Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(9):1180-1191. doi:10.7150/ijbs.21722 This issue Cite
Research Paper
Antisense lncRNA FOXF1-AS1 Promotes Migration and Invasion of Osteosarcoma Cells Through the FOXF1/MMP-2/-9 Pathway
1. Department of Orthopaedic Surgery, Shanghai Tenth People's Hospital Affiliated to Tongji University, Shanghai 200072, PR China;
2. Institute of Bone Tumor Affiliated to Tongji University, School of Medicine, Shanghai 200072, PR China.
* Zhu Kun-Peng, Zhang Chun-Lin, and Ma Xiao-Long contributed equally to this study and share first authorship. These authors are considered Co-first authors.
Received 2017-7-2; Accepted 2017-7-26; Published 2017-9-5
Abstract
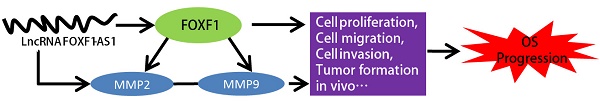
Osteosarcoma (OS) is the most common primary malignant bone cancer in children and adolescents. Long non-coding RNAs (lncRNAs) have been shown to play significant role in various cancers, including OS. In a previous study, we have reported that a novel antisense lncRNA FOXF1-AS1, also known as FENDRR, could sensitize doxorubicin-resistance of OS cells through down-regulating ABCB1 and ABCC1. Here in, the critical role of FOXF1-AS1 in regulating OS progression was further investigated. Firstly, we found that FOXF1-AS1 and its antisense transcript FOXF1 expression were positively up-regulated in OS tissues and cell lines and correlated with poor prognosis of OS patients. Besides, FOXF1-AS1 as well as FOXF1 silencing significantly inhibited cell proliferation, migration, invasion of OS cells and tumor growth both in vitro and vivo through decreasing the expression of MMP2 and MMP9, whereas enhanced expression of FOXF1-AS1 had the opposite effects. In addition, mechanistically, both of FOXF1-AS1 and FOXF1 could regulate the expression of MMP2 and MMP9 at mRNA and protein levels, whereas FOXF1-AS1 could influence the FOXF1expression but FOXF1 did not have the same effect on FOXF1-AS1. Rescue assay further showed that FOXF1-AS1 overexpression efficiently reversed the knockdown of MMP2 and MMP9 expression induced by si-FOXF1. Thus, we concluded that FOXF1-AS1 may promote migration and invasion of OS cells through the FOXF1/MMP-2/-9 pathway. Taken together, these findings demonstrated the underlying mechanism of FOXF1-AS1 in the regulation of OS progression and provide a novel potential target in the OS therapy.
Keywords: Osteosarcoma, LncRNA FOXF1-AS1, FOXF1, MMP, Progression.
Introduction
Osteosarcoma (OS) is the most common primary malignant bone cancer and the second leading cause of cancer-related death in children and adolescents [1]. The current treatment strategies include wide tumor excision combined with adjuvant chemotherapy and radiotherapy, which have greatly increased the 5-year survival rate to 70%-80% [2]. However, the prognosis of patients with tumor recurrence or lung metastases still remains poor [3, 4]. Thus, it is necessary to further identify the molecular mechanisms underlying osteosarcoma tumorigenesis and progression to find a new applicable treatment target.
Long noncoding RNA (lncRNA) is a novel class of non-protein coding transcripts longer than 200 nucleotides [5]. Recently, lncRNAs have been reported to play important regulatory roles in a variety of physiological and pathological processes, including carcinogenesis [6]. In cancer, these dysregulated lncRNAs always function through interacting with and further regulating the expression of the oncogenic or tumor suppressive genes [7-9]. Actually, many lncRNAs including the “star molecules” like GAS5, PVT1 and HOTAIR have been identified to be tumor-associated in osteosarcoma progression, metastasis and chemoresistance [10]. For example, Ye et al. reported that lncRNA GAS5 suppresses cell growth and EMT in osteosarcoma by regulating the miR-221/ARHI pathway [11]. Song et al. found that lncRNA PVT1 promotes tumor progression by regulating miR-497/HK2 axis in osteosarcoma [12]. Wang et al. showed that overexpression of lncRNA HOTAIR promotes tumor growth and metastasis in osteosarcoma [13]. However, the specific lncRNAs involved in the OS progression and its related regulatory mechanism still needed to be fully elucidated.
In the current study, we focused on the function and regulatory mechanism of a novel lncRNA, FOXF1-AS1, also known as FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA), in osteosarcoma progression. In a previous study, we screened the lncRNA and mRNA expression profiles of the doxorubicin-resistant human OS cell line MG63/DXR and its parental cell line MG63 ascertained by microarray analysis and further found the most down-regulated lncRNA FOXF1-AS1 with 20-fold change in the MG63/DXR cell could sensitize doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1 [14, 15]. However, the roles of lncRNA FOXF1-AS1 in OS progression remain unclearly defined. Based on the previous result, we further determined the vital role of lncRNA FOXF1-AS1 in the cell proliferation, migration, and invasion of OS in vitro and vivo. And we identified that lncRNA FOXF1-AS1 was a single antisense oligonucleotide RNA transcribed from the negative strand of Forkhead box protein F1 (FOXF1) and concordantly up-regulated with FOXF1 in osteosarcoma cell lines and tissues. Besides, our data showed that FOXF1-AS1 can serve as a biomarker to predict the prognosis of OS patients. In addition, we found that lncRNA FOXF1-AS1 promotes OS progression through the FOXF1/MMP-2/-9 pathway and FOXF1-AS1 may be a potential target of OS treatment.
Method and Materials
Cell lines and culture conditions
Six human osteosarcoma cell lines (SaoS2, HOS, KH-OS, MG63, 143B and U2-OS) were purchased from American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Gibco, Gran Island, NY, USA), 100 U/mL of penicillin and 100 mg/mL of streptomycin (Invitrogen). Normal osteoblast cells (hFOB1.19) obtained from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultured in Ham's F12/ DMEM supplemented with 10% FBS, 100 U/mL penicillin and 100 mg/mL streptomycin. Cultures were maintained at 37°C in a humidified CO2 (5%) atmosphere.
Clinical samples
A total of 82 primary osteosarcoma patients who underwent surgery at Department of Orthopedics, Shanghai Tenth People's Hospital between July 2006 and December 2016 were included in this study. The study was approved by the Ethics Committee of Shanghai Tenth People's Hospital and written informed consent was obtained from all the study participants. All the resected specimens were immediately frozen in liquid nitrogen and stored at-80°C until RNA extraction. All the cases had a definite pathological diagnosis and the clinical stages of these patients were determined according to the Enneking Stage. The clinical parameters of osteosarcoma patients in this study are presented in Table 1.
Clinical parameters of osteosarcoma patients enrolled in this study
| Pathological characteristics | Cases (n) | FOXF1-AS1 expression | FOXF1 expression | P value | ||||
|---|---|---|---|---|---|---|---|---|
| High(45) | Low(37) | P value | High(43) | Low(39) | P value | |||
| Gender | ||||||||
| Male | 52 | 28 | 24 | 0.82 | 27 | 25 | 0.84 | 0.15 |
| Female | 30 | 17 | 13 | 16 | 14 | |||
| Age | ||||||||
| ≥25 | 24 | 14 | 10 | 0.73 | 13 | 11 | 0.78 | 0.22 |
| <25 | 58 | 31 | 27 | 30 | 28 | |||
| Location | 0.07 | 0.68 | 0.09 | |||||
| Distal of Femur | 38 | 22 | 16 | 24 | 14 | |||
| Proximal of Tibia | 28 | 16 | 12 | 15 | 13 | |||
| Other | 16 | 7 | 9 | 4 | 12 | |||
| Lung Metastasis | 0.012 | 0.009 | ||||||
| Yes | 26 | 21 | 5 | 20 | 6 | <0.05 | ||
| No | 56 | 24 | 32 | 23 | 33 | |||
| Clinical stage | 0.025 | 0.032 | ||||||
| I+IIA | 28 | 11 | 17 | 8 | 20 | |||
| IIB/III | 54 | 34 | 20 | 35 | 19 | |||
Quantitative real-time polymerase chain reaction
Total RNA was extracted from tissues and cells with Trizol reagent (TAKARA) according to the product description. All mRNAs were reverse transcribed according to the protocol of the PrimeScript® RT Master Mix Perfect Real Time (TAKARA). qPCR was performed using SYBR Geen Master Mix (Applied Biosystems) on an Applied Biosystems 7900HT Real-Time System. The primers were shown in Table 2.
Primers used for PCR validation
| Gene | Forward and Reverse primer |
|---|---|
| FOXF1-AS1 | F:5' TTCATCGGCTGCGTATTCG 3' R:5' TTGCCTTCTAGTCGCCTCC 3' |
| FOXF1 | F:5' AAGCCGCCCTATTCCTACATC 3' R:5' GCGCTTGGTGGGTGAACT 3' |
| MMP2 | F:5' CTGCGGTTTTCTCGAATCCATG 3' R:5' GTCCTTACCGTCAAAGGGGTATCC 3' |
| MMP9 | F:5' GAGGCGCTCATGTACCCTATGTAC 3' R:5' GTTCAGGGCGAGGACCATAGAG 3' |
| GAPDH | F:5' CATGAGAAGTATGACAACAGCCT 3' R:5' AGTCCTTCCACGATACCAAAGT 3' |
Western blotting analysis
Cells were collected and lysed using RIPA protein extraction reagent (Beyotime, Beijing, China) supplemented with a protease inhibitor cocktail (Roche, Pleasanton, USA). Proteins were loaded onto the SDS-PAGE gels for electrophoresis, transferred to PVDF membranes and blocked in 5% milk prior to incubation with the indicated primary antibodies and the secondary antibodies. Autoradiograms were quantified by densitometry using GAPDH as a control. Antibodies against FOXF1, MMP2 and MMP9 were purchased from Cell Signaling Technology (Danvers, MA, USA).
Immunochemistry (IHC)
Paraffin-embedded tissue sections from osteosarcoma specimens were given a heat pretreatment of 60°C for one hour, then dewaxed in xylene, re-hydrated in an ethanol series (100-50%) and treated in 0.01 mol/L citrate buffer (pH 6.0) for antigen retrieval. The antibody used in the experiment included goat anti-FOXF1 polyclonal antibody (Danvers, MA, USA), rabbit-anti-goat IgG (Danvers, MA, USA). The following experimental procedure was according to the manufacturer's instructions.
Plasmid construction and cell transfection
MG63 and 143B cells were transiently transfected with siRNAs after being sowed into the 6-well plates overnight. A scrambled negative control, a plasmid overexpressing FOXF1-AS1, and an empty vector, were cultured as well using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) and FuGENE® HD Transfection Reagent (Roche, Germany) according to the manufacturer's instructions, respectively. 48h after transfection, the cells were harvested to detect the overexpression or knockout efficiency via qRT-PCR. Two different siRNAs against FOXF1-AS1 were designed and synthesized by GenePharma (Shanghai, China). The target sequences for the si-FOXF1-AS1 and the synthetic FOXF1-AS1 sequence (3099bp) were described in the previous study (15). The siRNA sequence against FOXF1 (si-FOXF1) was 5'- GCTCAACGAGTGCTTCATCAAGCTACCCA -3'.
Cell proliferation assay
Cell viability was determined with CCK-8 assay. 1×103 cells were seeded into a 96-well plate with quadruplicate repeat for each condition. Cells were incubated in 10 % CCK-8 (DOJINDO, Japan) diluted in normal culture medium at 37 °C until visual color conversion occurred. Proliferation rates were determined at 24, 48, 72h after transfection. The absorbance of each well was measured with a microplate reader set at 570nm.
Wound healing assay and cell invasion assay
A total of 1×105 stable transfected cells were seeded onto six-well plates and cultured overnight. Wounds were created by scratching cell layer with a sterile plastic pipette tips and washed with culture medium. Cells were further cultured with medium containing 1% FBS in 48h.
For the invasion assays, a 24-well transwell chamber (corning, 8 mm) with the upper chamber coated with Matrigel (BD Bioscience) was used. 1.0×105 cells in 100uL serum free DMEM medium were seeded in the top chamber, 500uL medium containing 10% FBS was placed to the lower chamber. After incubation for 48 h, cells on the upper membrane surface were wiped off using a cotton swab and the cells that had traversed the membrane were staining by crystal violet and counted.
Xenograft transplantation
Female nude (BALB/c) mice (4 weeks old) were purchased. Mice were divided into several groups according to the completely randomized method. MG63 cells stably expressing si-FOXF1-AS1 or negative control were propagated and 1×107 cells were inoculated subcutaneously into the right side of the posterior flank of mice. Tumor growth was examined at the indicated time points and tumor volumes were measured. After 7 weeks, the mice were killed and tumors were removed and weighed. All animal procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Shanghai Tenth People's Hospital. The detailed procedure has been described in the previous study [15, 16].
Statistical analysis
All statistical analyses were performed using SPSS 22.0 software (IBM) and Graph pad Prism 5.0. Differences between groups were analyzed using the Student's t test or one-way ANOVA. Overall survival and free-recurrence survival were calculated by Kaplan-Meier survival analysis and compared using the log-rank test. p values<0.05 were considered statistically significant.
Results
lncRNA FOXF1-AS1 was up-regulated in the osteosarcoma tissues and cell lines, correlated with lung metastasis and worse prognosis
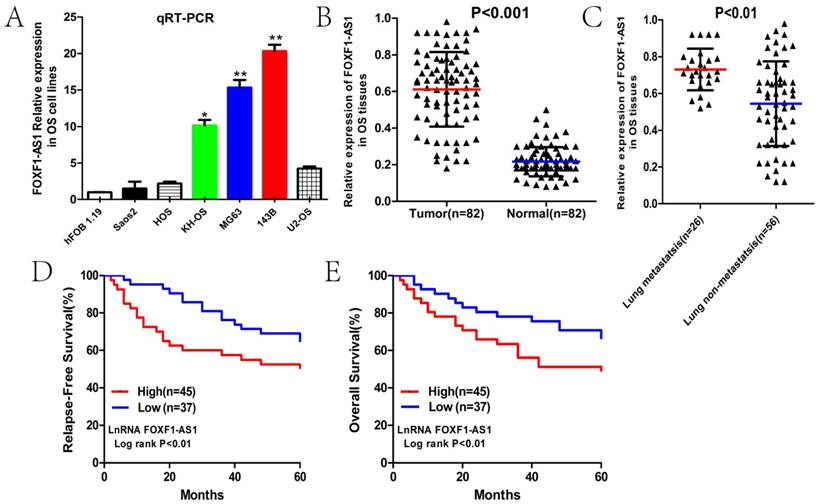
First, we measured the expression level of FOXF1-AS1 in six human OS cell lines and the normal osteoblast cell line hFOB1.19 by qRT-PCR. The results showed that FOXF1-AS1 level was much higher in SaoS2, HOS, KH-OS, U2-OS, MG63, and 143B cells, compared with hFOB1.19 cells (Fig. 1A). Of them, MG63 and 143B cells with a higher expression of FOXF1-AS1 were selected as research represents in the following studies.
qRT-PCR analysis was then used to test the FOXF1-AS1 expression levels in 82 pairs of OS and paracancerous tissues to further study the relationship of FOXF1-AS1 with clinical progression and outcome of OS patients. FOXF1-AS1 expression was conspicuously up-regulated in OS tissues compared with normal tissues (Fig. 1B). Subsequently, we classified our patients into lung metastasis and lung non-metastasis groups and found that the expression of FOXF1-AS1 in the lung metastasis group at early stage was obviously higher than that in the lung non-metastasis group (Fig. 1C). In addition, the median expression level was used as the cut-off. Kaplan-Maier analysis showed that patients with higher expression of FOXF1-AS1 had shorter relapse-free and overall survival time than those with lower FOXF1-AS1 expression (Fig. 1D and E). Collectively, these results outlined FOXF1-AS1 as possible markers of clinical outcome in OS patients.
LncRNA FOXF1-AS1 was up-regulated in the osteosarcoma tissues and cell lines, correlated with lung metastasis and worse prognosis. (A) Expression level of FOXF1-AS1 in six human OS cell lines and normal osteoblast cell line hFOB1.19. (B) Expression level of FOXF1-AS1 in 82 pairs of OS and paracancerous tissues. (C) Expression level of FOXF1-AS1 in OS tissues of lung metastasis and lung non-metastasis group at early stage. (D-E) Patients with high FOXF1-AS1expression had a shorter free-relapse and overall survival time than those with low FOXF1-AS1 expression. *P < 0.05, **P < 0.01.
FOXF1-AS1 promoted OS cell proliferation, migration, invasion and tumor growth in vitro and vivo. (A) FOXF1-AS1 expression in MG63 and 143B cells after si-RNA or vector transfection. (B) CCK-8 assays were performed to detect the function of FOXF1-AS1 on OS cell proliferation ability. (C) Transwell assays were performed to measure the invasion ability of OS cells after transfection. (D) Wound healing assays were performed to examine the migration capacity of OS cells after transfection. (E) General imaging of nude mice and resected tumors in the four groups when exposed to the same treatment. (F) The nude mice were sacrificed in the 7th week. Tumors formed in the FOXF1-AS1 group grew much faster compared with the FOXF1-AS1-NC group and the volumes of transplanted tumors and weights of nude mice were smaller in the si-FOXF1-AS1 group when compared with the si-NC group. *P < 0.05, **P < 0.01.
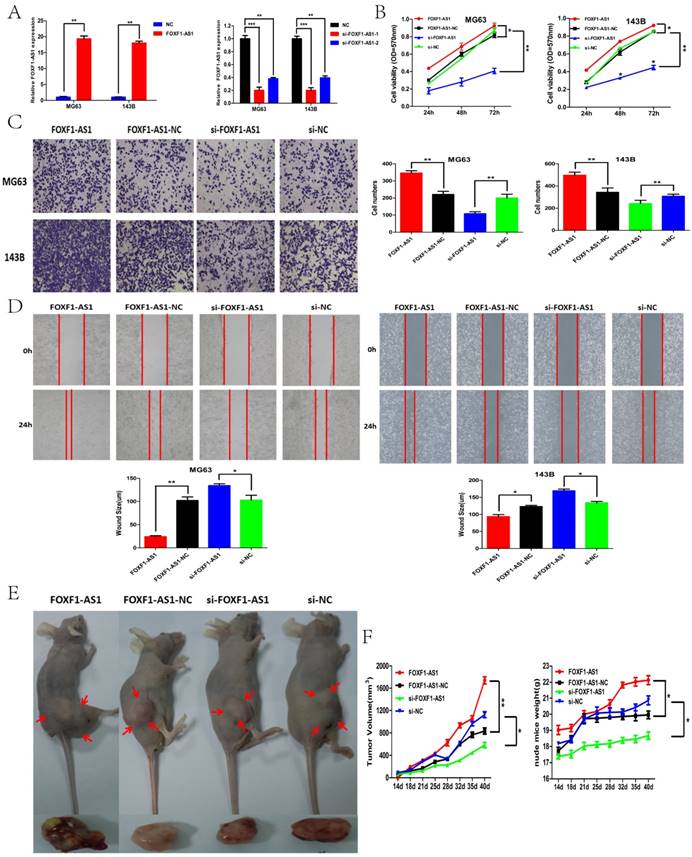
FOXF1-AS1 promoted OS cell proliferation, migration, invasion and tumor growth in vitro and vivo
We further analyzed the biological functions of FOXF1-AS1 in OS cells. The stably transfected cell lines with overexpression or knockdown of FOXF1-AS1 were established in MG63 and 143B cells and the transfected efficiency of FOXF1-AS1 was subsequently confirmed by qPCR analysis (Fig. 2A). Cell proliferation was measured by using CCK-8 in the 24, 48 and 72 hours after transfection. Results showed that cell proliferation rate in the FOXF1-AS1 group were significantly higher compared with the FOXF1-AS1-NC group, whereas obviously lower in the si-FOXF1-AS1 group than the si-NC group (Fig. 2B). Meanwhile, transwell assay and wound healing assay were performed to investigate the role of FOXF1-AS1 in the regulation of cell migration and invasion in OS cells and the results demonstrated that forced FOXF1-AS1 expression significantly increased the invasion ability, while FOXF1-AS1 knockdown showed a conspicuously reduced cell invasion compared with the control cells (Fig. 2D).
To further determine the effects of FOXF1-AS1 on tumorigenesis in vivo, we analyzed the effects of FOXF1-AS1 on the OS growth in nude mice. MG63 cells transfected with FOXF1-AS1, FOXF1-AS1 -NC, si-FOXF1-AS1, or si-NC were subcutaneously injected into athymic mice and were allowed to develop measurable tumors, respectively. As was shown in the figure 2E, from 2th to 7th week, it is obvious that tumors formed in the FOXF1-AS1 group grew much faster compared with the FOXF1-AS1 -NC group and the volumes of transplanted tumors and weights of nude mice were smaller in the si-FOXF1-AS1 group when compared with the si-NC group (Fig. 2F). Taken together, these results demonstrated that FOXF1-AS1 may have oncogenic properties to promote OS growth and progression in vitro and vivo.
FOXF1 expression was positively correlated with FOXF1-AS1 and poor prognosis of OS patients
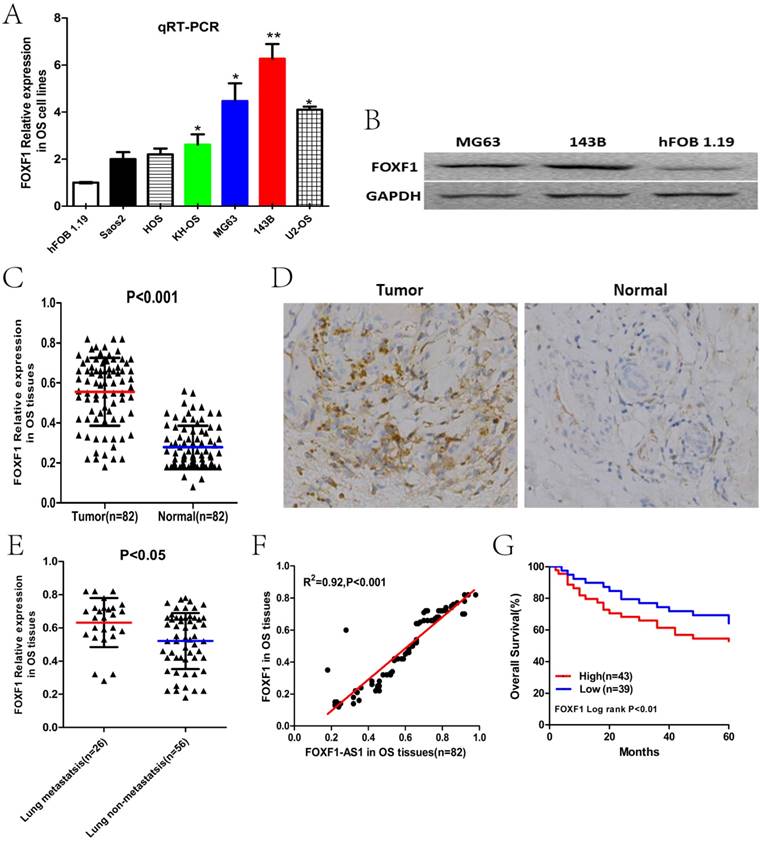
To investigate the role of FOXF1, the cognate sense transcript of FOXF1-AS1, in OS carcinogenesis, we first examined the expression level of FOXF1 in OS cell lines and tissues previously described by qRT-PCR, WB and IHC. The results showed that FOXF1 was significantly up-regulated OS cell lines and tumor tissues (Fig. 3A-D). Subsequently, we also found that the expression of FOXF1 in the lung metastasis group at early stage was obviously higher than that in the lung non-metastasis group (Fig.3E). Besides, according to the correlation analysis, we found that FOXF1 expression was positively correlated with FOXF1-AS1 in OS tissues (R2=0.92, P<0.001), which may suggest the possible regulating relationship between them (Fig. 3F). In addition, Kaplan-Maier analysis also showed that patients with high FOXF1 expression had lower overall survival time than those with low FOXF1 expression (Fig.3G). These results indicated a potential oncogenic role of FOXF1 in the OS development, consistent with the role of lncRNA FOXF1-AS1 in OS.
siRNA-mediated knockdown of FOXF1 suppressed OS cell proliferation, migration, invasion and tumor growth in vitro and vivo
We then tested whether FOXF1 was functionally involved in OS progression. MG63 and 143B cells stably transfected with si-FOXF1 or si-NC were established (Fig. 4A). CCK-8 assay showed that cell proliferation rate was significantly decreased in the si-FOXF1 group, when compared to the si-NC group (Fig. 4B).Meanwhile, transwell and wound healing assay demonstrated that the numbers of migrating and invading cells in the si-FOXF1 group were significantly decreased compared with the si-NC group (Fig. 4C and D).Consistent with the results in vitro, the tumor volumes and nude mice weights of the si-FOXF1 group of MG63 cells were significantly smaller than that of the control group (Fig.4E and F). These results indicated that the down-regulation of FOXF1 expression was sufficient in inhibiting tumor growth and progression of OS cells in vitro and vivo.
FOXF1-AS1 positively regulated the FOXF1/MMP-2/-9 pathway
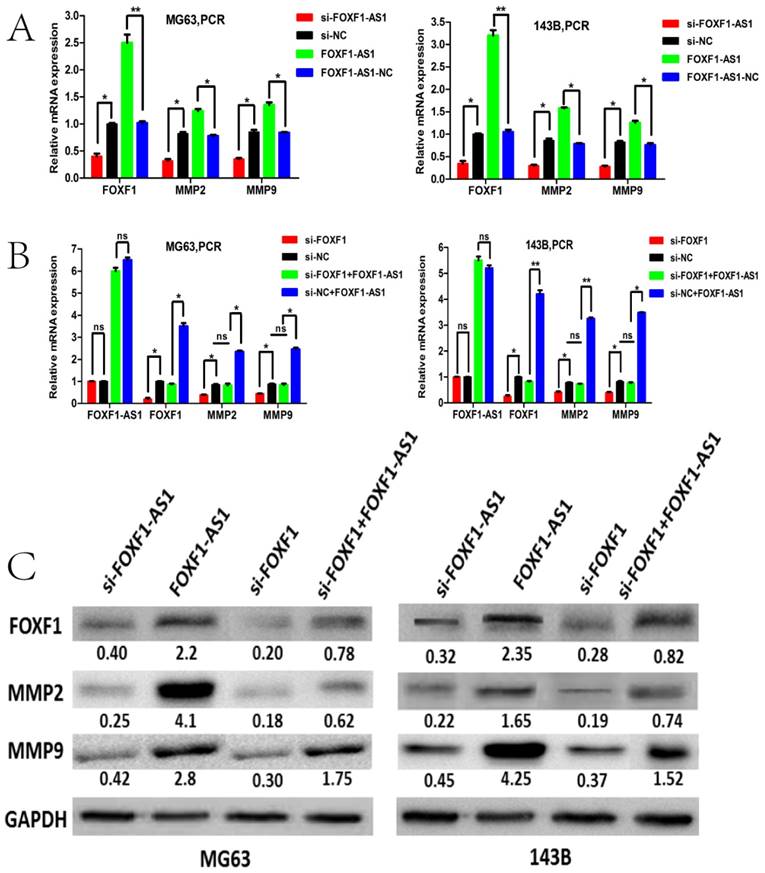
To identify whether FOXF1 was the target gene of lncRNA FOXF1-AS1, we first investigated FOXF1-AS1 and FOXF1 mRNA expression levels in the si-FOXF1-AS1 and si-FOXF1 transfected MG63 (143B) cells by qRT-PCR. Results demonstrated that FOXF1-AS1 knockdown significantly reduced, whereas FOXF1-AS1 overexpression significantly increased the FOXF1 mRNA levels. Western blotting also demonstrated the same changing trend of FOXF1 protein levels with the FOXF1-AS1 knockdown or overexpression. In contrast, FOXF1 knockdown did not affect the mRNA expression of lncRNA FOXF1-AS1 (Fig. 5A and B).
Since MMPs are well known to be involved in the breakdown of the extracellular matrix during OS cells invasion, we then asked whether lncRNA FOXF1-AS1 could promote OS cell migration and invasion through regulating the expression of MMPs. We then assessed the MMP2 and MMP expression by qPCR and WB in the MG63 and 143B cells with knockdown or overexpression of FOXF1-AS1. Interestingly, we found that the mRNA and protein expression level of MMP2 and MMP9 decreased when FOXF1-AS1 was knockdown and increased when FOXF1-AS1 was overexpressed in the two cells. Besides, we found that the mRNA and protein expression level of MMP2 and MMP9 also decreased when FOXF1 was knockdown by si-RNA. However, MMP2 and MMP9 expression did not change in the MG63 and 143B cells when overexpressing the FOXF1-AS1 with down-regulating the FOXF1 by siRNA at the same time (Fig.5C). These data suggested that lncRNA FOXF1-AS1 may contribute to OS progression through the FOXF1/MMP-2/-9 pathway.
FOXF1 expression was positively correlated with FOXF1-AS1 and poor prognosis of OS patients. (A) Expression level of FOXF1 in six human OS cell lines and normal osteoblast cell line hFOB1.19 examined by PCR. (B) Expression level of FOXF1 in MG63, 143B cells and normal osteoblast cell line hFOB1.19 examined by WB. (C) Expression level of FOXF1 in 82 pairs of OS and paracancerous tissues examined by PCR. (D) Immunohistochemical staining of FOXF1 protein level in OS and paracancerous tissues. (E) Expression level of FOXF1 in OS tissues of lung metastasis and lung non-metastasis group at early stage. (F) A positive correlation was found between FOXF1-AS1 and FOXF1 expression in OS tissues. (G) OS patients with lower expression of FOXF1 had longer overall survival time than those with higher expression. *P < 0.05, **P < 0.01.
Discussion
Accumulating evidence has revealed that dysregulated lncRNAs may pinpoint the spectrum of disease progression and independently predict patient outcome in various cancers, including lung cancer [17], breast cancer [18], glioma [19], osteosarcoma [20], and so on. It is an urgent need to understand the molecular mechanism of lncRNAs to explore effective therapeutic target for the treatment of osteosarcoma. Actually, recent studies have reported that several lncRNAs were involved in the progression of OS by regulating cell development, differentiation, proliferation, invasion, and apoptosis. For example, Zhao et al. found that lncRNA CCAT1/miR-148a axis promotes osteosarcoma proliferation and migration through regulating PIK3IP1 [21]. Liu et al. reported that lncRNA MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p [22]. Han et al. showed that lncRNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s [23].
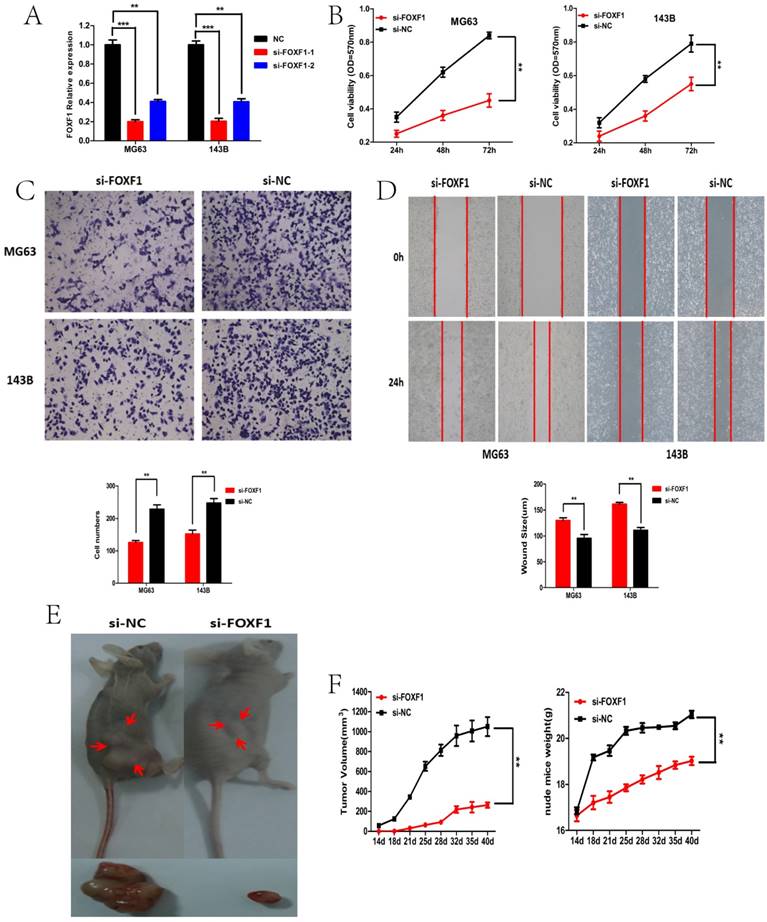
siRNA-mediated knockdown of FOXF1 suppressed OS cell proliferation, migration, invasion and tumor growth in vitro and vivo. (A) FOXF1expression in MG63 and 143B cells after si-RNA or si-NC transfection. (B) CCK-8 assays were performed to examine cell proliferation rate of OS cells transfected with si-FOXF1. (C) Transwell assays were performed to identify the invasion capacity of OS cells transfected with si-FOXF1. (D) Wound healing assays were performed to examine the migration capacity of OS cells transfected with si-FOXF1. (E) General imaging of nude mice and resected tumors in the si-FOXF1 and si-NC transfected groups when exposed to the same treatment. (F) The nude mice were sacrificed in the 7th week. Tumors formed in the si-FOXF1 group grew slower compared with the si-NC group and the volumes of transplanted tumors were smaller in the si-FOXF1 group when compared with the si-NC group. *P < 0.05, **P < 0.01.
FOXF1-AS1 positively regulated the FOXF1/MMP-2/-9 pathway. (A) qRT-PCR was performed to study the interaction between the FOXF1、MMP2 and MMP9 expression levels in MG63 and 143B cells with FOXF1-AS1 overexpression or knockdown. FOXF1-AS1 positively regulated the mRNA level of FOXF1、MMP2 and MMP9 expression. (B) FOXF1 positively regulated the mRNA level of MMP2 and MMP9 expression other than FOXF1-AS1 expression in MG63 and 143B cells. Overexpression of FOXF1-AS1 could rescue the knockdown effect on MMP2 and MMP9 mRNA expression by si-FOXF1. (C) FOXF1-AS1 positively regulated the protein level of FOXF1、MMP2 and MMP9 expression in MG63 and 143B cells. FOXF1 positively regulated the protein level of MMP2 and MMP9 expression and overexpression of FOXF1-AS1 could rescue the knockdown effect on MMP2 and MMP9 protein expression by si-FOXF1.*P < 0.05, **P < 0.01.
FOXF1-AS1, also known as FENDRR, is a 3099-bp lncRNA, located on chromosome 3q13.31, divergently transcribed from the FOXF1 promoter and co-expressed with FOXF1 [24]. Fendrr, homologous to FENDRR in human, was first identified by Grote et al. He found that fendrr plays important roles in the progress of heart and body wall development in mouse and it could bind to PRC2 as well as TrxG/MLL to regulate chromatin structure and gene activity [25]. Then, Szafranski reported that FENDRR was related to ACD/MPV, a lethal lung developmental disorder [26]. Besides, several reports have identified the involvement of FOXF1-AS1 in the carcinogenesis. Xu et al. found that low FOXF1-AS1 expression in gastric cancer tissues predicted poor prognosis and FOXF1-AS1 overexpression inhibited the cell invasion and migration by decreasing the expression of FN1 and MMP-2/MMP-9 [27]. Miao et al. demonstrated that loss of lncRNA FOXF1-AS1 regulated EMT, stemness and metastasis of non-small cell lung cancer cells via EZH2 and down-regulation of FOXF1, which may show the tumor-suppressor role of FOXF1-AS1 in cancer [28].
Nevertheless, the expression and underlying mechanism of FOXF1-AS1 involved in OS is still unclear. Just in a previous study, we identified that lncRNA FOXF1-AS1 (FENDRR) was the most down-regulated of 20-fold change in the doxorubicin-resistant OS cell lines and promoted doxorubicin resistance in OS by inducing the expression of classical multi-drug resistance related genes ABCB1 and ABCC3[14, 15]. In this study, we first identified that FOXF1-AS1 was overexpressed in osteosarcoma tissues and cell lines, correlated with poor prognosis. The biological function of FOXF1-AS1 was subsequently investigated in our study. Our data indicated that knockdown of FOXF1-AS1 inhibited and overexpression of it promoted OS cell proliferation, migration and invasion, tumor growth both in vitro and vivo. Furthermore, we found that FOXF1-AS1 regulated migration and invasion of OS cells through up-regulating the expression of FOXF1 as well as matrix metalloproteinase II and IX (MMP2 and MMP9), key proteinases during cancer invasion[29, 30]. These results suggested that lncRNA FOXF1-AS1 functions as an oncogene and plays a critical role in OS progression, which is different from its tumor-suppressor function in the gastric and lung cancer.
LncRNA FOXF1-AS1 is one of antisense long noncoding RNAs and its conjugate gene FOXF1 (forkhead transcription factor 1) belongs to the 43 members of transcription factor FOX gene family [31]. A large number of studies have shown that FOXF1 protein regulates cellular proliferation, differentiation, metabolic homeostasis and plays an important role in tumorigenesis [32], but its functional role as an oncogene or tumor-suppressor gene was still controversial. Several reports have identified the oncogenic role of FOXF1 gene in various cancers. For instance, Nilsson G et al. showed that FOXF1 promotes breast cancer cell migration by up-regulating lysyl oxidase and suppressing Smad2/3 signaling [33]. Fulford L et al. found that FOXF1 promotes prostate cancer by stimulating the mitogen-activated protein kinase ERK5 [34]. Milewski D et al. reported that FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor [35]. In contrast, other published also showed the converse role of FOXF1 as a tumor-suppressor gene. For example, Tamura M et al. showed that FOXF1 is a novel target gene of the p53 family and suppresses cancer cell migration and invasiveness [36]. Lo PK et al. identified the epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer [37]. Zhao et al. reported that decreased FOXF1 promotes hepatocellular carcinoma tumorigenesis, invasion, and stemness and is associated with poor clinical outcome [38]. All these studies suggest that FOXF1 may act as an oncogene or tumor suppressor depending on the tissue and specific type of cancer.
However, there is seldom report about FOXF1 in osteosarcoma. In the present study, we first identified that FOXF1 was increased in OS cell lines and tissues and positively correlated with the expression of FOXF1-AS1 in the same tissues. FOXF1 also could act as an independent prognostic factor of OS patients, with a longer survival time of lower expression. Furthermore, functional analysis found that FOXF1 also promotes proliferation, migration and invasion of OS cells and tumor growth both in vitro and vivo through up-regulating MMP2 and MMP9. Besides, we found that FOXF1-AS1 could influence the FOXF1expression but FOXF1 did not have the same effect on FOXF1-AS1 and FOXF1-AS1 overexpression efficiently reversed the knockdown of MMP2 and MMP9 mRNA and protein expression induced by si-FOXF1, which suggested the oncogenic role of lncFOXF1-AS1/FOXF1/MMP-2/-9 pathway in OS progression.
However, the specific mechanism of lncRNA FOXF1-AS1 regulating the FOXF1 expression still needed to be further identified. Considering the characteristic of antisense lncRNA, we firstly analyzed the gene sequence of the antisense lncRNA FOXF1-AS1 and its conjugate gene FOXF1, but we did not find an overlapping region to possibly form a RNA-RNA double-strand structure between them, which is different from the antisense-lncRNA KRT7-AS [39], BACE1-AS [40], FOXC2-AS1 [16] and so on. Whereas, on basis of the subcellular location of FOXF1-AS1 by RNA FISH and nuclear and cytoplasmic fractions described in our previous study [15], we speculated the cytoplasmic lncRNA FOXF1-AS1 may regulate the expression of FOXF1 and MMPs at the posttranscriptional level as an endogenous competing RNA to sponge the related miRNAs. However, the hypothesis needed to be verified by the bioinformatics analysis and dual luciferase reporter assay.
In summary, we reported that highly expressed FOXF1-AS1 is an oncogenic lncRNA that exerts a crucial role in the OS progression. Besides, our study revealed that antisense lncRNA FOXF1-AS1 promotes proliferation, migration and invasion of OS through the FOXF1/MMP-2/-9 pathway, thus probably providing a novel therapeutic target for the treatment of OS.
Abbreviations
OS: Osteosarcoma; lncRNA: long non-coding RNA; FOXF1: Forkhead transcription factor 1; qRT-PCR: quantitative real-time polymerase chain reaction; WB: Western blotting; NC: Negative control; si-: siRNA; MMP: Matrix metalloproteinase; FENDRR: FOXF1 adjacent non-coding developmental regulatory RNA; PRC2: Polycomb repressive complex 2; TrxG/MLL: Trithorax group/MLL protein complexes; ACD/MPV: Alveolar capillary dysplasia with misalignment of pulmonary veins; EMT: Epithelial-mesenchymal transition; GAS5: Growth arrest specific 5; PVT1: Pvt1 oncogene; HOTAIR: HOX transcript antisense RNA; CCAT1: Colon cancer associated transcript 1; MALAT1: Metastasis associated lung adenocarcinoma transcript 1.
Acknowledgements
This project was supported by a Grant from the National Natural Science Foundation of China (No.81572630), Shanghai Pujiang Program of Shanghai Science and Technology Commission (NO.13PJD023) and Shanghai Jiaotong University Medical-Engineering Cross Research Fund (NO.YG2012MS49).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: A Meta-Analysis and Review of the Literature. Am J Orthop (Belle Mead NJ). 2015;44:547-53
2. Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65-92
3. Vijayamurugan N, Bakhshi S. Review of management issues in relapsed osteosarcoma. Expert Rev Anticancer Ther. 2014;14:151-61
4. Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16:543-56
5. Malih S, Saidijam M, Malih N. A brief review on long noncoding RNAs: a new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumour Biol. 2016;37:1479-85
6. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-66
7. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097-109
8. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-61
9. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452-63
10. Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7:e2389
11. Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial-Mesenchymal Transition in Osteosarcoma by Regulating the miR-221/ARHI Pathway. J Cell Biochem. 2017 May 18. doi: 10.1002/jcb.26145
12. Song J, Wu X, Liu F, Li M, Sun Y, Wang Y. et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017 S0006-291X:31143-9
13. Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K. et al. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells. 2015;38:432-40
14. Zhu KP, Zhang CL, Shen GQ, Zhu ZS. Long noncoding RNA expression profiles of the doxorubicin-resistant human osteosarcoma cell line MG63/DXR and its parental cell line MG63 as ascertained by microarray analysis. Int J Clin Exp Pathol. 2015;8:8754-73
15. Zhu KP, Ma XL, Zhang CL. LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. Oncotarget. 2017 May 18. doi: 10.18632/oncotarget.17985
16. Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66-75
17. Yang J, Lin J, Liu T, Chen T, Pan S, Huang W. et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung cancer. 2014;85:110-5
18. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A cytoplasmic NF-kB interacting long noncoding RNA blocks IkB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370-81
19. Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122-8
20. Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR. et al. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200-6
21. Zhao J, Cheng L. Long non-coding RNA CCAT1/miR-148a axis promotes osteosarcoma proliferation and migration through regulating PIK3IP1. Acta Biochim Biophys Sin (Shanghai). 2017;49:503-12
22. Liu K, Huang J, Ni J, Song D, Ding M, Wang J. et al. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell cycle. 2017;16:578-87
23. Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7:770-83
24. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-14
25. Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579-85
26. Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O. et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23-33
27. Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L. et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63
28. Miao L, Huang Z, Zengli Z, Li H, Chen Q, Yao C. et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget. 2016;7:68339-49
29. Zhang J, Zhu X, Li H, Li B, Sun L, Xie T. et al. Piperine inhibits proliferation of human osteosarcoma cells via G2/M phase arrest and metastasis by suppressing MMP-2/-9 expression. Int Immunopharmacol. 2015;24:50-8
30. Roomi MW, Kalinovsky T, Rath M, Niedzwiecki A. In vitro modulation of MMP-2 and MMP-9 in pediatric human sarcoma cell lines by cytokines, inducers and inhibitors. Int J Oncol. 2014;44:27-34
31. Wei HJ, Nickoloff JA, Chen WH, Liu HY, Lo WC, Chang YT. et al. FOXF1 mediates mesenchymal stem cell fusion-induced reprogramming of lung cancer cells. Oncotarget. 2014;5:9514-29
32. Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV. Forkhead transcription factor FoxF1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget. 2016;7:1912-26
33. Nilsson G, Kannius-Janson M. Forkhead Box F1 promotes breast cancer cell migration by upregulating lysyl oxidase and suppressing Smad2/3 signaling. BMC cancer. 2016;16:142
34. Fulford L, Milewski D, Ustiyan V, Ravishankar N, Cai Y, Le T. et al. The transcription factor FOXF1 promotes prostate cancer by stimulating the mitogen-activated protein kinase ERK5. Sci Signal. 2016;9:ra48
35. Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B. et al. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor. Oncogene. 2017;36:850-62
36. Tamura M, Sasaki Y, Koyama R, Takeda K, Idogawa M. Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Oncogene. 2014;33:4837-46
37. Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ. et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70:6047-58
38. Zhao ZG, Wang DQ, Hu DF, Li YS, Liu SH. Decreased FOXF1 promotes hepatocellular carcinoma tumorigenesis, invasion, and stemness and is associated with poor clinical outcome. Onco Targets Ther. 2016;9:1743-52
39. Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z. et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927-36
40. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE. et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723-30
Author contact
![]() Corresponding author: Zhang Chun-Lin, MD, Department of Orthopaedic Surgery, Shanghai Tenth People's Hospital Affiliated to Tong ji University, 301, Yan-chang Middle Road, Shanghai 200072, China. E-mail:shzhangchunlin123com. Fax: +86 13761904091.
Corresponding author: Zhang Chun-Lin, MD, Department of Orthopaedic Surgery, Shanghai Tenth People's Hospital Affiliated to Tong ji University, 301, Yan-chang Middle Road, Shanghai 200072, China. E-mail:shzhangchunlin123com. Fax: +86 13761904091.

 Global reach, higher impact
Global reach, higher impact