mice with lncHR1 expression (lncHR1TG) group were selected through the quantification of lncHR1 by qRT-PCR. As showed in Fig 5A, lncHR1TG group was ~4.5-fold greater comparing to WT group. In order to consistent with steatosis cells, we adopt a high-fat diet fed mice in the groups of transgenic and wild-type mice.
The HFD showed that, compared with before induction, weight gain of wild-type mice and transgenic mice were more than doubled (Fig S5). The results suggested that the obese mice were induced successfully through a high-fat diet fed. Also, hepatic SREBP-1c, FAS and ACCα protein level in the lncHR1TG group were significantly lower than those in the WT group (Fig 5B). Results of lipoprotein metabolism in serum indicated that serum TG, VLDL and LDL were decreased in the lncHR1TG group comparing to WT group (Fig 5C-E).
Then, we evaluated the effect of lncHR1TG group on hepatic TG and lipid metabolism. Results showed that liver TG was decreased in the lncHR1TG group (Fig 5F). The ratio of liver weight to body weight in the lncHR1TG group significantly was declined comparing to the WT group at the ninth week (Fig 5G). LncHR1TG group gained less weight than the WT group with significantly different at every week (Fig 5H). Thus, maintaining expression lncHR1 is useful for limiting liver lipid accumulation in the case of HFD induction. In brief, lncHR1 inhibited SREBP-1c protein level, limited TG accumulation and affected lipoprotein metabolism in mice with the lncHR1TG group.
Discussion
The mammalian genome transcribes several thousand lncRNAs but how lncRNAs function is just being understood. So, mechanisms of action for only a few are described. Evidence suggests that lncRNAs may contribute to control of immunity, tumorigenesis, cell proliferation, differentiation and apoptosis. However, systematic identification and characterization of lncRNAs involved in hepatitis viral infection and hepatic lipid metabolism are lacking.
We report the discovery of a novel lncRNA molecule lncHR1, which suppresses the SREBP-1c expression and participates in lipid metabolism regulation. Our data suggest that TG and LD accumulation can be inhibited by lncHR1 in both cultured hepatoma cells and in transgenic mice.
Biological characteristics of lncHR1. (A) Structural diagram of lncHR1 gene. (B) Endogenous lncHR1 expression was measured in 11 cell lines using absolute qRT-PCR. (C) Intracellular localization of lncHR1 was visualized in Huh7 cells with RNA-FISH assays (blue for nucleus and green for lncHR1). Scale bars, 13 μm.
LncHR1 negatively regulates endogenous SREBP-1c and FAS. (A) LncHR1 overexpression plasmid was transfected into Huh7 cells in a dose dependent manner and intracellular lncHR1 was quantified by qRT-PCR. An empty vector was a negative control. The data were normalized to relative fold changes. (B) Post-transfection of lncHR1 overexpression plasmid for 48 h, endogenous SREBP-1c mRNA (up) and protein (down) were measured by qRT-PCR and Western blot, respectively. (C) LncHR1 knockdown plasmid (shRNA) was transfected into Huh7 cells in a dose dependent manner and intracellular lncHR1 lever was quantified by qRT-PCR. (D) Forty-eight hours post transfection of lncHR1 knockdown plasmid, endogenous SREBP-1c mRNA (up) and protein (down) levels were measured by qRT-PCR and Western blot, respectively. (E) HCV was inoculated into Huh7 cells 24 h before transfection with either control vector or lncHR1 overexpression plasmid and 48 h post-transfection, endogenous SREBP-1c protein was measured with Western blot. Actin was used as an equal loading control. (F) Endogenous FAS protein expression was measured in Huh7 cells with overexpressed (up) or decreased (down) lncHR1. Empty vectors were negative controls. Data are means ± SD. *p < 0.05, **p < 0.01
LncHR1 suppressed TG accumulation and LD formation in steatotic hepatocytes. Huh7 cells were transfected with lncHR1 overexpression (A) or knockdown (B) plasmids, and then treated with OA at 24 h post-transfection. An empty vector was used as negative control. Endogenous SREBP-1c protein was measured with Western blot (A, B) and intracellular TG was quantified (C, D). Intracellular LD formation in Huh7 cells was visualized with oil red O staining. The nuclei were indicated by DAPI staining. Scale bars, 30 μm. (E, F). Volume of LD in each treatment was determined with Image J software (G, H). Data are means ± SD. *p < 0.05, **p < 0.01, *** p < 0.001.
Genomic studies have identified numerous genes involved in the lipid metabolism that can be affected by HCV infection. Currently, few studies about the function of liver disease-related lncRNA are available. HULC is the first documented liver-specific lncRNA [24], that is specifically and highly expressed in HCC compared to normal liver tissue [24]. Recently, Li and colleagues revealed a mouse-specific and liver-enriched lncRNA, lncLSTR, which regulates TG clearance during lipid homeostasis [25]. However, the relationship of lncRNAs associated with hepatitis viral infection and hepatic lipid metabolism in human cells are unknown. Interestingly, our efforts to screen HCV infection-regulated host lncRNAs revealed that lncHR1 is a negative regulator of SREBP-1c expression and TG accumulation. Alignment sequence analysis indicated that lncHR1 is a human-specific lncRNA. To our knowledge, lncHR1 is the first lncRNA reported to regulate lipid synthesis via SREBP-1c.
Hepatic lipid abnormalities are a characteristic feature of HCV infection. In experimental studies using cultured cells, the expression of transfected HCV core proteins has been found to increase lipid accumulation [26]. Studies examining the effect of HCV infection on SREBP-1c have shown that the genes are transcriptionally induced, and their proteolytic cleavage is stimulated [22]. Then, in another study, the expression of HCV core protein was found to activate the FAS [27] .Our results indicated that lncHR1 inhibited the expression of SREBP-1c by HCV infection. This suggests that lncHR1 is able to inhibit viral infection and protect the host.
LncHR1 regulated lipid metabolism in HFD-induced transgenic mice. (A) LncHR1 expression in the lncHR1TG group was ~9-fold greater than the WT group (n= 6/group). (B) In liver tissues, SREBP-1c, FAS and ACCα level in both groups were measured with Western blot. β-actin was used as an equal loading control. (C-E) Serum TG, VLDL and LDL were measured. (F) Liver TG were measured in the lncHR1TG group and WT group (n=6/group). (G) At week 9 after HFD induction, the ratio of liver to body weight in transgenic mice was calculated in both groups (n=6/group). (H) Weight gain of two groups was measured weekly up to the 9th week. Data are means ± SD. *p < 0.05, **p < 0.01.
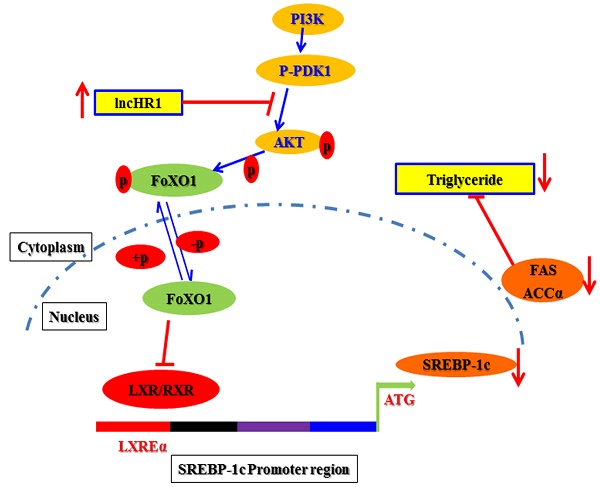
Former experiments verified that lipid metabolism genes SREBP-1c, whose expression was upregulated during early HCV infection [28]. It has been shown that PI3K/AKT signalling pathway is also stimulated by HCV infection and the result of this activation is the increased expression of target genes involved in hepatic lipid biosynthesis, such as FAS and ACCα [22]. Our part of results suggested that lncHR1 could regulate AKT/FoxO1 phosphorylation and then affect the location of FoxO1 (the detailed results haven't shown in paper). So, the detailed molecular mechanism of lncHR1 regulation of SREBP-1c expression needs to be further studied.
SREBP-1c regulates lipogenic gene expression, enhances FA synthesis, and accelerates TG accumulation [29]. Our observations confirmed that lncHR1 indeed suppressed the expression of SREBP-1c and FAS in vitro. In the liver, FA was synthesized by catalytic synthesis of TG, which can be stored in LD into the hepatocytes or secreted as TG-enriched lipoproteins into the bloodstream. We reported in vitro that lncHR1 could suppress TG synthesis and decrease LD formation in OA-induced hepatic cell.
At present, there is no research on the establishment of lncRNA animal model. We established an lncHR1 transgenic mouse model and had lncHR1 high expression mice. The data indicated that in the lncHR1TG group of HFD-induction, basal hepatic SREBP-1c FAS, and ACCα were decreased. Then, the blood biochemical indicators of the lipoprotein may reflect the accumulation of lipids in the body. Theoretically, less serum lipoprotein could be caused by decreased TG synthesis and increased FA oxidation. Our data show that serum TG, VLDL and LDL were decreased in the lncHR1TG group. So we can speculate that lncHR1 can prevent the excessive synthesis of TG caused by high fat diet. A main form of lipids within hepatocytes, mostly in the form of triglycerides, is a prerequisite for the development of non-alcoholic steatohepatitis. In our study, the result showed that hepatic TG and the ratio of liver /body weight were reduced in the lncHR1TG group. And the results suggest that lncHR1 may serve as a new molecule for the treatment of TG accumulation in liver. These data indicate that lncHR1 may be a protective host non-coding RNA, which can be up-regulated in some diseases and avoids improper activation of the lipid synthesis pathway.
Thus, lncHR1 affects cellular lipid metabolism by regulating critical lipogenic transcriptional factors SREBP-1c. Whether other cellular lncRNAs perform functions similar to lncHR1 is unclear but lncHR1 may have therapeutic potential for treating hyperlipidemia-associated diseases.
Supplementary Material
Supplementary figures and tables.
Abbreviations
HCC: Hepatocellular carcinoma; lncRNA: long non-coding RNA; SREBP-1c: sterol regulatory element binding protein 1c; OA: Oleic acids; LXRα: liver X receptor α; DAPI: 4', 6-diamidino-2-phenylindole; HFD: high-fat diet; DIO mice: diet-induced obesity mice; FAS: fatty acid synthase; ACCα: Acetyl-CoA carboxylase α; RT-PCR: reverse transcription-polymerase chain reaction; qRT-PCR: quantitative real-time polymerase chain reaction; LD: lipid droplet; TG: triglyceride.
Acknowledgements
This work was supported by the following funding sources, National Natural Science Foundation of China (81672030), National Basic Research Program of China (2015CB554301), CAMS Initiative for Innovative Medicine (2016-I2M-3-020) and National Natural Science Foundation of China (81471954).
Author's contribution
Duan Li and Wei Yang designed and conducted experiments. Wei Yang and Min Cheng supervised all the experiments. Yuqiang Niu, Xiaojing Chi, Xiuying Liu, Jingjing Fan, Heng Fan provided technical and material support. Duan Li, Wei Yang and Yongsheng Chang analyzed the data and prepared the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lee JS, Ward WO, Knapp G, Ren H, Vallanat B, Abbott B. et al. Transcriptional ontogeny of the developing liver. BMC Genomics. 2012;13:33
2. Negro F. Hepatitis C virus and liver steatosis: when fat is not beautiful. J Hepatol. 2004;40:533-5
3. Siagris D, Christofidou M, Theocharis GJ, Pagoni N, Papadimitriou C, Lekkou A. et al. Serum lipid pattern in chronic hepatitis C: histological and virological correlations. J Viral Hepat. 2006;13:56-61
4. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330-42
5. Goodridge AG. Dietary regulation of gene expression: enzymes involved in carbohydrate and lipid metabolism. Annu Rev Nutr. 1987;7:157-85
6. Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331-40
7. Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987-92
8. Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I. et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819-30
9. Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125-31
10. Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72-82
11. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
12. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-27
13. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-504
14. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298-307
15. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38
16. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
17. Li X, Wu Z, Fu X, Han W. Long Noncoding RNAs: Insights from Biological Features and Functions to Diseases. Med Res Rev. 2013;33:517-53
18. Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. Journal of Molecular Medicine. 2014;92:337-46
19. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y. et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302-11
20. Cheng M, Si Y, Yang Y, Liu X, Gong Q, Zhao J. et al. Recombinant human interleukin 28B: anti-HCV potency, receptor usage and restricted cell-type responsiveness. J Antimicrob Chemother. 2012;67:1080-7
21. Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J. et al. High-Throughput Profiling of Alpha Interferon- and Interleukin-28B-Regulated MicroRNAs and Identification of let-7s with Anti-Hepatitis C Virus Activity by Targeting IGF2BP1. Journal of Virology. 2013;87:9707-18
22. Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-30
23. Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV. et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362-7
24. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106
25. Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H. et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455-67
26. Abid K, Pazienza V, de Gottardi A, Rubbia-Brandt L, Conne B, Pugnale P. et al. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744-51
27. Jackel-Cram C, Babiuk LA, Liu Q. Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol. 2007;46:999-1008
28. Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R. et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669-74
29. Latasa MJ, Moon YS, Kim KH, Sul HS. Nutritional regulation of the fatty acid synthase promoter in vivo: sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc Natl Acad Sci U S A. 2000;97:10619-24
Author contact
![]() Corresponding author: Tel: +86 10 67872436; Fax: +86 10 67872436; Email: wyangpumc.edu.cn
Corresponding author: Tel: +86 10 67872436; Fax: +86 10 67872436; Email: wyangpumc.edu.cn

 Global reach, higher impact
Global reach, higher impact