10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(5):545-560. doi:10.7150/ijbs.18649 This issue Cite
Review
Multifaceted Regulation of Gene Expression by the Apoptosis- and Splicing-Associated Protein Complex and Its Components
Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, Guwahati-781039, Assam, India.
Received 2016-12-7; Accepted 2017-2-24; Published 2017-4-10
Abstract
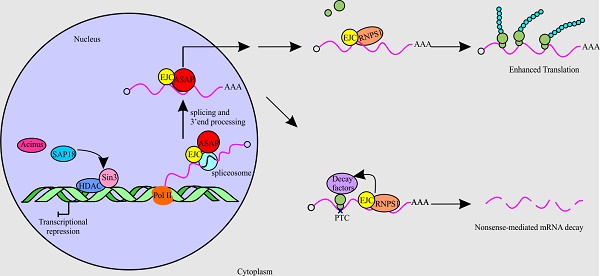
The differential deposition of RNA-binding proteins (RBPs) on pre-mRNA mediates the processes of gene expression. One of the complexes containing RBPs that play a crucial part in RNA metabolism is the apoptosis-and splicing-associated protein (ASAP) complex. In this review, we present a summary of the structure of ASAP complex and its localization. Also, we discuss the findings by different groups on various functions of the subunits of the ASAP complex in RNA metabolism. The subunits of the ASAP complex are RNPS1, Acinus and SAP18. Originally, the ASAP complex was thought to link RNA processing with apoptosis. Further studies have shown the role of these components in RNA metabolism of cells, including transcription, splicing, translation and nonsense-mediated mRNA decay (NMD). In transcription, RNPS1 is involved in preventing the formation of R-loop, while Acinus and SAP18 suppress transcription with the help of histone deacetylase. On the one hand, individual components of the ASAP complex, namely RNPS1 and Acinus act as splicing activators, whereas on the other hand, in-vitro assay shows that the ASAP complex behaves as splicing repressor. In addition, the individual members of the ASAP complex associates with the exon junction complex (EJC) to play roles in splicing and translation. RNPS1 increases the translation efficiency by participating in the 3'end processing and polysome association of mRNAs. Similarly, during NMD RNPS1 aids in the recruitment of decay factors by interacting with EJC.
Keywords: ASAP, EJC, NMD, RBP, RNA metabolism, splicing, transcription, translation, apoptosis.
Introduction
In eukaryotic cells, expression of genes starts in the nucleus when transcription yields pre-mRNAs. Transcription is then followed by translation, but before the translation begins, these pre-mRNAs have to undergo wide range of post-transcriptional processing, including splicing, 5' end capping and 3' end polyadenylation. Even though transcription and post-transcriptional processing involve different machineries but evidence suggest, transcription of intron-containing genes is coupled with pre-mRNA processing [1]. All these coordinated processes of RNA metabolism are mediated by a huge number of RNA-binding proteins (RBPs), which are mostly associated with trans-acting RNA to form RNA-protein complex called ribonucleoprotein particles (RNPs). Although all RBPs interact with pre-mRNA, different RBPs have diverse specificity and affinity for RNA sequences. In addition, different RBPs recruit particular subsets of auxiliary proteins on pre-mRNA, thus further changing the RBP composition on mRNAs. Altogether these remodelling of RBPs determine the fate of each mRNA [2].
One of the protein complexes that is a part of the RBPs is the apoptosis-and splicing-associated protein (ASAP) complex. The ASAP complex was initially isolated from HeLa cell nuclear extract, which comprises of subunits, namely RNA-binding protein with serine-rich domain 1 (RNPS1), apoptotic chromatin condensation inducer in the nucleus (Acinus) and Sin3-associated protein of 18 kDa (SAP18) [3]. ASAP dynamically interface with a multiprotein complex termed, exon junction complex (EJC), which gets placed at a distance of 20-24 nt upstream of exon-exon junction on mRNA concomitant with splicing. It serves as binding site for proteins like ASAP, splicing, export and nonsense-mediated mRNA decay (NMD) factors. The EJC has four subunits in its core, namely the RNA helicase eukaryotic initiation factor 4A3 (eIF4A3), Y14, MAGOH and metastatic lymph node 51 (MLN51) [4].
The binding of ASAP with EJC is believed to facilitate the functions of ASAP subunits in RNA metabolism starting from transcription, splicing and translation to NMD. The ASAP complex has also been found to function in apoptosis and hence got its name after its role in apoptosis as well as in splicing at the time of discovery. In the presence of apoptotic stimuli like staurosporine, ASAP complex accelerates the progression of cell death and also gets disassembled during apoptosis. Thus, the ASAP complex is also presumed to play an essential part in the regulation of splicing during apoptosis [3].
In this review, we discuss the structural aspects of the ASAP complex. We also describe the individual and complex-associated functions of RNPS1, Acinus and SAP18 in RNA metabolism, including transcription, splicing, translation, NMD and apoptosis.
Structure of the ASAP complex
The ASAP complex came into the picture while identifying protein complexes containing SAP18. SAP18 was earlier known to be a part of Sin3 histone deacetylase (Sin3-HDAC) complex. With the intention to find additional protein complexes associated with SAP18, biochemical fractionations of HeLa nuclei extract were carried out and SAP18 specific antibodies utilised in Western blotting led to the finding of a novel complex termed as ASAP. Mass spectrometry data identified the subunits of the ASAP complex as 50 kDa protein RNPS1, 220 kDa isoform of Acinus (Acinus-L) and 18 kDa protein SAP18 [3].
RNPS1 is a general activator of pre-mRNA splicing and its amino acid sequence is related to serine/arginine-rich (SR) protein family [5, 6]. SR proteins have domains rich in arginine and serine dipeptides and also have an RNA-binding domain known as RNA recognition motif (RRM). The SR proteins are required in the regulation of pre-mRNA splicing, mRNA export and translation [7]. Similar to members of the SR protein family, RNPS1 amino acid sequence comprises of an N-terminal serine-rich domain (S domain), a middle RRM domain and a carboxy-terminal with arginine/serine/proline-rich (RS/P) domain. The RRM domain encompasses conserved RNP-1 and RNP-2 sub-motifs and a conserved decapeptide box (Figure 1A) [6]. The RRM domain of RNPS1 is reported to be more closely related to the RRM of hnRNP C, a constituent of heterogeneous nuclear RNP particles, which functions in mRNA splicing, as compared to the RRM of other SR proteins [5]. Intriguingly, RNPS1 mediates protein-protein interaction with its RRM domain which is usually utilized by other SR proteins to bind nucleic acids [8]. Individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) analysis has shown that RNPS1 mainly binds 20-24 nt upstream of exon-exon junctions of mRNAs. These mRNAs predominantly encode proteins involved in RNA processing, cell cycle control, or chromosome organization. Although the binding is non-sequence specific but recent study indicates minor sequence preferences for motifs like 5'-GAAGA-3'[9]. Interestingly, this motif 5'-GAAGA-3' resembles the binding site preference of an SR protein, SRSF1 (ASF/SF2). This motif serves as an exonic splicing enhancer (ESE); ESEs are cis-acting sites on exons of pre-mRNA that stimulate the splicing of an upstream weak intron [10]. Evidence that RNPS1 and SRSF1 interacts as demonstrated via assays like the yeast two-hybrid system and mass spectrometry [11, 12] indicates that RNPS1 bound to GAAGA motif aids in the recruitment of SR protein (SRSF1) or it might be vice-versa i.e. recruitment of SRSF1 may tether RNPS1 to ESE site. The sequential recruitment of either RNPS1 or SRSF1 to this GAAGA site remains to be explored.
Acinus, the next subunit of the ASAP complex, is a nuclear factor, which upon activation by caspase-3, functions in apoptotic chromatin condensation during apoptosis. It also functions in transcriptional regulation and RNA processing. Acinus exists in three isoforms: Acinus-L, Acinus-S and Acinus-S'. Acinus-L is the longest isoform with 1341 amino acids, while Acinus-S and Acinus-S' contain 583 and 568 amino acids, respectively. Moreover, Acinus-S contains unique sequence MLSESKEG at the N-terminus followed by residues 767-1341 of Acinus-L. Similarly, Acinus-S' contains residues 774-1341 of Acinus-L [13]. Acinus-S' amino acid sequence also includes an RRM domain, an RNPS1-SAP18 binding (RSB) motif and C-terminal end rich in Arg/Ser, Arg/Glu and Arg/Asp repeats [8, 14]. All the isoforms of Acinus share a common central RRM domain, RSB motif and a C-terminal domain. The isoforms vary only in their N-terminal domain. While N-termini of Acinus-S and Acinus-S' have no conserved domains, the N-terminal domain of Acinus-L contains an additional SAP [after SAF-A/B, Acinus and PIAS (STAT inhibitors)] motif and an RS domain [15] (Figure 1B). Nevertheless, the isoforms of Acinus have no functional differences [13, 14, 16, 17] and are capable of forming the ASAP complex, as well as interact with the EJC complex [3, 18]. The SAP motif of Acinus-L plays a critical role in binding AT-rich chromosomal regions termed as scaffolds or matrix attachment regions (SARS/MARS) [19-21]. SARS/MARS attach chromatin loops to nuclear scaffold/matrix and are the sites of gene regulation [22, 23]. As a result, the binding of the SAP motif with SARS/MARS could bring Acinus-L in the vicinity of target genes and facilitates interaction with other regulatory proteins to increase their regulatory effect on transcription and splicing [24]. It has been shown that Acinus mediates binding of the ASAP complex to RNA and it mostly binds to exonic regions [8, 25]. When Acinus is bound to exons, it can bind to either canonical or non-canonical EJC positions and is associated with eIF4A3 at canonical EJC positions. Whereas Acinus also binds to the 3'end of the intronic region spanning the intron-exon boundary, this binding is independent of the core EJC. Introns that are bound by Acinus are shorter, have higher GC content and bear weak 5' and 3' splice sites [25].
Schematic representation of the domain structure of apoptosis-and splicing-associated protein (ASAP) complex components. (A) RNA-binding protein with serine-rich domain 1 (RNPS1) includes serine-rich (S) domain (residues 69-121), RNA recognition motif (RRM) (residues 162-243) and C-terminal arginine/serine/proline-rich (RS/P) domain (residues 268-305). RRM domain contains RNP-1 (residues 203-210), RNP-2 (residues 163-168) sub-motifs and conserved decapeptide box (Cons. Box) (residues 221-230). (B) The three isoforms of apoptotic chromatin condensation inducer in the nucleus (Acinus-L, S and S') are indicated. Acinus-L includes P-loop, SAP motif, arginine-serine (RS) domain, RRM domain (residues 987-1093) and RNPS1-SAP18 binding (RSB) motif (residues 1210-1237). The N-terminal domain of Acinus-S contains the unique sequence followed by residues 767-1341 of Acinus-L and Acinus-S' contains residues 774-1341 of Acinus-L. (C) Sin3-associated protein of 18 kDa (SAP18) includes an ubiquitin-like (UBL) domain (residues 20-143).
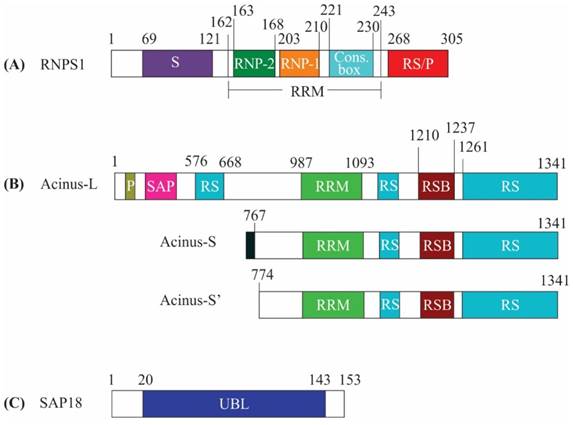
SAP 18 was originally isolated as a part of Sin3-HDAC complex and was found to be involved in deacetylation of histones as well as in transcriptional repression [26, 27]. The solution structure of SAP18 exhibits the presence of a conserved ubiquitin-like (UBL) domain (Figure 1C) and the ubiquitin-like structure contains antiparallel five stranded β sheet curled around a central α helix [28]. It has been observed that ubiquitin-like fold-containing proteins mostly act as a platform for the assembly of multiprotein complexes [29]. In line with the above, SAP18 stably associate with RNPS1:Acinus heterodimer to form the ASAP complex, whereas is incapable of interacting with RNPS1 and Acinus in isolation [18].
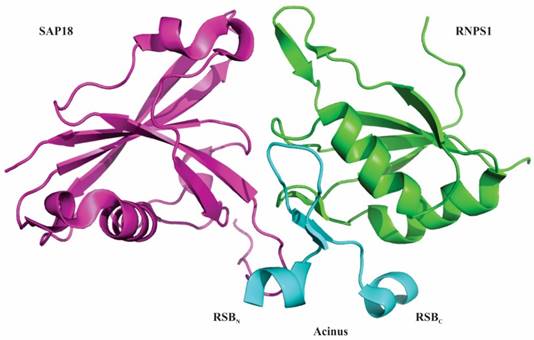
In 2012 Murachelli et al. reported the crystal structure of a minimal ASAP complex. They used 656-683 residues of Drosophila.melanogaster Acinus, 159-244 residues of human RNPS1 and 14-143 residues of mouse SAP18. While human Acinus rapidly degrades when purified from bacterial cultures, D. melanogaster Acinus was stable and could therefore be used during large-scale purifications. The crystal structure revealed that the interacting platform of the ASAP complex constitutes the RRM domain of RNPS1, the UBL domain of SAP18 and the RSB motif of Acinus [8]. The RSB motif of Acinus contains β-hairpin fold that is flanked by amino-terminal helical segment (RSBN) and carboxy-terminal helical segment (RSBC). The amino-terminal helical segment of RSB motif interacts with SAP18 and its carboxy-terminal helical segment interacts with RNPS1 (Figure 2) [8]. It has also been found that RNPS1 and SAP18 interact with Pinin, a splicing coactivator. This interaction uses an RSB motif present in Pinin to form an alternative protein complex known as PSAP [8, 30, 31]. Interestingly, incubation of PSAP complexes with excessive amount of Acinus did not generate ASAP complexes, implying that the RSB motif of Pinin has binding affinity towards RNPS1 and SAP18 higher than Acinus. The solvent accessible part of the RSB motif of Acinus and Pinin is conserved with a basic Lys-Thr-Lys/Arg sequence, indicating the use of similar conserved sequences by ASAP and PSAP to bind other proteins and RNA [8]. Both, Acinus and Pinin have been suggested to associate with the EJC to control the process of transcription as well as splicing [3, 14, 30, 32-35] and the recruitment might depend on the splicing of subset of transcripts. Therefore, the recruitment of either ASAP or PSAP complexes via EJC before or during splicing could be mutually exclusive events [8].
Structure of ASAP complex. RNA-binding protein with serine-rich domain 1 (RNPS1) is represented in green, apoptotic chromatin condensation inducer in the nucleus (Acinus) is in blue and Sin3-associated protein of 18 kDa (SAP18) is in purple. The interacting surface of ASAP complex comprises of the RNA recognition motif (RRM) of RNPS1, the ubiquitin-like (UBL) domain of SAP18 and RNPS1-SAP18 binding (RSB) motif of Acinus. The RSB motif of Acinus (Drosophila melanogaster) folds into a β-hairpin flanked by short amino and carboxy-terminal helical segments. The amino-terminal helical segment (RSBN) of RSB motif interacts with SAP18 and its carboxy-terminal helical segment (RSBC) interact with RNPS1. Atomic coordinates of ASAP complex with PDB accession code 4A8X were modelled with PyMol (http://www.pymol.org/) according to Murachelli et al. 2012.
Localization of the ASAP complex
Protein localization studies demonstrate the distribution of proteins in various cellular compartments and thus, help in gaining insight into their potential functions. For instance, it is believed that proteins that localize in nuclear speckles are involved in pre-mRNA processing. These factors are then recruited to perichromatin fibrils or the periphery of nuclear speckles, which are the regions of transcription and splicing [36, 37].
Studies have shown that RNPS1 colocalizes with SR proteins in nuclear speckles, characteristics of most of the pre-mRNA processing factors [6]. Both RS/P and S domains are necessary for localization of RNPS1 in nuclear speckles [35]. Acinus-S and Acinus-S' also localizes to nuclear speckles and the RS domain along with RD/E repeats of Acinus-S' are essential for localization, whereas Acinus-L localizes diffusely in the nucleoplasm and is dependent on the SAP motif of Acinus-L [24, 38]. Interestingly, it has been seen that sub-nuclear localization of RNPS1 is guided by Acinus isoforms. In the absence of Acinus isoforms, RNPS1 localizes partially in nuclear speckles and partially in the nucleoplasm. Whereas coexpression of RNPS1 and Acinus-S' colocalize both proteins in nuclear speckles, alternatively coexpression of RNPS1 and Acinus-L direct both RNPS1 and Acinus-L in the nucleoplasm [24]. SAP18 also colocalizes with Acinus to nuclear speckles and mutation in the ubiquitin-like fold of SAP18 hinders nuclear speckle localization [32]. Intriguingly, the interaction of RNPS1 and SAP18 with the SR protein SRSF11 (p54 or SRp54) and other splicing factors is thought to be required for their proper nuclear speckle localization [32, 35]. Immunofluorescence data supports the colocalization of ASAP complex components in nuclear speckles suggesting the assembly of ASAP complex in nuclear speckles [3].
Roles of the ASAP components in mRNA metabolism
The ASAP protein components SAP18 and RNPS1 are shuttling proteins and alternate between the nucleus and the cytoplasm, whereas Acinus is a nuclear-restricted protein. Consequently, Acinus is involved in mRNA metabolism pertaining to the nucleus. The stable association of SAP18 requires the Acinus:RNPS1 heterodimer. Since Acinus dissociates before mRNA export, it is likely that SAP18 also dissociates soon after mRNA export. Thus, SAP18 is likely involved in mRNA metabolism in the nucleus [18]. In contrast, RNPS1 remains associated with the EJC in the spliced mRNA as well as with CBP80, the large subunit of nuclear 7-methyl-G cap binding complex in the cytoplasm, but it does not associate with eukaryotic translation initiation factor 4E (eIF4E), which recruits ribosomes to the 5'-cap structure. Altogether, these data implies that RNPS1 remain bound to the spliced mRNA in the cytoplasm until the initial round of translation and therefore is involved in mRNA metabolism both in nucleus and cytoplasm [39].
Transcription
During transcription, RNA polymerase catalyses the synthesis of RNA from DNA template. However, the nascent RNA transcript that exits out of the RNA polymerase could anneal back with the template DNA to form the DNA:RNA hybrid termed as R-loop. Persistent R-loops cause deleterious effects like DNA double-strand breaks as well as genomic instability. Therefore, cells need to prevent their formation or remove R-loops that have already been formed (reviewed in [40]).
RNPS1, Acinus and SAP18 in transcription
The assembly of RBP complexes on nascent RNA transcripts function in preventing R-loops and studies have shown that RNPS1 and SRSF1 are crucial proteins in this context [41]. Depletion of SRSF1 leads to R-loop formation, hypermutation, high molecular weight (HMW) DNA fragmentation and G2 cell cycle arrest [42]. Intriguingly, overexpression of RNPS1 suppresses the effects of SRSF1 depletion, suggesting that it complements the lack of SRSF1 in mRNP formation. One another possibility is that RNPS1 influences other unknown proteins, which in turn complements the role of SRSF1 in averting R-loop formation [41].
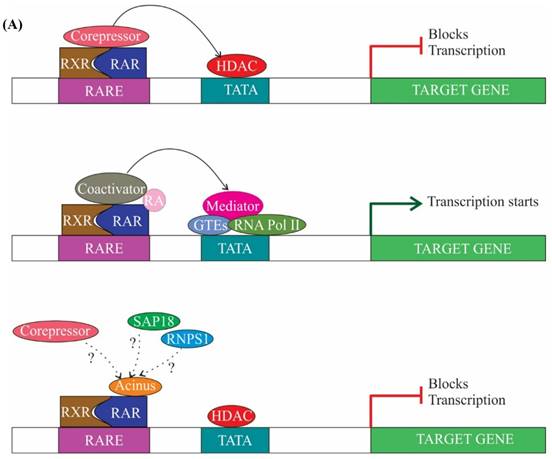
Transcription is largely regulated by transcription factors, which associate with cis-acting regulatory sequences in/near the promoter region and facilitate assembly of transcriptional machinery. Transcription factors can function in ligand dependent or ligand independent manner. Ligand-dependent transcription requires the binding of a ligand to activate the transcription factor. For example, expression of genes which have a retinoic acid response element (RARE) in their promoter region can be regulated by both ligand-dependent and ligand-independent means. In the ligand-dependent manner, a ligand named retinoic acid (RA) associates with a specific nuclear receptors termed RA receptors (RARs), which are heterodimerized with retinoid X receptors (RXRs) at the RARE site of genes. Binding of ligand causes conformational alteration in RARs resulting in dissociation of corepressors and successive recruitment of activators and expression of genes (Figure 3A) (reviewed in [43, 44]). It has been demonstrated that Acinus-S' interacts via its carboxy-terminal domain with RARs to repress both ligand-dependent and ligand-independent expression of RAR-regulated genes (Figure 3A). Although the exact mechanism behind the repression by Acinus-S' is not known, but it seems that histone deacetylation is partly responsible for the repression activity [14].
The other subunit of the ASAP complex, SAP18, is also a component of Sin3 complex, which acts as a global transcriptional repressor [26, 45]. Sin3 complex interacts with HDAC and mediates transcriptional repression (Figure 3B) [26]. However, it is not clear whether SAP18 independently (without forming ASAP) or as a part of the ASAP complex interacts with the components of Sin3 complex to mediate recruitment of HDAC and repression of transcription. Briefly, the above studies give an impression of RNPS1 involvement in suppression of R-loop formation [41], and the function of Acinus and SAP18 in repression of transcription [14, 26]. Acinus interacts with SAP18, which in turn interacts with Sin3 complex and recruits HDAC. Hence, the function of Acinus and SAP18 in transcription repression might be inter-correlated.
Splicing
Splicing of pre-mRNA is catalysed by the spliceosome, which undergoes sequential re-arrangements during splicing. At first, the U1 snRNP binds to the 5' splice site and along with other non-snRNPs form the early (E) complex. The E complex then recruits U2 snRNP to the branch site and forms the A complex (prespliceosome complex) [46]. The formation of the A complex commits the pre-mRNA splicing to a pair of 5' and 3' splice sites [47]. Subsequently, recruitment of U4/U5/U6 tri-snRNPs generate the pre-catalytic B complex and rearrangements like dissociation of U1 and U4 snRNPs together with U2/U6 and U6/pre-mRNA base pairing lead to the generation of an active spliceosome (B* complex). The B* complex functions in catalysis of the first step of splicing giving rise to C complex. The C complex then undergoes conformational rearrangements and activated C complex catalyses the final step yielding the spliced mRNA [46].
Presence of ASAP components in spliceosomal complex
Identifying the protein components of affinity purified spliceosome A complex by liquid chromatography ESI tandem mass spectrometry (LC-ESI-MSMS) revealed the presence of RNPS1 and Acinus [48]. RNPS1 besides being interacting with spliceosome A complex, also enhances the generation of ATP-dependent spliceosome A complex [49]. Additionally, it has been observed that RNPS1 interacts with Pinin and SRSF11, which in turn interact with U2 snRNP proteins and U2 snRNP auxiliary factor 65 kDa (U2AF65), respectively [35, 50, 51]. It is known that U2AF65 binds to the polypyrimidine tract (PPT) of introns and recruits U2 snRNP to the branch site stimulating the generation of spliceosome A complex [52]. Thus, supporting the observation that RNPS1 is one of the factors that influences the formation of A complex [35]. Another study reported the presence of RNPS1, Acinus and SAP18 in spliceosome B and C complexes as well [53].
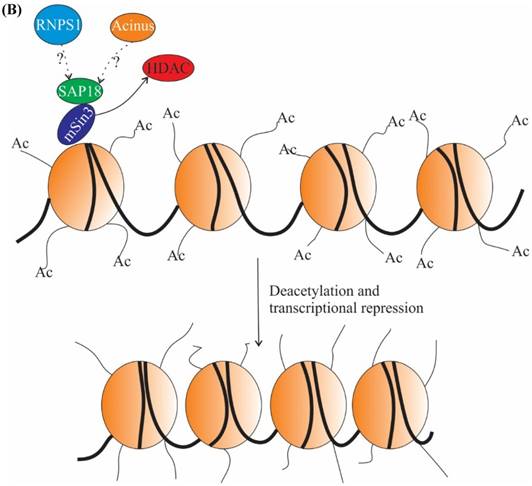
The role of ASAP components in transcription. (A) In absence of ligand retinoic acid (RA), corepressor complex binds to RA receptors (RARs) which are heterodimerized with retinoid X receptors (RXRs), resulting in repression of transcription by the recruitment of histone deacetylation complex (HDAC) (top panel). In presence of ligand RA, coactivator complexes are recruited and subsequently, transcription is activated (middle panel). Acinus-S' interacts with RARs and represses transcription. The repression may be brought about by HDAC, corepressor and other unknown mechanisms (bottom panel). (B) SAP 18 interacts with mSin3 complex. mSin3 in turn interacts with HDAC and promotes deacetylation and repression of transcription. Undotted arrows indicate known interactions, whereas dotted arrows indicate unknown interactions. RARE, retinoic acid response element; GTEs, general transcription factors; Ac, acetylation.
Role of RNPS1 in splicing
Studies have shown that RNPS1 activates constitutive splicing of pre-mRNA. For example, in the presence of limiting amounts of splicing factor SRSF1, RNPS1 stimulates constitutive splicing of β-globin pre-mRNA. It also influences the constitutive splicing of transcripts like HIV-tat, immunoglobulin μ-chain (IgM), Drosophila ftz and δ-crystalline [6]. However, RNPS1 influences primarily the major splicing pathway as it stimulates splicing of pre-mRNA with GU-AG introns and has no impact on AT-AC intron-containing pre-mRNA [6]. Supporting the line of evidence, knockdown of RNPS1 via siRNA instigates retention of introns in transcripts like MDM2 (murine double minute2) and AURKB (Aurora B kinase) [54].
Acinus role in splicing
Acinus-L and Acinus-S' promote both constitutive as well as alternative splicing of pre-mRNAs with weak 5' splice site but have no impact on strong 5' splice site [24]. It has been shown that the retinoic acid (RA)-dependent minigene with a weak splice site is efficiently spliced by Acinus, whereas splicing of a RA-responsive minigene bearing a strong 5' splice site is not augmented. This splicing activity by Acinus is demonstrated through its RRM domain, which is crucial for its association with RA. The RA-dependent splicing activity of Acinus in RA-responsive genes is most probably due to the release of corepressor complexes and recruitment of coactivators in the presence of RA. In the absence of RA, the unliganded RA receptor RAR/RXR heterodimer at the RARE promoter site interacts with corepressor complexes, causing compaction of chromatin and thus, hampering recruitment of both transcriptional and splicing machinery [24]. Interestingly, it has been observed that RNPS1 represses the action of Acinus-S' in non-RA-dependent pre-mRNA splicing, but only partially in RA-dependent pre-mRNA splicing. The possible mechanism behind repression is competition between Acinus and RNPS1 for splicing factors such as SR proteins recruited by RNA polymerase II and general transcription factors. As RNPS1 silences the splicing activity of Acinus-S', RNPS1 is more likely to have a higher affinity for splicing factors than Acinus-S'. However, distinct transcription activators and coactivators recruit specific splicing factors; similarly in RA-dependent splicing, different subsets of splicing factors might be recruited. As a consequence, competition between Acinus-S' and RNPS1 is abated in ligand-dependent pre-mRNA splicing and splicing activity of Acinus-S' is only partly repressed [24]. In summary, it can be speculated that competition between RNPS1 and Acinus modulates splicing of pre-mRNA transcripts. In addition, the presence of particular cis-acting RNA sequences like weak splice-sites could also decide the recruitment of different ASAP components.
Involvement of SAP18 in splicing
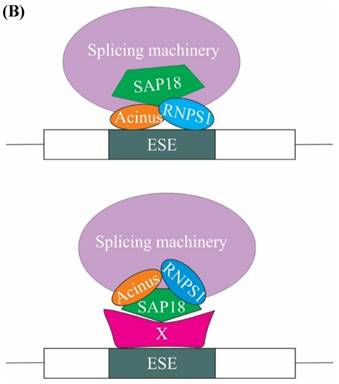
A subsequent study showed that SAP18 functions in modulation of splicing by exon inclusion activity. It has been hypothesized that exon inclusion activity mediated by SAP18 could be the result of either 1) SAP18 acting as a scaffold protein for bringing together the ASAP complex and spliceosomal machinery on the exonic splicing enhancers (ESEs) present on the pre-mRNA or 2) SAP18 functions as an adaptor molecule, recruited to the ESE by a hypothetical protein, SAP18 then eventually recruits RNPS1, Acinus and other splicing factors (Figure 4B) [32].
ASAP's function in splicing
Surprisingly, the ASAP (ASAP-L and ASAP-S) as a complex was reported to repress RNA processing mediated by splicing factors like SRSF1, SRSF2 and RNPS1; albeit RNPS1 mediated splicing was less sensitive to ASAP complexes [3]. The contradictory role of individual ASAP components as splicing activator and the ASAP complex as splicing repressor could be due to the variable interacting partners associated with RNPS1, Acinus and SAP18 in vivo. The function of the ASAP complex as repressor of splicing was studied in vitro and till date similar in vivo studies are still missing. These would be required to understand the effect of dynamics of interacting partners on the subunits of ASAP complex.
Concerted function of ASAP components and EJC in splicing
Components of the ASAP complex, RNPS1, Acinus and SAP18, does not only form ASAP complex but is also involved in generation of PSAP complex and peripheral components of the EJC. Specifically, many functional roles of RNPS1 with EJC have been demonstrated. For example, it was recently shown that the core EJC (eIF4A3; Mago, homolog of human MAGOH; Y14) and RNPS1 are crucial for Wingless (Wg)/Wnt signalling by regulating the splicing of gene discs large 1 (dlg1) [55]. Wnt signalling is essential for cell proliferation, cell migration, cell polarity, pattern formation, stem cell maintenance and adult tissue homeostasis. In drosophila, the glycoprotein Wnt binds to the Frizzled (Fz) family of receptors and a co-receptor called Arrow (Arr) (LRP5/6 in vertebrates) to form Wnt/Fz/Arr trimeric complex that activates the phosphoprotein Dishevelled (Dsh). The activated Dsh destabilizes APC/Axin/GSK3 (adenomatosis polyposis coli, Axin and glycogen synthase kinase 3) degradation complex and inhibits degradation of a transcription factor, Armadillo (Arm) (β-catenin in vertebrates). The protected Armadillo translocate to the nucleus and triggers transcription of a subset of genes like the TCF/LEF (T cell factor/lymphocyte enhancer binding factor) family of transcription factors [56]. In the lysosome, Dlg1 is thought to protect Dsh from degradation by reducing the ubiquitination modification on Dsh. The proteins of the core EJC and RNPS1 positively regulate the splicing of cell polarity determinant Dlg1 transcript, thus controlling Wnt signalling. In connection with this, knockdown of the core EJC or RNPS1, reduced the expression of Dlg1 and Dsh, resulting in reduced Wnt signalling [55].
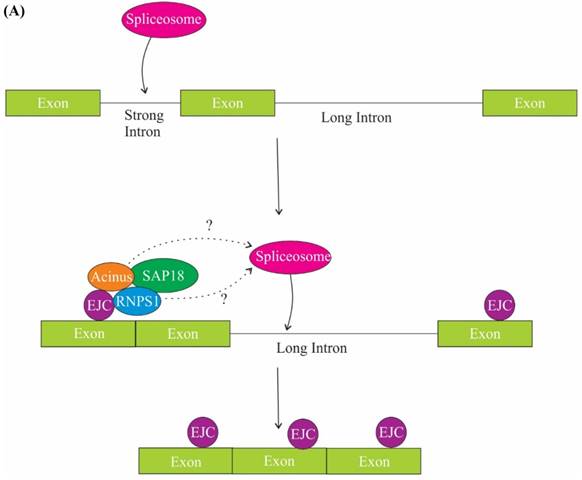
The role of ASAP components in splicing. (A) One of the mechanisms of regulation of splicing by RNPS1 and Acinus is splicing of long sub-optimal introns. Splicing of strong adjacent introns deposit EJC at exon junctions and EJC in turn recruits RNPS1 and Acinus in the close proximity of weak intron, thus facilitating spliceosome assembly and splicing of the long weak intron. (B) One of the models explaining the role of SAP 18 in splicing regulation is that SAP18 serve as a scaffold protein for the assembly of ASAP complex and spliceosomal machinery via its ubiquitin-like fold on exonic splicing enhancers (ESEs). The other model is that SAP18 is recruited to the ESE by a hypothetical protein X, SAP18 then functions as an adaptor molecule and recruits RNPS1, Acinus and other splicing factors.
In Drosophila, loss of core EJC (eIF4A3, Mago and Y14) or RNPS1 also result in exon skipping of heterochromatin genes containing large intron (greater than 250 bp), thus indicating a role of the core EJC and RNPS1 in exon definition [57, 58]. Exon definition is a phenomenon where splicing machinery identifies splice sites across small exons rather than long introns [10, 59]. There are several possible explanations for the role of RNPS1 and EJC in exon definition of large intron-containing genes. In one possibility, EJC is likely to act as an adaptor for SR factors, which assist in exon definition. The second possibility is that EJC masks the binding site for splicing factors or RNA motifs present in pre-mRNA that is involved in splice site choice and thus helps in exon retention [60]. The third possibility presumes that EJC stabilizes the interaction of spliceosomal complex with splice sites [57]. Moreover, the spliceosomal complex recognizes only those splice sites which are found across an optimal intron length (less than 200 bp) and fails to discern splice sites across long introns. Thus, the deposition of EJC complex on exons adjacent to long introns helps spliceosome to splice out long introns [61]. Nevertheless, long introns in Drosophila also bear repetitive splice sites and its presence probably act as an alternative to EJC-dependent exon definition mechanism [62].
The core EJC (Mago and Y14), RNPS1 and Acinus also play a role in repressing transposable elements in both Drosophila germline and surrounding somatic follicle cells by regulating splicing of piwi transcripts. Piwi is a constituent of the piRNA pathway that silences transposable elements primarily in gonads [63, 64]. The piwi pre-mRNA has suboptimal splice site in its fourth intron due to the presence of a weak PPT and the large size of the fourth intron. Splicing of this weak fourth intron is facilitated by RNPS1, Acinus and EJC and depends upon splicing of the adjacent strong flanking introns. Knockdown of core EJC (Mago and Y14), Acinus or RNPS1 leads to the retention of the fourth intron and decreases piwi protein level, inhibiting the piRNA pathway [65, 66]. Thus, a model has been suggested in which splicing of strong flanking introns lead to the deposition of EJC at exon junctions, which recruits RNPS1 together with Acinus in the close vicinity of weak intron, thus facilitating spliceosome assembly to splice out weak intron (Figure 4A) [65, 66]. However, introns bearing weak PPT can be spliced independent of strong flanking introns, but only if their length is less than 90 nt [67]. Collectively, it can be perceived that introns less than 200 bp are spliced independent of exon definition, whereas introns greater than 250 bp in length may require exon definition for splicing [57].
Translation efficiency
Splicing of pre-mRNA together with end modifications generate competent mRNA for translation. The process of splicing not only prepares an mRNA for translation but also augments its expression as compared to intronless mRNAs. The higher gene expression is due to the deposition of various RNA-binding proteins on spliced mRNA that regulate efficient mRNA processing, mRNA export and translation of the spliced mRNA [68-71]. The processing of the mRNA at the 5' and 3' end is of key importance in stimulating translational efficiency, as mRNA stability largely depends on the existence of cap at the 5' end and the length of polyadenylations present at the 3' end. The polyadenylation also has implication in the export of mRNA from the nucleus to the cytoplasm, whereas deadenylation from the end results in mRNA decay [72].
RNPS1 and EJC enhance 3' end processing and translation efficiency
The fact that a variety of proteins that interact with EJC remain associated with spliced mRNA until the first round of translation led to the finding that RNPS1 is also involved in translation. In this regard, it has been shown that tethering of RNPS1 and SRm160 (Serine/Arginine Repetitive Matrix 1) to the intronless CAT and β-globin mRNA notably enhanced the 3' end processing and mRNA steady state levels, resulting in enhanced expression [73]. Additionally, RNPS1 and SRm160 when tethered to a dsx (doublesex) reporter transcript elicit 3' end cleavage in vivo [74]. This might be due to the recruitment of the 3' end cleavage machinery, as SRm160 is known to interact with cleavage polyadenylation specificity factor 160 kDa subunit (CPSF-160) [75]. CPSF-160 functions in 3' end processing of pre-mRNAs by binding to AAUAAA signal sequence in pre-mRNA and interacting with poly (A) polymerase and other processing factors to stimulate 3' end cleavage and polyadenylation [76]. In addition, combinatorial RNAi study revealed genetic interaction between SRm160 ortholog of Caenorhabditis elegans (RSR-1) and Cleavage stimulation factor 50 kDa subunit (CstF-50), which is also involved in 3' end processing of pre-mRNA [74]. Altogether these implies that the interaction of RNPS1 with SRm160 might support 3' end processing, which increases mRNA levels and hence availability for translational machinery (Figure 5). Another study showed that tethering of RNPS1 to intronless Renilla luciferase mRNA enhanced translational efficiency. The enhanced translational efficiency correlates to the increased association of spliced mRNAs with polysomes, implying that RNPS1 along with other EJC proteins (Y14, MAGOH, Up-frameshift (Upf) proteins) also promote polysome association [77]. Taken together these reports suggest that deposition of EJC on spliced mRNA augments gene expression. However, the involvement of Acinus and SAP18 in translation have not yet been documented.
Nonsense-Mediated mRNA Decay
NMD or mRNA surveillance is a process, which degrades aberrant mRNAs with premature termination codons (PTCs) that could result in deleterious truncated proteins, thus maintaining mRNA quality control. NMD also maintains homeostasis by regulating the expression of normal genes. NMD is instigated when a PTC is present more than 50 nt upstream of the last exon-exon junction in the mRNA [78-80]. This underlines the importance of splicing for NMD activation [81, 82]. The core NMD factors conserved in eukaryotes are Upf1, Upf2, and Upf3 [83-87]. Notably, Upf2 and Upf3 occur in complex with the EJC on spliced mRNA [88-90]. When translating ribosome stalls at the PTC, Upf1, an ATP-dependent RNA helicase is recruited along with other factors like protein kinase SMG1 complex (comprises of SMG1, SMG8 and SMG9) and eukaryotic release factors eRF1 and eRF3 to form a complex termed as SURF (SMG1-Upf1-eRF1-eRF3) [91]. DEAD box RNA helicase, DHX34 then recruits Upf1 to Upf2 and Upf3 bound EJC complex generating the decay inducing (DECID) complex. As soon as SURF complex associates with EJC on spliced mRNAs, SMG1 phosphorylates Upf1 [91, 92]. Phosphorylated Upf1 further recruits SMG-6 and the SMG-5:SMG-7 complex, which remodels the mRNP by dephosphorylating Upf1 and thereby promotes NMD (Figure 6) [93]. SMG-6 is an endonuclease, which likely contributes to the cleavage of the aberrant RNA [94-96]. Intriguingly, NMD can also be activated without the assembly of EJC, albeit less efficiently than EJC-dependent NMD [97, 98].
Role of RNPS1 in Nonsense-Mediated mRNA Decay
In mammals, two distinct routes of NMD have been observed. One of the routes is Upf2 dependent while the other is Upf2 independent [99]. Tethering of RNPS1 to a reporter mRNA elicits Upf2-dependent NMD. RNPS1 has been shown to interact with Upf3b and Upf2 and possibly recruits other factors involved in NMD (Figure 6) [99, 100]. Different routes of NMD activation might regulate a subset of NMD substrates and their NMD efficiency [101]. Interestingly, the efficiency of NMD varies between different cell types [102-106] and the cellular concentration of RNPS1 is a limiting factor in NMD efficiency [107]. However, it is unclear, whether RNPS1 only functions in recruitment of Upf proteins or if it has any other role in NMD. Asides RNPS1, the involvement of other subunits (Acinus and SAP18) of the ASAP complex in NMD is uncertain.
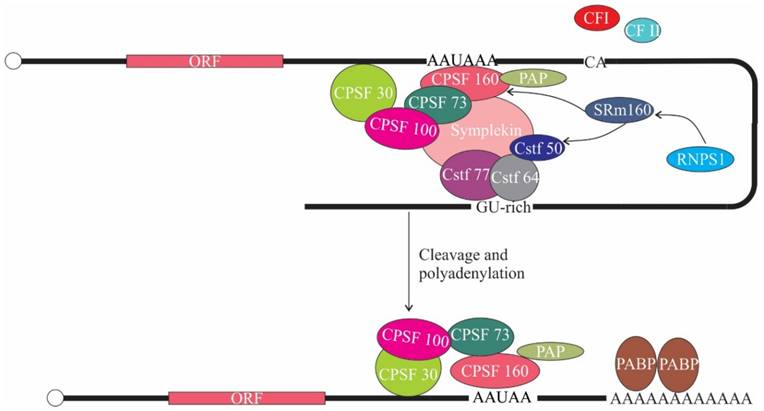
The role of ASAP components in translation. SRm160 interacts with Cleavage polyadenylation specificity factor-160 (CPSF-160) and Cleavage stimulation factor-50 (CstF-50). RNPS1 in turn interacts with SRm160. CPSF binds to AAUAAA signal sequence in pre-mRNA and associates with poly(A) polymerase (PAP) and CstF, thereby promoting 3' end cleavage and polyadenylation. CF I, cleavage factor I; CF II, cleavage factor II; PABP, poly(A) binding protein.
The role of ASAP components in NMD. During translation when ribosome encounters premature termination codon (PTC), it gets stalled at the PTC and as a result Up-frameshift 1 (Upf1) is recruited along with other factors like protein kinase SMG1 complex and eukaryotic release factors eRF1, eRF3 and DEAD box RNA helicase (DHX34) to form the surveillance (SURF) complex. DHX34 then stimulates interaction of Upf1 with Upf2-Upf3, which is bound to the EJC core and RNPS1. DHX34 also triggers ribosome disassembly and release of release factors in an ATP hydrolysis-dependent manner, remodelling SURF complex to Decay inducing (DECID) complex. This leads to phosphorylation of Upf1 and recruitment of SMG-6 and SMG-5: SMG-7 complex. SMG-6 is an endonuclease and subsequently, mRNA decay gets activated. m7Gppp, 7-methylgyanosine.
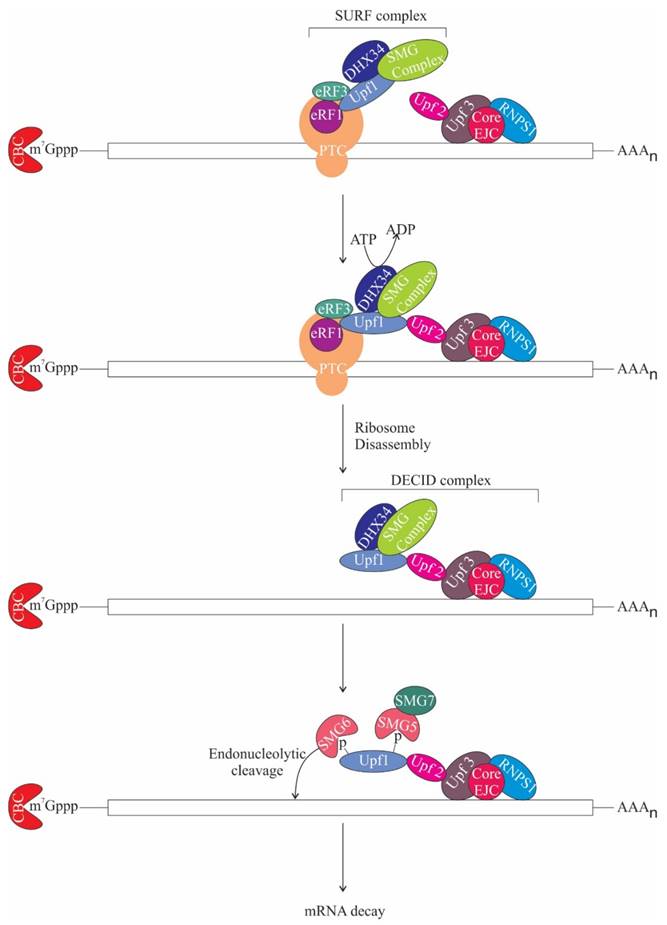
Furthermore, in humans, severity of genetic diseases is likely to be related to differential NMD efficiency together with exon skipping and alternative splicing [108, 109]. For example, in humans, the presence of nonsense mutation in dystrophin gene activates NMD, consequently causing deficiency of dystrophin protein leading to an acute manifestation of muscular dystrophy, termed Duchenne muscular dystrophy. However, variability in NMD efficiency as well as exon skipping and alternative splicing generate partially functional C-terminally truncated dystrophin protein, thus lowering the severity of disease, termed as Becker muscular dystrophy [102].
Apoptosis
Apoptosis is a type of “programmed” cell death that takes place during the course of development, aging, homeostasis and immune reactions in a cell. Cytoplasmic and nuclear morphological changes that accompany apoptosis are cell shrinkage, membrane blebbing, breakdown of the nuclear lamina, oligonucleosomal DNA fragmentation and condensation of chromatin. During apoptosis, the family of cysteinyl aspartate-specific proteases (caspases) get activated and cleave specific proteins to trigger cell death (reviewed in [110]).
Functions of ASAP components in apoptosis
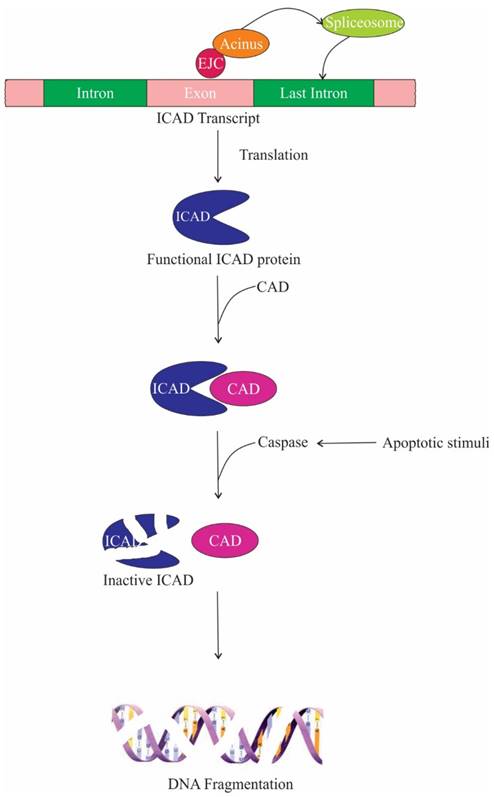
One of the target proteins of caspase-3 is Acinus, which gets activated by cleavage and induces chromatin condensation without oligonucleosomal DNA fragmentation [13]. On the contrary, Joselin et al. showed that when cells are subjected to apoptotic stimuli, knockdown of Acinus isoforms do not hinder chromatin condensation, rather prevent oligonucleosomal DNA fragmentation [17]. Two parallel or sequential pathways lead to chromatin condensation and DNA fragmentation during apoptosis. In one of these pathways, CAD (caspase-activated deoxyribonuclease) digests nuclear DNA. CAD is expressed as a complex with its inhibitor/chaperone, ICAD (inhibitor of caspase-activated deoxyribonuclease); ICAD is cleaved by caspases upon apoptotic stimuli releasing CAD. The second pathway is caspase-independent, in which apoptosis-inducing factor (AIF), a mitochondrial intermembrane flavoprotein, causes cleavage of nuclear DNA into high molecular weight DNA fragment and also condenses chromatin at the nuclear periphery [111]. Hence, it is conceivable that the inhibition of oligonucleosomal DNA fragmentation in Acinus depleted cells is due to the repression of nucleolytic activity of CAD. The repression is brought about by producing non-functional ICAD protein. Upon depletion of Acinus, the last intron of ICAD is retained, encoding a shorter ICAD protein isoform, which is incapable of functioning as a chaperone for CAD and thus impeding the DNA fragmentation during apoptosis. Therefore it can be speculated that Acinus regulates splicing of ICAD transcript and promotes DNA fragmentation upon apoptotic stimuli (Figure 7) [25]. In addition, it has been observed that caspase-3 cleaves the SAP motif of the Acinus-L during apoptosis, which compromises the DNA binding affinity of Acinus-L. Since the SAP motif is required to bind with SARS/MARS for chromosomal organization, loss of Acinus-L could induce chromatin degradation [15, 19]. The ASAP complex has also been implicated in accelerating cell death upon induction of apoptosis [3].
Constituents of the ASAP complex are also involved in the regulation of apoptosis via generating distinct isoforms of apoptotic regulators through alternative splicing. It has been shown that siRNA-mediated knockdown of ASAP components (RNPS1, Acinus, SAP18) or EJC members (eIF4A3, Y14) increased the level of pro-apoptotic isoforms of various apoptotic regulators like Bcl-x, Bim and Mcl1 that instigate apoptosis [112]. The molecular mechanism behind the regulation remains to be determined. Nevertheless, it has been observed that the members of ASAP and EJC regulate splicing by binding to distinct cis-acting elements present in the pre-mRNA of these apoptotic regulators, which are considerably different from the conventional assembly site of the EJC. For example, RNPS1 binds to SB1, an element present more than 250 nt upstream of Bcl-xs (pro-apoptotic splice variant) 5' splice site, whereas eIF4A3 and Y14 associate with B2 element located downstream of the Bcl-xs 5' splice site. This suggest that the binding is distinct from the canonical site of EJC deposition. This raises the question, what could trigger the deployment of ASAP and EJC components for controlling the production of pro-apoptotic and anti-apoptotic splice variants? One of the probable explanations could be that NMD regulates homeostasis and degrades erroneous mRNAs. Therefore, in order to maintain normal NMD activity, the cell ensures that there are adequate amounts of proteins participating in NMD. Hence, depletion of ASAP and EJC components lead to alteration in the alternative splicing of apoptotic regulators, which triggers the production of pro-apoptotic isoforms and thus, apoptosis of the cell [112].
Other Cellular Roles
Genome-wide RNA binding profile of Acinus has shown its binding to many mRNAs that encode proteins with functions in cell cycle regulation. Loss of Acinus hindered cell cycle progression through the S phase and the cells remained in G1 phase [25]. Cyclin A is one of the proteins that is required for the progress of cell cycle through S and M phase by forming complex with cyclin-dependent kinases CDK2 and CDK1, respectively [113]. Cyclin A1 is highly expressed in male germ cells, hematopoietic stem cells and is also found to be elevated in leukaemia cell lines [114, 115]. Expression of Cyclin A1 is upregulated when Acinus is phosphorylated, consequently increasing cell proliferation. However, depletion of Acinus and serine/arginine (SR) protein-specific kinase 2 (SRPK2), that phosphorylates Acinus, decreased the steady-state level of Cyclin A1 and reduce the growth rate of leukaemia cells [116]. Phosphorylation is an essential modification of RS domain-containing splicing factors like Acinus that promotes specific protein-protein interactions between splicing factors [117]. Hence, regulation of mRNA processing of Cyclin A1 by Acinus might be a possible reason behind its role in S and G2/M phase progression [116].
The role of ASAP components in apoptosis. Acinus aids in the recruitment of splicing machinery for the faithful splicing of the last intron of inhibitor caspase-activated deoxyribonuclease (ICAD). Functional ICAD acts as a chaperone for caspase-activated deoxyribonuclease (CAD), thus during apoptotic stimuli caspase cleaves ICAD releasing functional CAD, which then promotes DNA fragmentation.
Conclusions
RNPS1, Acinus and SAP18, serve a crucial role in RNA metabolism starting from transcription to NMD. During transcription, as the assembly of RBPs on nascent RNA prevents R-loop formation and RNPS1 together with SRSF1 are believed to be crucial proteins in this context. Therefore, it can be speculated that RNPS1 also functions as binding platform for the assembly of mRNPs and this is further supported by the RNA binding properties of RNPS1.
In the process of splicing, all the ASAP constituents play a significant function in both constitutive splicing as well as alternative splicing [6, 65, 112]. However, there are many questions that are left unanswered in this context. One of the main questions is whether ASAP components associate with EJC in order to carry out their functions in splicing. Next, how the ASAP components participate in splicing. It can be hypothesized that ASAP components act as a recruitment factor of spliceosomal machinery through protein-protein interactions with splicing factors. Furthermore, it is unclear whether involvement of ASAP complex components in splicing is substrate specific or splice site specific. Although all the ASAP components act as splicing activator but on the other hand it is peculiar that when RNPS1, Acinus and SAP18 interact to form the ASAP complex then ASAP-L and ASAP-S inhibit RNA splicing activity of splicing factors (SRSF2, SRSF1 and RNPS1) [3]. It would be worthwhile to understand the real mechanism behind such differences.
The detail mechanism by which ASAP complex regulates RNA metabolism remains to be elucidated fully. It is not yet determined whether components of ASAP complex solely or as a part of the ASAP complex takes part in the regulation of RNA metabolism. Additionally, question like how ASAP components are recruited to their targets still remains to be investigated. It would be fascinating to know how ASAP members are recruited to the spliceosome and to the spliced mRNA. A multitude of proteins interact with spliced mRNA to form mRNPs and the dynamics of mRNPs decide the fate of each mRNA. Therefore, it would be interesting to discover the interacting proteins of the ASAP complex. Many areas are yet to be explored to get a vivid picture of ASAP complex dynamics within the cell. Further research in this area will provide more insights into its role in RNA metabolism, recruitment to the target transcripts, interacting partners and pathological implications.
Acknowledgements
We acknowledge the facilities provided by the Indian Institute of Technology Guwahati, Assam, India. We also acknowledge funding from Indian Institute of Technology Guwahati project grant (BTSUGIITG-01) to K.K.S. We thank Dr. Neils H. Gehring for his valuable suggestions on the review.
Abbreviations
Acinus: apoptotic chromatin condensation inducer in the nucleus; ASAP: apoptosis-and splicing-associated protein; CAD: caspase-activated deoxyribonuclease; CPSF-160: cleavage polyadenylation specificity factor 160 kDa subunit; CstF-50: cleavage stimulation factor 50 kDa subunit; Dlg1: discs large 1; Dsh: dishevelled; EJC: exon junction complex; eIF4A3: RNA helicase eukaryotic initiation factor 4A3; ESE: exonic splicing enhancers; HDAC: histone deacetylase; hnRNP: heterogeneous ribonucleoprotein particle; ICAD: inhibitor of caspase-activated deoxyribonuclease; mRNA: messenger ribonucleic acid; mRNPs: messenger ribonucleoprotein particles; NMD: nonsense-mediated mRNA decay; piRNA: piwi-interacting RNA; PPT: polypyrimidine tract; PSAP: protein complex of Pinin, SAP18 and RNPS1; PTC: premature termination codons; RA: retinoic acid; RAR: retinoic acid receptor; RARE: retinoic acid response element; RBP: RNA-binding protein; RD/E: arginine/aspartate/glutamate-rich; RNA: ribonucleic acid; RNPS1: RNA-binding protein with Serine-rich domain 1; RRM: RNA recognition motif; RS: arginine/serine-rich; RSB: RNPS1-SAP18 binding motif; RS/P: arginine/serine/proline-rich; RXR: retinoid X receptors; S: serine-rich; SAP: SAF-A/B, Acinus and PIAS (STAT inhibitors) motif; SAP18: Sin3-associated protein of 18 kDa; SARS/MARS: scaffolds or matrix attachment regions; snRNPs: small nuclear ribonucleoprotein particles; SR: serine/arginine-rich; SRm160: Serine/Arginine Repetitive Matrix 1; SRPK2: serine/arginine (SR) protein-specific kinase 2; U2AF65: U2 snRNP auxiliary factor 65 kDa; UBL: ubiquitin-like; Upf: up-frameshift protein; 3'UTR: 3' untranslated region; Wnt: wingless
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163-75
2. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS letters. 2008;582:1977-86
3. Schwerk C, Prasad J, Degenhardt K, Erdjument-Bromage H, White E, Tempst P. et al. ASAP, a Novel Protein Complex Involved in RNA Processing and Apoptosis. Molecular and Cellular Biology. 2003;23:2981-90
4. Hir HL, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17:41-54
5. Badolato J, Gardiner E, Morrison N, Eisman J. Identification and characterisation of a novel human RNA-binding protein. Gene. 1995;166:323-7
6. Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. The EMBO Journal. 1999;18:4560-70
7. Shepard PJ, Hertel KJ. The SR protein family. Genome Biology. 2009;10:242
8. Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat Struct Mol Biol. 2012;19:378-86
9. Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume A-M. et al. Exon Junction Complexes Show a Distributional Bias toward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins. Cell Reports. 2016;16:1588-603
10. Zheng Z-M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. Journal of biomedical science. 2004;11:278-94
11. Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M. et al. Dynamic Protein-Protein Interaction Wiring of the Human Spliceosome. Molecular Cell. 2012;45:567-80
12. Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S. et al. Large-scale mapping of human proteins-protein interactions by mass spectrometry. Molecular Systems Biology. 2007;3:89
13. Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168-73
14. Vucetic Z, Zhang Z, Zhao J, Wang F, Soprano KJ, Soprano DR. Acinus-S′ Represses Retinoic Acid Receptor (RAR)-Regulated Gene Expression through Interaction with the B Domains of RARs. Molecular and Cellular Biology. 2008;28:2549-58
15. Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends in Biochemical Sciences. 2000;25:112-4
16. Hu Y, Yao J, Liu Z, Liu X, Fu H, Ye K. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. The EMBO Journal. 2005;24:3543
17. Joselin AP, Schulze-Osthoff K, Schwerk C. Loss of Acinus Inhibits Oligonucleosomal DNA Fragmentation but Not Chromatin Condensation during Apoptosis. Journal of Biological Chemistry. 2006;281:12475-84
18. Tange TØ, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869-83
19. Göhring F, Schwab BL, Nicotera P, Leist M, Fackelmayer FO. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. The EMBO Journal. 1997;16:7361-71
20. Kipp M, Göhring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M. et al. SAF-Box, a Conserved Protein Domain That Specifically Recognizes Scaffold Attachment Region DNA. Molecular and Cellular Biology. 2000;20:7480-9
21. Renz A, Fackelmayer FO. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Research. 1996;24:843-9
22. Nickerson JA, Blencowe BJ, Penman S. The Architectural Organization of Nuclear Metabolism. Ronald B, Kwang WJ, editors. International Review of Cytology: Academic Press. 1996:67-123
23. Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Critical reviews in eukaryotic gene expression. 2000;10:73-90
24. Wang F, Soprano KJ, Soprano DR. Role of Acinus in regulating retinoic acid-responsive gene pre-mRNA splicing. J Cell Physiol. 2015;230:791-801
25. Rodor J, Pan Q, Blencowe BJ, Eyras E, Cáceres JF. The RNA-binding profile of Acinus, a peripheral component of the exon junction complex, reveals its role in splicing regulation. RNA. 2016;22:1411-26
26. Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone Deacetylases and SAP18, a Novel Polypeptide, Are Components of a Human Sin3 Complex. Cell. 1997;89:357-64
27. Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M. et al. SAP30, a Novel Protein Conserved between Human and Yeast, Is a Component of a Histone Deacetylase Complex. Molecular Cell. 1998;1:1021-31
28. McCallum SA, Bazan JF, Merchant M, Yin J, Pan B, de Sauvage FJ. et al. Structure of SAP18: A Ubiquitin Fold in Histone Deacetylase Complex Assembly. Biochemistry. 2006;45:11974-82
29. Kiel C, Serrano L. The Ubiquitin Domain Superfold: Structure-based Sequence Alignments and Characterization of Binding Epitopes. Journal of Molecular Biology. 2006;355:821-44
30. Li C, Lin R-I, Lai M-C, Ouyang P, Tarn W-Y. Nuclear Pnn/DRS Protein Binds to Spliced mRNPs and Participates in mRNA Processing and Export via Interaction with RNPS1. Molecular and Cellular Biology. 2003;23:7363-76
31. Costa E, Canudas S, Garcia-Bassets I, Pérez S, Fernández I, Giralt E. et al. Drosophila dSAP18 is a nuclear protein that associates with chromosomes and the nuclear matrix, and interacts with pinin, a protein factor involved in RNA splicing. Chromosome Research. 2006;14:515-26
32. Singh KK, Erkelenz S, Rattay S, Dehof AK, Hildebrandt A, Schulze-Osthoff K. et al. Human SAP18 mediates assembly of a splicing regulatory multiprotein complex via its ubiquitin-like fold. RNA. 2010;16:2442-54
33. Wang P, Lou P-J, Leu S, Ouyang P. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochemical and Biophysical Research Communications. 2002;294:448-55
34. Joo J-H, Taxter TJ, Munguba GC, Kim YH, Dhaduvai K, Dunn NW. et al. Pinin modulates expression of an intestinal homeobox gene, Cdx2, and plays an essential role for small intestinal morphogenesis. Developmental biology. 2010;345:191-203
35. Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and Its Associated Factors: a Versatile Alternative Pre-mRNA Splicing Regulator In Vivo. Molecular and Cellular Biology. 2004;24:1174-87
36. Spector DL. Nuclear domains. Journal of Cell Science. 2001;114:2891
37. Spector DL, Lamond AI. Nuclear Speckles. Cold Spring Harbor Perspectives in Biology. 2011;3:a000646
38. Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic Analysis of Interchromatin Granule Clusters. Molecular Biology of the Cell. 2004;15:3876-90
39. Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. The EMBO Journal. 2002;21:3536-45
40. Aguilera A, García-Muse T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Molecular Cell. 2012;46:115-24
41. Li X, Niu T, Manley JL. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108-15
42. Li X, Manley JL. Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genomic Instability. Cell. 2005;122:365-78
43. Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1-16
44. Chambon P. A decade of molecular biology of retinoic acid receptors. The FASEB Journal. 1996;10:940-54
45. Ayer DE, Lawrence QA, Eisenman RN. Mad-max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767-76
46. Will CL, Lührmann R. Spliceosome Structure and Function. Cold Spring Harbor Perspectives in Biology. 2011;3:a003707
47. Lim SR, Hertel KJ. Commitment to Splice Site Pairing Coincides with A Complex Formation. Molecular Cell. 2004;15:477-83
48. Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J. et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. The EMBO Journal. 2007;26:1737-48
49. Trembley JH, Tatsumi S, Sakashita E, Loyer P, Slaughter CA, Suzuki H. et al. Activation of Pre-mRNA Splicing by Human RNPS1 Is Regulated by CK2 Phosphorylation. Molecular and Cellular Biology. 2005;25:1446-57
50. Zhang WJ, Wu JY. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Molecular and Cellular Biology. 1996;16:5400-8
51. Brandner JM, Reidenbach S, Kuhn C, Franke WW. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the ″speckle″ type. European Journal of Cell Biology. 1998;75:295-308
52. Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS Region with Pre-mRNA Branch Point and Promotion of Base Pairing with U2 snRNA. Science. 1996;273:1706
53. Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846-50
54. Fukumura K, Wakabayashi S, Kataoka N, Sakamoto H, Suzuki Y, Nakai K. et al. The Exon Junction Complex Controls the Efficient and Faithful Splicing of a Subset of Transcripts Involved in Mitotic Cell-Cycle Progression. International Journal of Molecular Sciences. 2016;17:1153
55. Liu M, Li Y, Liu A, Li R, Su Y, Du J. et al. The exon junction complex regulates the splicing of cell polarity gene dlg1 to control Wingless signaling in development. eLife. 2016;5:e17200
56. MacDonald BT, He X. Frizzled and LRP5/6 Receptors for Wnt/β-Catenin Signaling. Cold Spring Harbor Perspectives in Biology. 2012;4:a007880
57. Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P. et al. The Exon Junction Complex Controls the Splicing of mapk and Other Long Intron-Containing Transcripts in Drosophila. Cell. 2010;143:251-62
58. Roignant J-Y, Treisman JE. Exon Junction Complex Subunits Are Required to Splice Drosophila MAP Kinase, a Large Heterochromatic Gene. Cell. 2010;143:238-50
59. De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdisciplinary Reviews: RNA. 2013;4:49-60
60. Yu Y, Maroney PA, Denker JA, Zhang XHF, Dybkov O, Lührmann R. et al. Dynamic Regulation of Alternative Splicing by Silencers that Modulate 5′ Splice Site Competition. Cell. 2008;135:1224-36
61. Fox-Walsh KL, Dou Y, Lam BJ, Hung S-p, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16176-81
62. Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of Large Introns in Drosophila by Recursive Splicing at Nonexonic Elements. Genetics. 2005;170:661-74
63. Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313-21
64. Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R. et al. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell. 2009;137:522-35
65. Malone CD, Mestdagh C, Akhtar J, Kreim N, Deinhard P, Sachidanandam R. et al. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes & Development. 2014;28:1786-99
66. Hayashi R, Handler D, Ish-Horowicz D, Brennecke J. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes & Development. 2014;28:1772-85
67. Guo M, Lo PC, Mount SM. Species-specific signals for the splicing of a short Drosophila intron in vitro. Molecular and Cellular Biology. 1993;13:1104-18
68. Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Molecular and Cellular Biology. 1988;8:4395-405
69. Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618-30
70. Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607-17
71. Ryu WS, Mertz JE. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. Journal of Virology. 1989;63:4386-94
72. Wahle E, Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiology Reviews. 1999;23:277-95
73. Wiegand HL, Lu S, Cullen BR. Exon Junction Complexes Mediate the Enhancing Effect of Splicing on mRNA Expression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11327-32
74. McCracken S, Longman D, Johnstone IL, Cáceres JF, Blencowe BJ. An Evolutionarily Conserved Role for SRm160 in 3′-End Processing That Functions Independently of Exon Junction Complex Formation. Journal of Biological Chemistry. 2003;278:44153-60
75. McCracken S, Lambermon M, Blencowe BJ. SRm160 Splicing Coactivator Promotes Transcript 3′-End Cleavage. Molecular and Cellular Biology. 2002;22:148-60
76. Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3'-end formation. Genes & Development. 1995;9:2672-83
77. Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes & Development. 2004;18:210-22
78. Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nature reviews Molecular cell biology. 2012;13:700-12
79. Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay — Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2013;1829:612-23
80. Popp MW-L, Maquat LE. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annual review of genetics. 2013;47:139-65
81. Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445-56
82. Brocke KS, Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE. The human intronless melanocortin 4-receptor gene is NMD insensitive. Human Molecular Genetics. 2002;11:331-5
83. Lykke-Andersen J, Shu M-D, Steitz JA. Human Upf Proteins Target an mRNA for Nonsense-Mediated Decay When Bound Downstream of a Termination Codon. Cell. 2000;103:1121-31
84. Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and Characterization of Human Orthologues to Saccharomyces cerevisiae Upf2 Protein and Upf3 Protein (Caenorhabditis elegans SMG-4). Molecular and Cellular Biology. 2001;21:209-23
85. Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10928-32
86. Applequist SE, Selg M, Raman C, Jäck HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Research. 1997;25:814-21
87. Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Current Opinion in Cell Biology. 2005;17:316-25
88. Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F. et al. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10050-5
89. Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b Form an NMD-Activating Complex. Molecular Cell. 2003;11:939-49
90. He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Molecular and Cellular Biology. 1997;17:1580-94
91. Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S. et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Development. 2006;20:355-67
92. Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y. et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes & Development. 2009;23:1091-105
93. Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Research. 2012;40:1251-66
94. Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. The EMBO Journal. 2006;25:5117-25
95. Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609-17
96. Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49-55
97. Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3[prime]-UTR promotes aberrant termination and triggers nonsense- mediated mRNA decay. Nature. 2004;432:112-8
98. Muhlrad D, Parker R. Aberrant mRNAs with extended 3' UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299-307
99. Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW. et al. Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements. Molecular Cell. 2005;20:65-75
100. Lykke-Andersen J, Shu M-D, Steitz JA. Communication of the Position of Exon-Exon Junctions to the mRNA Surveillance Machinery by the Protein RNPS1. Science. 2001;293:1836-9
101. Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J. Communication with the Exon-Junction Complex and Activation of Nonsense-Mediated Decay by Human Upf Proteins Occur in the Cytoplasm. Molecular Cell. 2007;27:780-92
102. Kerr TP, Sewry CA, Robb SA, Roberts RG. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Human Genetics. 2001;109:402-7
103. Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A. et al. Mutations in the JARID1C Gene, Which Is Involved in Transcriptional Regulation and Chromatin Remodeling, Cause X-Linked Mental Retardation. American Journal of Human Genetics. 2005;76:227-36
104. Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Human Molecular Genetics. 2003;12:217-25
105. Resta N, Susca FC, Di Giacomo MC, Stella A, Bukvic N, Bagnulo R. et al. A homozygous frameshift mutation in the ESCO2 gene: Evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol. 2006;209:67-73
106. Hutchinson S, Furger A, Halliday D, Judge DP, Jefferson A, Dietz HC. et al. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: a potential modifier of phenotype? Human Molecular Genetics. 2003;12:2269-76
107. Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Research. 2007;35:4542-51
108. Frischmeyer PA, Dietz HC. Nonsense-Mediated mRNA Decay in Health and Disease. Human Molecular Genetics. 1999;8:1893-900
109. Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801-8
110. Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicologic pathology. 2007;35:495-516
111. Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43-50
112. Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M. et al. Proteins Associated with the Exon Junction Complex Also Control the Alternative Splicing of Apoptotic Regulators. Molecular and Cellular Biology. 2012;32:954-67
113. Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. The EMBO Journal. 1992;11:961-71
114. Wolgemuth DJ, Lele KM, Jobanputra V, Salazar G. The A-type cyclins and the meiotic cell cycle in mammalian male germ cells. International Journal of Andrology. 2004;27:192-9
115. Yang R, Nakamaki T, Lübbert M, Said J, Sakashita A, Freyaldenhoven BS. et al. Cyclin A1 Expression in Leukemia and Normal Hematopoietic Cells. Blood. 1999;93:2067-74
116. Jang S-W, Yang S-j, Ehlén Å, Dong S, Khoury H, Chen J. et al. SRPK2 Promotes Leukemia Cell Proliferation by Phosphorylating Acinus and Regulating Cyclin A1. Cancer research. 2008;68:4559-70
117. Wang H-Y, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC. et al. SRPK2: A Differentially Expressed SR Protein-specific Kinase Involved in Mediating the Interaction and Localization of Pre-mRNA Splicing Factors in Mammalian Cells. The Journal of Cell Biology. 1998;140:737-50
Author contact
![]() Corresponding author: kusumsinghernet.in; Tel.: +91-(0)-361-2583206
Corresponding author: kusumsinghernet.in; Tel.: +91-(0)-361-2583206

 Global reach, higher impact
Global reach, higher impact