Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(8):1008-1018. doi:10.7150/ijbs.19891 This issue Cite
Review
Linker Histone in Diseases
Beijing Key Laboratory of Protein Posttranslational Modifications and Cell Function, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Peking University Health Science Center, #38 Xueyuan Road, Beijing 100191, China.
Received 2017-3-2; Accepted 2017-5-30; Published 2017-7-18
Abstract
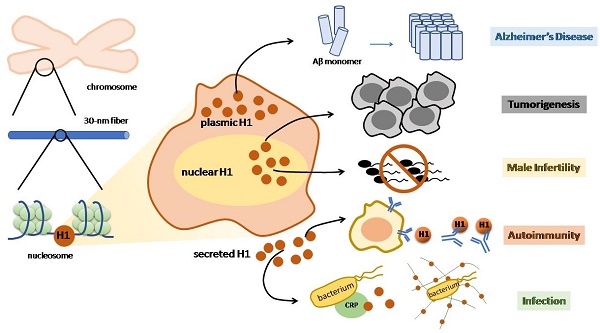
The linker histone is a protein that binds with the nucleosome, which is generally considered to achieve chromatin condensation in the nucleus. Accumulating evidences suggest that the linker histone is essential in the pathogenesis of several diseases. In this review, we briefly introduce the current knowledge of the linker histone, including its structure, characteristics and functions. Also, we move forward to present the advances of the linker histone's association with certain diseases, such as cancer, Alzheimer's disease, infection, male infertility and aberrant immunity situations, focusing on the alteration of the linker histone under certain pathological conditions and its role in developing each disease.
Keywords: Linker Histone, Nucleosome, Chromatin, Cancer, Alzheimer's Disease.
Introduction
To form a chromosome, the first stage of DNA packaging is to form nucleosomes. A nucleosome is a basic repeating unit of chromatin, which includes an octamer of eight core histones, a linker histone and a certain length of DNA. The DNA in a nucleosome unit includes a core DNA that wraps the octamer with the length of ~147 bp, and a linker DNA of 20-60bp. The eight core histones of the octamer have been well studied, including two of each H2A, H2B, H3 and H4 [1]. However, the fifth histone, the linker histone (H1), has been proven the most difficult one to understand, either structurally or functionally [2]. Recent studies have revealed several important characteristics of the linker histone and more evidences accumulate to indicate its functions. In this review, we focus on the structural and functional characteristics of linker histone, and discuss its association with several important diseases, offering new insights of novel targets for treatments.
Current Knowledge of Linker Histone
Linker histone structure
The structure of the linker histone includes three domains, a N-terminal domain, a globular domain and a C-terminal domain. The N-terminal domain is the shortest domain of H1, with a length of between 20 and 35 amino acids. It is divided into two sub-regions. The distant region is enriched in hydrophobic residues while a highly basic region is close to the globular domain [3]. The globular domain is highly conserved with about 80 amino acids. It possesses three helices, three loops and two short beta sheets in hairpin [4, 5]. The C-terminal domain has approximately 100 amino acids with high variation among species.
Linker histone variants
In human, 11 H1 histone variants have been reported so far, seven somatic ones including H1.1-H1.5, H1.0 and H1.10 and four germ line-specific ones including testis-specific variants (TS)H1.6, (TS)H1.7, (TS)H1.9 and oocyte-specific variant (OO)H1.8 [6]. Among the somatic histone H1 variants, H1.1 to H1.5 are replication-dependent while H1.0 and H1.10 are replication-independent through the cell cycle. H1.2 to H1.5 are expressed ubiquitously, whereas H1.1 is expressed in only certain types of cells and tissues. H1.0 accumulates in terminal differentiated cells. Regarding gene location, H1.1 to H1.5-encoding genes are clustered in a region of chromosome 6, together with the core histone genes. H1.10 and H1.0 gene are located on chromosome 3 and 22 respectively [6]. Series of experiments demonstrate that each variant exhibits its own characteristics and functions, including the expression pattern, chromatin affinity and knock-out phenotypes (Table 1) [7, 8]. However, knocking out or overproducing certain H1 variants could be compensated by other H1 variants' alteration, thus making it more complicated to understand their precise functions and the importance of H1 variants [6].
Linker histone, nucleosome and chromatin formation
It is generally acknowledged that the linker histone binds to and protects DNA, specifically the linker DNA, a “string” linking up nucleosomes. Micrococcal nuclease digestion revealed that each linker histone interacts with 10 bp of each linker DNA [9, 10]. Besides, Thoma et al. showed that the linker histone-containing chromatin exhibits a “zig-zag” fiber structure pattern, indicating that the linker histone is involved in determining the angle between the axis of the fiber and the flat faces of the nucleosomal disks, as well as the higher level of chromatin compaction [11, 12]. These evidences suggest that the linker histone is able to protect linker DNA as well as to determine the structure of the chromatin, and therefore it is of great significance to find out how linker histone binds to the nucleosome.
To confirm the possibility and feasibility of linker histone interacting and binding to the structure is the first step. Meyer et al. have done the work to prove that the space between the nucleosome surface and linker DNA is large enough to contain the globular domain [13]. In addition, the orientation of linker DNA also determines if the cavity is big enough to accommodate the linker histone. Shukla et al. reported that modified H2A altered the entry/exit angle of nucelosomal DNA, which hinders the binding of H1 to the nucleosome [14]. A similar work by Shed et al. reported that mutant nucleosome with H2A variant exhibits extended length of linker DNA, which results in a far weaker contact with linker histone for its excessive distance [15]. Despite that, steric restriction still exists for the interaction between linker histone and the nucleosome. Other proteins, such as PARP-1, can also bind to the nucleosome, hindering the binding site on nucleosome and interrupt linker histone's binding [16]. To sum up, the structure of the nucleosome core as well as the linker DNA has provided the linker histone with an available site for stable binding, as long as there is no other protein binding or linker DNA alteration to hinder the process.
Characteristics of Linker Histone Variants
| Unified Name | Gene Location(human) | Expression Pattern | Cell Cycle Dependence | Genomic Distribution Pattern (human) [21] | Knock-out Phenotype | |
|---|---|---|---|---|---|---|
| H1.1 | 6p21.3 | Somatic | replication- dependent | (IMR90) enriched in intergenic regions and at active chromosome 19; present at promoters | / | |
| H1.2 | 6p21.3 | Somatic | (IMR90) depleted in TSS of active genes, intergenic regions and CpG islands; enriched at LADs. | (IMR90) enriched at chromosome X; | (T47D, MCF7) Cell cycle arrest in G1 [81]; (T47D) reduction in nucleosome spacing [81]; | |
| H1.3 | 6p21.3 | Somatic | / | (IMR90) No significant difference [81]; | ||
| H1.4 | 6p21.3 | Somatic | / | (T47D, MCF10A) Deleterious effect with an increase in the subG1 peak [81]; | ||
| H1.5 | 6p22.1 | Somatic | (IMR90) differentiation-dependent enrichment | (IMR90) SIRT1 down-regulation and lower H3K9me2 level [24]; | ||
| (TS)H1.6 | 6p21.3 | Testis | / | / | ||
| H1.0 | 22q13.1 | Somatic | replication- independent | (T47D) enriched at the nucleolus | (hESC) Impaired differentiation [82]; (IMR90) No significant difference [81]; | |
| (TS)H1.7 | 12q13.1 | Testis | / | / | ||
| (OO)H1.8 | 3q22.1 | Oocyte | / | / | ||
| (TS)H1.9 | 17q21.33 | Testis | / | / | ||
| H1.10 | 3q21.3 | Somatic | (T47D) enriched in active chromatin | / | ||
IMR90: a human lung fibroblast cell line; T47D: a human breast cancer cell line; MCF7: a human breast adenocarcinoma cell line; MCF10A: a human non-tumor breast epithelial cell line; hESC: human embryonic stem cell.
With the possibility and feasibility of such interaction being confirmed, studies concerning the exact location of the linker were carried out. Among them, two models received the most attention. Muyldermans's model suggests that H1 binds symmetrically to the dyad axis, while Wolffe's model indicates an asymmetrical pattern that the linker histone binds off the axis [17, 18]. Evidences accumulated to support either model, yet controversy remained. Until recently, an important study has successfully observed its exact location by using cryogenic electron microscopy and finally confirmed the asymmetrical binding pattern [19]. In this study, Song et al. illustrated that the globular domain of the linker histone deviates from the dyad axis of the nucleosome. The asymmetry of the two sides of the nucleosome, like a “head” and a “tail” of the coin, sets a specific twist of each nucleosome to form the higher-order structure of the chromatin. Moreover, nucleosomes are packed in a two-start stack in the 30-nm chromatin fiber. Adjacent nucleosome cores are in either head-to-head interaction or tail-to-tail interaction pattern, resulting in the self-association of the linker histones, which plays an important part in the stabilization of the 30-nm fiber [19, 20]. Altogether, the linker histone binds asymmetrically with the nucleosome, which helps to form and determine a high-order structure of the chromatin.
Linker Histone Distribution
Understanding the distribution of the linker histone is quite important to reflect possible functions of the linker histone. The first total H1 map was made by a genome-wide study using chromatin immunoprecipitation (ChIP), exhibiting that H1 level is dramatically decreased near the transcription start site of active genes and intergenic regions but not in repressed promoters [16]. Furthermore, with the success in obtaining specific antibodies of the linker histone, the precise mapping of H1 variants in the genome has started to emerge, aiding the understanding of the specific functions of certain variants. For example, H1.2~H1.5 are enriched in transcription start site of active genes and intergenic regions [21]. H1.2 and H1.3 are depleted in GC- and gene-rich regions and active promoters in knock-in mouse ESCs [22]. Another study demonstrated that H1.10 is enriched in active chromatin in human breast cancer while H1.0 mostly distributed at the nucleolus [23].
Generally speaking, each H1 variant maintains a specific and fixed distribution pattern, yet the pattern can be slightly different depending on the differentiation state of the cell. A case in point is H1.5. Li et al. investigated the genomic distribution of H1.5 in various cell lines [24]. The study illustrated that during the process of cell differentiation from hESCs to neural progenitor cells and further to neurons and astrocytes, there is a significant increase in H1.5 binding. A similar pattern is also observed in the process of primary keratinocyte turning into a more differentiated stratified layer [24]. This evidence suggests that H1.5 genomic distribution is in association with the differentiation state of the cell.
However, linker histone is not restricted in the nucleus. The earliest evidence dated back to 1989 when Smith et al. demonstrated that the linker histone interacted with epidermolytic toxin in the cytoplasmic region [25]. Since then, evidences accumulated to indicate linker histone's presence in the cytoplasm, including Zlatanova et al. achieving to isolate linker histone from mouse liver cell cytoplasm [26]. Further investigation revealed that the linker histones are presented on the surface of viable cell as well as outside the cell, which leads us to a more sophisticated understanding of linker histone's distribution and its underlying function [27].
The Functions of Linker Histone
It is difficult to assign a precise description on linker histone's function. In early experiments, scientists even cast doubt on whether linker histones are essential in organisms. For example, H1.0-depleted mice showed no obvious phenotype [28]. Also, the studies on both H1-deleted Tetrahymena strain and Hho1p knock-out yeast showed no significant change on growth rate [29, 30]. As research progressed, scientists have proposed certain functions that have been observed on the linker histone.
Inside the Nucleus: Transcriptional Activity and DSB Repair
It is long been implicated that the linker histone represses transcription. The linker histone is capable of binding to the nucleosomes and further, compacting the chromatin from the “beads on string” structure to the 30-nm fiber [2, 18]. Based on such knowledge, it is quite natural to speculate that the linker histone's effects on transcriptional activity is to repress gene expression since the linker histone binds to and hinders the binding site on DNA for transcription factors and cofactors. For example, the linker histone is required for the basal repression of ngoA in growing cells by binding to the nucleosome to inhibit the interaction between USF (upstream stimulatory factor) and its target DNA [31, 32]. Also, Ura and colleagues' study suggests another underlying mechanism that the linker histone can inhibit the factors from binding to the DNA by reducing the mobility of the nucleosome [33].
To make it more complex, the linker histone does not always exhibit repressing effect. According to the study of Shen et al, the linker histone is able to directly activate CyP gene in starved cells [31]. Moreover, the post-translational modification (PTM) of the linker histone, including acetylation, methylation, phosphorylation, ADP-ribosylation and so on, also regulates the transcriptional activities of certain genes due to its rich-lysine C-terminal domain [34]. For example, Pin1, a proline-isomerase, binds to and dephosphorylates the linker histone, which leads to the destabilizing of H1 to the chromatin and thus may alter the transcriptional activity of the related genes [35]. Another example is that GCN5 can acetylate H1.4K36 to promote the transcription of CFOS, EGR1 and CJUN. The acetylated linker histone recruits TAF-1, a subunit of the general transcription factor TFIID, and thus promotes the transcription of the target genes [36]. Therefore, it is only safe to conclude that the linker histone, as well as its PTM, exerts certain influence on the transcriptional activity of certain genes. Its specific function and its underlying mechanism, either activating or repressing the gene transcription, remains to be confirmed.
More recently, a new study points out that the linker histone is also associated with DNA double strain break (DSB) repair. Under the circumstance of DSB, K63-linked polyubiquitylation of H1 is markedly upregulated, suggesting that the linker histone plays a possible role in the repair process. Downregulating overall H1 inhibits the accumulation of K63-linked ubiquitin and the recruitment of E3 ubiquitylation ligase RNF168 [37]. Further investigation found out that the linker histone polyubiquitylation is dependent on RNF8 (E3 ubiquitylation ligase) and UBC13 (E2 ubiquitin-conjugating enzyme). Afterwards, RNF168 binds with K63-linked ubiquitin of H1 and triggers the recruitment of downstream factors to DSB sites to proceed the repair process. Therefore, the linker histone is essential in connecting the process of the recruitment of RNF168 by UCB12 and RNF8, serving as a key substrate of RNF8 and providing K63-linked ubiquitin as a binding platform for RNF168 to the DSB site [37]. This study reveals the function of histone H1 on DSB repair.
Extranuclear and extracellular functions
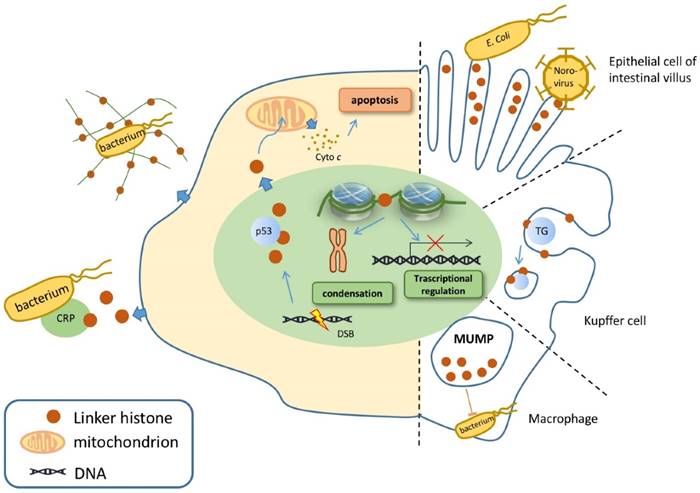
While linker histones are mostly located inside the nuclear, certain subtypes are identified outside [26]. Linker histones are able to relocate in the extranuclear area, mainly in the cytoplasm and on the cell membrane. Therefore, these relocated linker histones certainly possess unique characteristics that enable them to perform various functions outside the nucleus (Figure 1).
Linker histone is associated with the process of apoptosis. In face of DSB, the linker histones are released in a p53-dependent manner, resulting in the releasing of cytochrome c from mitochondria, which eventually leads to cell apoptosis (Figure 1) [38]. Yet how strong is the association between H1 and cell apoptosis remains controversial because linker histone over-expressed cells don't exhibit significant changes on gene expression, cell cycle progression or the overall chromatin structure [39-41].
Linker histone can exert its influence on innate immune system. The linker histone was also found in the granules of macrophages, named as murine microbicidal protein 1 and 2 (MUMP-1&2) initially [42]. MUMP exhibits antimicrobial activity against a large variety of microbes, including Salmonella typhimurium, E. coli, and Staphylococcus aureus [42] (Figure 1). In addition, linker histone acts as a pattern recognition receptor on nonspecific cytoxic cells (NCC) of catfish, which also possess antimicrobial activity against several bacteria [27].
Besides exerting influence in the cytoplasm, the linker histone also function on the surface of the cell. Macrophages of the liver, also known as Kupffer cells, express linker histone on the cell membrane, which enables the linker histone to bind with thyroglobulin and consequently to be internalized and cleared inside the macrophage [43].
Another case in point is that a cytoplasmic pool of linker histones is identified on the tip of the villus cell of intestine epithelium [44]. These linker histones are able to interact with Escherichia coli, which suggests that linker histone is associated with E. coli's colonization as well as the onset of bacterial diarrhea [45]. In addition, a type of virus Norovirus is also identified to bind to H1, which assigns the linker histone to the association with the onset of viral diarrhea. Yet this interaction seems to be specific, since Poliovirus or hepatitis E virus is unable to bind with the linker histone in vitro (Figure 1) [46].
While the function of defending against microbes outside the nucleus is already intriguing, scientists step further to demonstrate that the linker histone also exerts its antibacterial function outside the cell. In face of an explosion of interleukin-8 or endotoxin, neutrophils release antimicrobial agents within minutes, including core histones, linker histones and elastase, strung together by a web of DNA, forming the structure of neutrophil extracellular traps (NET). Further investigation revealed that the linker histone is the major component of NET [42]. NET is able to trap pathogens and to restrict the deadly proteins that are released by neutrophils from harming surrounding host cells [47]. Surprisingly, linker histone does more than trapping microbes. Fragments of H1 can interact with bacterial membrane lipids, increasing its membrane permeability and disrupting cell membrane, and thus achieve to kill pathogens [42, 48]. Linker histone's another extracellular function is demonstrated by its ability to bind to C reaction protein (CRP). When the linker histone does not bind to DNA, it has two binding sites for CRP, with one calcium-dependent site and the other calcium-independent site. CRP is able to bind to microbes and undermine their membrane, resulting in the clearance of microbes [49]. Thus, linker histone is also associated with CRP-dependent microbe clearance (Figure 1). To conclude, linker histone exhibits important extranuclear and extracellular functions mainly on the innate immune system.
Linker Histone Associated Diseases
Linker Histone in Cancer Cells
Quantity Alterations
The transcription level of the linker histone is difficult to detect mainly because its mRNA lacks a long poly A tail [50]. Not until recently did scientists reveal that the mRNA level of H1 gene is altered in cancer cells. For example, ovarian adenocarcinoma cancer cells exhibit a 40% reduction in overall linker histone mRNA level compared with benign tumors [51].
More importantly, not only does the overall H1 level in cancer cells alter, the expression pattern of certain variants also differs. In benign prostate epithelial cells and low grade cancer cells, H1.5 exhibits low expression level and a weaker reactivity. However, in higher grade or even metastatic prostate cancer cells, the expression level of H1.5 greatly increased [52]. Thus, it can be speculated that H1.5 is correlated to prostate cancer cells' aggressiveness and differentiation status. The same pattern of H1.5 is observed in other types of cancer cells [53]. For example, low grade pulmonary neuroendocrine tumors show low level of H1.5 while high grade cancer exhibits a strong staining pattern of H1.5 [54]. Therefore, it is reasonable to postulate that the replication-dependent subtypes might exhibit the similar pattern and consequently can be of use as diagnostic biomarkers. In differentiated normal cells, H1.0 is expressed at high levels while its expression level is down-regulated in various cancer cells. Moreover, H1.0 level is anti-correlated with the presence of the proliferating cell marker Ki67, particularly in high grade cancer cells [55].
Nuclear, cytoplasmic and extracellular functions of linker histone. In the nucleus, linker histone is responsible for chromatin condensation and transcriptional activity regulation. In the cytoplasm, linker histones are secreted to trigger the process of apoptosis when DNA double string break occurs. Certain cells, such as macrophages, contain linker histones granules that show antimicrobial activity. Extracellular linker histone forms NET to trap pathogens, or binds with CRP to kill the target pathogen. On the cell membrane of Kupffer cell, linker histone binds TGs and internalizing them.
To sum up, it is obvious that the expression level of H1 changes in cancer cells compared with normal cells and benign tumors. Certain subtypes exhibit specific but aberrant patterns. Therefore, the detection of H1 subtypes is of high clinical diagnostic value to distinguish malignant tumors from the benign ones.
Recurrent Mutations
Except for alteration in expression level, H1 gene mutation can also lead to tumorigenesis. Several recurrent mutations in linker histone genes are detected in certain types of cancer. For example, HIST1H1 B-E genes are identified as recurrent mutated genes in follicular lymphoma [56-58]. HIST1H1 mutations have also been identified in chronic lymphocytic leukemia, diffuse large B-cell lymphoma and colorectal cancer [57, 59-61].
There are some mechanisms to explain how the mutations in genes lead to cancer-promoting phenotypes. One possible explanation is that the mutation alters protein-protein interaction as well as its post-translational modification. Another possible mechanism is that the mutated linker histone fails to bind to linker DNA and therefore leads to a less compacted chromatin. This postulation is confirmed by a research showing that amino acid changes in linker histone indeed is able to lead to affinity alteration with DNA in follicular lymphoma [58].
Linker Histone was Mediated by Cancer-Related Proteins
Linker histone also plays an important part in the pathway of tumorigenesis (Figure 2). Several studies have confirmed that the linker histone can be mediated by various cancer-related proteins, interrupting the regular cell cycle and affecting the cell dividing process. For example, under the condition of genotoxic stress, CHD8 recruited the linker histone to the promoter of p53, which resulted in the chromatin condensation. p53 is a major tumor suppressor. With the linker histone condensing the chromatin of p53 promoter, its transcriptional activity is repressed, thereby inhibiting the functions of p53 and increasing the possibility of cell tumorigenesis (Figure 2) [62].
Phosphatase and tensin homolog (PTEN) is a tumor suppressor that interacts with the linker histone. These two proteins cooperatively repress H4K16 acetylation and promote the condensation of chromatin. In the absence of PTEN, linker histones dissociate from the chromatin. Furthermore, microarray analysis of PTEN-/- cells shows upregulated transcriptional activity of several oncogenes, including KRAS, BRAF, and AKT1. Thus, it can be concluded that PTEN promotes the linker histones attaching to the chromatin to enhance its condensation, resulting in the repression of cancer-promoting genes in normal cells; however, in cancer cells, mutated PTEN resulted in linker histones dissociating from the chromatin and thus upregulated the expression of certain cancer-promoting proteins (Figure 2) [63].
Linker histone in the pathway of tumorigenesis. Linker histone can respectively bind with CHD8, PTEN and MTA1. H1 binding with CHD8 represses the transcription of p53. H1 normally binds PTEN to promote chromatin condensation and repress the transcription of oncogenes; yet mutated PTEN represses H1 binding and leads to the chromatin relaxation, and promotes the transcription of oncogene in tumor cells. Also, H1 binding with MTA1 alters the transcriptional activity of several genes. These changes might result in the tumorigenesis of the cells through different pathways.
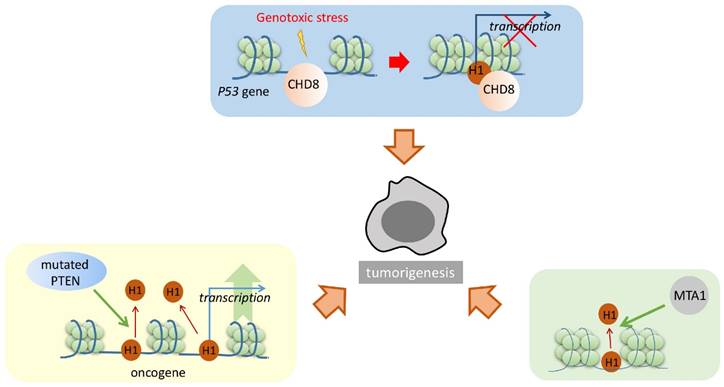
Metastasis-associated gene 1 (MTA1) also modulates the interaction between the linker histone and the chromatin. Although no physical interaction has been directly detected between the linker histone and MTA1, higher level of MTA1 actually leads to the disruption of the linker histone binding to the chromatin, therefore resulting in the alteration of certain gene expression levels (Figure 2) [56]. Similarly, such alteration will result in cancer-promoting phenotypes [53].
How H1 influences the process of tumorigenesis is still unknown. The linker histone appears as an “effector” of certain important cancer-related proteins. The affinity of the linker histone to the nucleosome can be modulated by these cancer-related proteins. The proteins either increase the binding or dissociate from it, therefore achieving to alter the expression level of several important cancer-related proteins, which eventually leads to change the growing pattern or the cell division pattern.
Linker Histone and Alzheimer's Disease
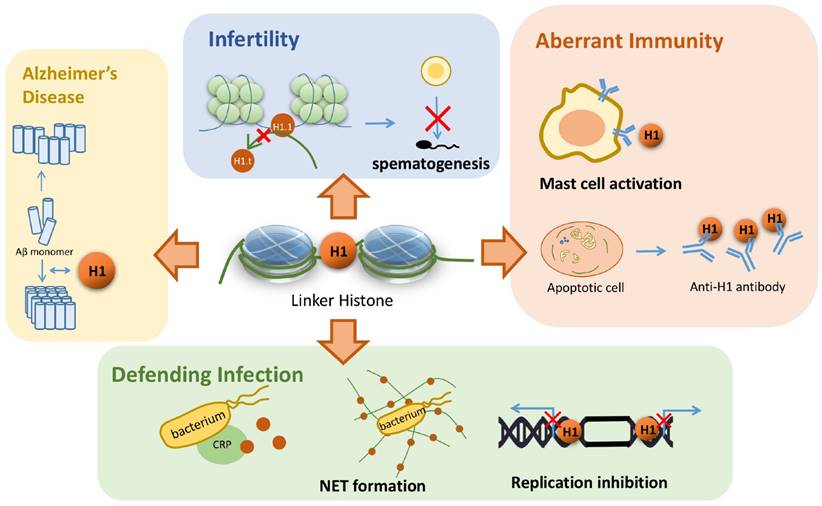
The existence of amyloid plaques in neurons is one of the most important characteristics in Alzheimer's disease. Duce et al. found that the linker histone is also in the amyloid plaque by immunohistochemistry [64]. Such a discovery raises scientists' interests to explore the relationship between the linker histone and the pathogenesis of Alzheimer's disease. Further investigation revealed that the linker histone can change the conformation of its own C terminal domain into an all beta structure with the trigger of detergents, which is ready for forming ribbon-like fibers. This is the evidence in vivo to suggest that the linker histones can form amyloid-like fibers, which further relates the linker histone to Alzheimer's disease [65]. More recently, another study found out that the linker histone can interact with beta-amyloid peptide, which results in both proteins' conformation changes. Subsequently, a great number of beta-amyloid peptides form laminar aggregates and thick bundles, in contrast to the aggregates of only 3~4 amyloid fibrils in the absence of the linker histones (Figure 3) [66]. Altogether, these studies suggest that the linker histones are capable of forming the amyloid-like fibers themselves. Besides, H1 also changes the conformation of beta-amyloid fibrils and results in the over-aggregation of these amyloid fibrils. These aggregated fibrils further form larger bundles and amyloid aggregates, which results in the development of Alzheimer's disease.
Linker histone's association with several diseases. Linker histone is able to assist the binding and organization of Aβ monomer and form a larger bundle with more fibrils, which might trigger Alzheimer's disease. In the process of spermatogenesis, the failure to replace a subtype of linker histone H1.1 can result in male infertility. Linker histones can induce the occurrence of type I hypersensitivity and is also associated with SLE. Linker histone also defends the organism from pathogen infection by forming NET or binding with CRP. It can also inhibit the replication of viral DNA by hindering in the both ends of the replication fork.
Linker Histone and Infertility
The linker histone also plays an important part in the process of spermatogenesis. Mammal sperms are characterized with highly condensed nuclei, and to achieve such high condensation, linker histones go over certain changes to remodel chromatin. When pachytene spermatocytes undergo DNA replication, most of the somatic H1 variants are replaced by a testis specific subtype H1.6. Subsequently, the chromatin structure alters in a significant manner, showing that H1.1 and H1.6 is related to male infertility by affecting spermatogenesis (Figure 3) [67]. Thus, it is implicated that the linker histone, especially H1.6, exerts its influence on chromatin remodeling in the process of spermatogenesis, and shows its probable association with male infertility. A recent study has confirmed this speculation. Although single knockout mouse lines of H1.1 and H1.6 genes are fully fertile [68-70], five knockout mouse lines of triple genes, including Acr/H1.1/Smcp, Acr/Tnp2/Smcp, Tnp2/H1.1/Smcp, Acr/H1.6/Smcp and Tnp2/H1.6/Smcp, exhibit drastic reductions in sperm motility, decreased migration in the female reproductive tract and reduced fertilization [71]. Although the underlying mechanism remains to be solved, these three parameters strongly support that H1.1 and H1.6 replacing disorder might trigger the chromatin structure abnormality, hindering the process of spermatogenesis and thus results in male infertility.
Linker Histone and Infection
Linker histone's ability to bind with DNA not only serves to regulate the transcriptional activity of genes but also helps to defend virus replication in host cells in an indirect manner. In Human Papillomavirus type 11 (HPV-11) infected human cells, H1 is able to bind to viral genome and therefore inhibits the initiation and elongation of viral DNA replication since the replication fork is blocked. E1, a viral protein of HPV-11, is required to bind with the linker histones and to displace them from the virus genome to initiate the viral genome replication [72]. Similarly, simian virus 40 large tumor antigen, a viral protein, is required to bind with the linker histone in order to disrupt nucleosomal structure and initiate DNA replication [73]. Thus, it can be implicated that the presence of the linker histone is essential to interrupt viral replication.
An increasing extent of investigation on linker histone's extranuclear function leads to scientists' attention on its capability of microbe clearance. As mentioned above, H1 in the cytoplasm can bind specifically to certain microbes, such as E. coli [45], norovirus [46], and thus achieves to clear these harmful bacteria or virus. H1 also achieves to clear the microbes in extracellular matrix. Neutrophils can secrete DNA, histones and other related proteins to form NET, which traps harmful microbes and hinders its potential to spread through circulation or across tissues [42]. Secreted linker histone can also bind with CRP and then together they binds to the microbe, leading to the destruction of its cell wall and eventually its death (Figure 3) [49]. To conclude, linker histone exhibits its effect on the clearance of a wide range of pathogens through various pathways. Therefore, it is possible that the linker histone might serve as a potential therapeutic target against certain infection, or a promising way to regulate innate immunity.
Linker Histone and Aberrant Immunity
Linker histone is found to serve as an alarmin that sets off type I hypersensitivity. In ovalbumin-sensitized mice an increased level of circulating linker histone was observed. High level linker histone activates mast cells, leading to its degranulation, and eventually induces typical symptoms of allergic rhinitis [74]. Therefore, the linker histone plays a rather important pathophysiological role in mast cell-mediated type I hypersensitivity. This finding suggests linker histone to be a potential therapeutic strategy against allergic rhinitis, or furthermore, a wider spectrum of diseases concerning allergy.
Cells that undergo apoptosis generally form apoptotic bodies, containing DNA, histones and so on. However, an inflammatory response against apoptotic bodies will take place if the process of phagocytosis is impaired, thus provoking autoimmunity (Figure 3) [75]. Specifically, the phagocytes that engulf linker histones will turn into LE cells, which is one of the signs for active SLE [76]. For patients that develop SLE, serum antibodies that target the C-terminal of the linker histone are often detected with high specificity [77, 78]. Consequently, it can be concluded that abnormal apoptosis induces certain inflammatory response that triggers the generation of H1 antibody, which leads to the development of SLE [27]. Yet it is important to point out that other anti-nuclear antibodies in SLE patients also show high affinity with the linker histone, thus whether auto-antibodies of SLE start out to target the linker histone needs further research [79, 80].
To sum up, the linker histone serves as a trigger in the course of certain autoimmune diseases. H1 is easily targeted by the immune system once exposed extracellularly, resulting in the activation of mast cell or B cell, and finally the development of the disease.
Conclusion and Perspectives
Linker histone is mainly a nucleus protein that binds to the linker DNA as well as the nucleosome to enhance the condensation of the chromatin. It exerts its influence on the transcriptional activities of certain genes. Moreover, the linker histone can relocate to the cytoplasm and the cell membrane surface, or even secrete into the extracellular matrix. These extranucleus linker histones serve as receptors and play important roles in several important pathways. Although controversy remains to be solved, current knowledge about the linker histone is increasing to reveal its mysterious mask.
What is more exciting is that with the understanding of the linker histone's characteristics and functions deepens, its relationship with several diseases begins to become clear to us. The most investigated association is with cancer cells and tumorigenesis. The cell replication status and the invasiveness of tumor cells are often associated with H1 and H1 subtype level, which indicates its valuable clinical implication as a biomarker to differentiate malignant tumors from the benign ones. The linker histone also plays important roles in the pathway of tumorigenesis, exerting its influences as an effector to modulate the expression of certain cancer-related genes and thus achieves to alter the proliferation status of the cell. The identification of such an important role indicates its application as a potential target for cancer therapy. The linker histone also emerges to be related with several other diseases such as Alzheimer's disease, male infertility and autoimmune disease. The associations between the linker histone and these diseases are not fully understood. More researches are needed to investigate the role of the linker histone in these diseases: whether the alteration of the linker histones is the trigger or the result of the disease.
Although we currently do not possess the complete knowledge of linker histone and its association with diseases, its outline is becoming clear to us while details are needed to complete the whole picture. Hopefully, the improvement of technology and methods as well as an increasing number of clinical researches will aid our understanding of linker histone and furthermore discover effective treatments against diseases.
Abbreviations
MUMP, microbicidal protein; PARP-1, poly(ADP-ribose) polymerase 1; CyP, cytochrome P450; USF, upstream stimulatory factor; NCC, nonspecific cytoxic cell; NET, neutrophil extracellular trap; CRP, C reaction protein; PTEN, phosphatase and tensin homolog; MTA1, metastasis-associated gene 1; ACR, proacrosin; Smcp, sperm mitochondria-associated cysteine-rich protein; Tnp2, transition protein 2; HPV-11, Human Papillomavirus type 11; SLE, systemic lupus erythematosus.
Acknowledgements
This work was supported by National Natural Science Foundation of China Grants 81672778, 81372165, 31261140372; Beijing Natural Science Foundation 5142009.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Watson JD, Gann A, Baker TA, Bell SP, Levine M, Losick R. Molecular Biology of the Gene. Cold Spring Harbor Laboratory Press. 1987
2. Crane-Robinson C. Linker histones: History and current perspectives. Biochim Biophys Acta. 2016;1859:431-5
3. Roque A, Ponte I, Suau P. Interplay between histone H1 structure and function. Biochim Biophys Acta. 2016;1859:444-54
4. Cerf C, Lippens G, Muyldermans S, Segers A, Ramakrishnan V, Wodak SJ. et al. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: sequential assignment and secondary structure. Biochemistry. 1993;32:11345-51
5. Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219-23
6. Millan-Arino L, Izquierdo-Bouldstridge A, Jordan A. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta. 2016;1859:510-9
7. Higurashi M, Adachi H, Ohba Y. Synthesis and degradation of H1 histone subtypes in mouse lymphoma L5178Y cells. J Biol Chem. 1987;262:13075-80
8. Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem Cell Biol. 2001;79:289-304
9. Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524-31
10. Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393-404
11. Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403-27
12. Bednar J, Woodcock CL. Cryoelectron microscopic analysis of nucleosomes and chromatin. Methods Enzymol. 1999;304:191-213
13. Meyer S, Becker NB, Syed SH, Goutte-Gattat D, Shukla MS, Hayes JJ. et al. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011;39:9139-54
14. Shukla MS, Syed SH, Goutte-Gattat D, Richard JL, Montel F, Hamiche A. et al. The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 2011;39:2559-70
15. Syed SH, Boulard M, Shukla MS, Gautier T, Travers A, Bednar J. et al. The incorporation of the novel histone variant H2AL2 confers unusual structural and functional properties of the nucleosome. Nucleic Acids Res. 2009;37:4684-95
16. Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819-21
17. Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402-5
18. Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis EN. et al. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614-7
19. Song F, Chen P, Sun D, Wang M, Dong L, Liang D. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376-80
20. Zhu P, Li G. Higher-order structure of the 30-nm chromatin fiber revealed by cryo-EM. IUBMB Life. 2016;68:873-8
21. Izzo A, Kamieniarz-Gdula K, Ramirez F, Noureen N, Kind J, Manke T. et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142-54
22. Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z. et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417
23. Mayor R, Izquierdo-Bouldstridge A, Millan-Arino L, Bustillos A, Sampaio C, Luque N. et al. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J Biol Chem. 2015;290:7474-91
24. Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012;8:e1002879
25. Smith TP, John DA, Bailey CJ. Epidermolytic toxin binds to components in the epidermis of a resistant species. Eur J Cell Biol. 1989;49:341-9
26. Zlatanova JS, Srebreva LN, Banchev TB, Tasheva BT, Tsanev RG. Cytoplasmic pool of histone H1 in mammalian cells. J Cell Sci. 1990;96( Pt 3):461-8
27. Parseghian MH, Luhrs KA. Beyond the walls of the nucleus: the role of histones in cellular signaling and innate immunity. Biochem Cell Biol. 2006;84:589-604
28. Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ. et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173-8
29. Makarov VL, Dimitrov SI, Petrov PT. Salt-induced conformational transitions in chromatin. A flow linear dichroism study. Eur J Biochem. 1983;133:491-7
30. Makarov VL, Dimitrov SI, Tsaneva IR, Pashev IG. The role of histone H1 and non-structured domains of core histones in maintaining the orientation of nucleosomes within the chromatin fiber. Biochem Biophys Res Commun. 1984;122:1021-7
31. Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475-83
32. Juan LJ, Utley RT, Adams CC, Vettese-Dadey M, Workman JL. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994;13:6031-40
33. Ura K, Hayes JJ, Wolffe AP. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 1995;14:3752-65
34. Sarg B, Lopez R, Lindner H, Ponte I, Suau P, Roque A. Identification of novel post-translational modifications in linker histones from chicken erythrocytes. J Proteomics. 2015;113:162-77
35. Raghuram N, Strickfaden H, McDonald D, Williams K, Fang H, Mizzen C. et al. Pin1 promotes histone H1 dephosphorylation and stabilizes its binding to chromatin. J Cell Biol. 2013;203:57-71
36. Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J. et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012;26:797-802
37. Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B. et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389-93
38. Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y. et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673-88
39. Brown DT, Gunjan A, Alexander BT, Sittman DB. Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain. Nucleic Acids Res. 1997;25:5003-9
40. Brown DT, Alexander BT, Sittman DB. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 1996;24:486-93
41. Gunjan A, Alexander BT, Sittman DB, Brown DT. Effects of H1 histone variant overexpression on chromatin structure. J Biol Chem. 1999;274:37950-6
42. Hiemstra PS, Eisenhauer PB, Harwig SS, van den Barselaar MT, van Furth R, Lehrer RI. Antimicrobial proteins of murine macrophages. Infect Immun. 1993;61:3038-46
43. Brix K, Summa W, Lottspeich F, Herzog V. Extracellularly occurring histone H1 mediates the binding of thyroglobulin to the cell surface of mouse macrophages. J Clin Invest. 1998;102:283-93
44. Rose FR, Bailey K, Keyte JW, Chan WC, Greenwood D, Mahida YR. Potential role of epithelial cell-derived histone H1 proteins in innate antimicrobial defense in the human gastrointestinal tract. Infect Immun. 1998;66:3255-63
45. Zhu G, Chen H, Choi BK, Del Piero F, Schifferli DM. Histone H1 proteins act as receptors for the 987P fimbriae of enterotoxigenic Escherichia coli. J Biol Chem. 2005;280:23057-65
46. Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. Inhibition of attachment of virions of Norwalk virus to mammalian cells by soluble histone molecules. Arch Virol. 2003;148:1659-70
47. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS. et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532-5
48. Luders T, Birkemo GA, Nissen-Meyer J, Andersen O, Nes IF. Proline conformation-dependent antimicrobial activity of a proline-rich histone h1 N-terminal Peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob Agents Chemother. 2005;49:2399-406
49. Minota S, Morino N, Sakurai H, Yamada A, Yazaki Y. Interrelationship between autoepitope, DNA-binding domain, and CRP-binding domain on a histone H1 molecule. Clin Immunol Immunopathol. 1993;66:269-71
50. Doenecke D, Albig W, Bouterfa H, Drabent B. Organization and expression of H1 histone and H1 replacement histone genes. J Cell Biochem. 1994;54:423-31
51. Medrzycki M, Zhang Y, McDonald JF, Fan Y. Profiling of linker histone variants in ovarian cancer. Front Biosci (Landmark Ed). 2012;17:396-406
52. Khachaturov V, Xiao GQ, Kinoshita Y, Unger PD, Burstein DE. Histone H1.5, a novel prostatic cancer marker: an immunohistochemical study. Hum Pathol. 2014;45:2115-9
53. Scaffidi P. Histone H1 alterations in cancer. Biochim Biophys Acta. 2016;1859:533-9
54. Hechtman JF, Beasley MB, Kinoshita Y, Ko HM, Hao K, Burstein DE. Promyelocytic leukemia zinc finger and histone H1.5 differentially stain low- and high-grade pulmonary neuroendocrine tumors: a pilot immunohistochemical study. Hum Pathol. 2013;44:1400-5
55. Kostova NN, Srebreva LN, Milev AD, Bogdanova OG, Rundquist I, Lindner HH. et al. Immunohistochemical demonstration of histone H1(0) in human breast carcinoma. Histochem Cell Biol. 2005;124:435-43
56. Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S. et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487-98
57. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-53
58. Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176-81
59. Landau DA, Wu CJ. Chronic lymphocytic leukemia: molecular heterogeneity revealed by high-throughput genomics. Genome Med. 2013;5:47
60. Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879-84
61. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD. et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268-74
62. Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T. et al. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172-82
63. Chen ZH, Zhu M, Yang J, Liang H, He J, He S. et al. PTEN interacts with histone H1 and controls chromatin condensation. Cell Rep. 2014;8:2003-14
64. Duce JA, Smith DP, Blake RE, Crouch PJ, Li QX, Masters CL. et al. Linker histone H1 binds to disease associated amyloid-like fibrils. J Mol Biol. 2006;361:493-505
65. Roque A, Teruel N, Lopez R, Ponte I, Suau P. Contribution of hydrophobic interactions to the folding and fibrillation of histone H1 and its carboxy-terminal domain. J Struct Biol. 2012;180:101-9
66. Roque A, Sortino R, Ventura S, Ponte I, Suau P. Histone H1 Favors Folding and Parallel Fibrillar Aggregation of the 1-42 Amyloid-beta Peptide. Langmuir. 2015;31:6782-90
67. Doenecke D, Drabent B, Bode C, Bramlage B, Franke K, Gavenis K. et al. Histone gene expression and chromatin structure during spermatogenesis. Adv Exp Med Biol. 1997;424:37-48
68. Drabent B, Saftig P, Bode C, Doenecke D. Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem Cell Biol. 2000;113:433-42
69. Rabini S, Franke K, Saftig P, Bode C, Doenecke D, Drabent B. Spermatogenesis in mice is not affected by histone H1.1 deficiency. Exp Cell Res. 2000;255:114-24
70. Lin Q, Sirotkin A, Skoultchi AI. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol Cell Biol. 2000;20:2122-8
71. Nayernia K, Drabent B, Meinhardt A, Adham IM, Schwandt I, Muller C. et al. Triple knockouts reveal gene interactions affecting fertility of male mice. Mol Reprod Dev. 2005;70:406-16
72. Swindle CS, Engler JA. Association of the human papillomavirus type 11 E1 protein with histone H1. J Virol. 1998;72:1994-2001
73. Ramsperger U, Stahl H. Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J. 1995;14:3215-25
74. Nakano T, Kamei R, Fujimura T, Takaoka Y, Hori A, Lai CY. et al. Impact of Histone H1 on the Progression of Allergic Rhinitis and Its Suppression by Neutralizing Antibody in Mice. PLoS One. 2016;11:e0153630
75. Schiller M, Bekeredjian-Ding I, Heyder P, Blank N, Ho AD, Lorenz HM. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008;15:183-91
76. Feierl E, Smolen JS, Karonitsch T, Stummvoll GH, Ekhart H, Steiner CW. et al. Engulfed cell remnants, and not cells undergoing apoptosis, constitute the LE-cell phenomenon. Autoimmunity. 2007;40:315-21
77. Costa O, Tchouatcha-Tchouassom JC, Roux B, Monier JC. Anti-H1 histone antibodies in systemic lupus erythematosus: epitope localization after immunoblotting of chymotrypsin-digested H1. Clin Exp Immunol. 1986;63:608-13
78. Schett G, Smole J, Zimmermann C, Hiesberger H, Hoefler E, Fournel S. et al. The autoimmune response to chromatin antigens in systemic lupus erythematosus: autoantibodies against histone H1 are a highly specific marker for SLE associated with increased disease activity. Lupus. 2002;11:704-15
79. Touloupi E, Routsias JG, Tzioufas AG. Cross-recognition between histones and La/SSB may account for anti-DNA reactivity in SLE patients. Clin Exp Immunol. 2005;142:172-9
80. Boumba VA, Seferiadis K. Rabbit anti-HMG-17 antibodies recognize similar epitopes on the HMG-17 molecule as lupus autoantibodies. Relation with histone H1 defined epitopes. J Pept Sci. 2002;8:683-94
81. Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227
82. Terme JM, Sese B, Millan-Arino L, Mayor R, Izpisua Belmonte JC, Barrero MJ. et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem. 2011;286:35347-57
Author contact
![]() Corresponding author: Yang Yang, Department of Biochemistry and Molecular Biology, Peking University Health Science Center, #38 Xueyuan Road, Beijing 100191, China. Phone: 86-10-82801602; E-mail: yangshedu.cn
Corresponding author: Yang Yang, Department of Biochemistry and Molecular Biology, Peking University Health Science Center, #38 Xueyuan Road, Beijing 100191, China. Phone: 86-10-82801602; E-mail: yangshedu.cn

 Global reach, higher impact
Global reach, higher impact