10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(12):1470-1478. doi:10.7150/ijbs.21312 This issue Cite
Research Paper
CRISPR-offinder: a CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif
1. Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education & Key Laboratory of Swine Genetics and Breeding of Ministry of Agriculture, College of Animal Science and Technology, Huazhong Agricultural University, Wuhan 430070, P. R. China;
2. The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan 430070, P. R. China;
3. The International Peace Maternity & Child Health Hospital of China Welfare Institute (IPMCH), Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P. R. China;
4. Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P. R. China;
5. iGeneTech Bioscience Co., Ltd., Beijing 102206, P. R. China.
* The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint first authors.
Received 2017-6-4; Accepted 2017-8-28; Published 2017-11-1
Abstract
Designing efficient and specific CRISPR single-guide RNAs (sgRNAs) is vital for the successful application of CRISPR technology. Currently, a growing number of new RNA-guided endonucleases with a different protospacer adjacent motif (PAM) have been discovered, suggesting the necessity to develop a versatile tool for designing sgRNA to meet the requirement of different RNA-guided DNA endonucleases. Here, we report the development of a flexible sgRNA design program named “CRISPR-offinder”. Support for user-defined PAM and sgRNA length was provided to increase the targeting range and specificity. Additionally, evaluation of on- and off-target scoring algorithms was integrated into the CRISPR-offinder. The CRISPR-offinder has provided the bench biologist a rapid and efficient tool for identification of high quality target sites, and it is freely available at
Keywords: genome editing, user-defined protospacer adjacent motif, off-target.
Introduction
CRISPR/Cas9 nucleases are widely used for genome editing, but they frequently induce unwanted off-target mutations [1]. The specificity of the CRISPR system is largely determined by how specific the single-guide RNA (sgRNA) targeting sequence is when compared to the rest of the genome for the genomic target. Thus, how to improve the efficiency of CRISPR/Cas genome editing and reduce its off-target effects has been extensively explored in this field.
The CRISPR/Cas system undoubtedly holds great potential for genome editing. Target site cleavage by CRISPR technology requires a protospacer adjacent motif (PAM) immediately downstream or upstream of the protospacer element to which the sgRNA binds. However, Cas9 from different types of bacteria or variants recognizes different PAM sequences [2-7]. Recent studies have revealed the potential of the Cpf1 nuclease to complement and extend the existing CRISPR-Cas9 genome-editing tools [8]. Cpf1, a single RNA-guided endonuclease, lacks tracrRNA (trans-activating crRNA), which utilizes a T-rich PAM on the 5' side of the guide. Another report showed that C2c1, C2c2 and C2c3 systems can also mediate DNA or RNA interference in a 5'-PAM-dependent fashion analogous to Cpf1 [9]. These newly found engineered nucleases have expanded the range of genome editing experiments.
Since the development of CRISPR technology, a number of CRISPR design tools have been created, such as Cas-OFFinder [10], sgRNAcas9 [11], CFD_Scoring [12] and sgRNA.Scorer 2.0 [13]. However, most of these tools can only design sgRNAs for the CRISPR/Cas system. In this study, a user-friendly standalone program named “CRISPR-offinder” was developed to provide researchers a tool for quick design of sgRNAs with minimal off-target effects for different CRISPR systems, especially newly discovered ones, such as Cpf1 and C2c1. Additionally, the effects of sgRNAs/crRNAs (CRISPR-RNAs) with different lengths on CRISPR/Cas9 or Cpf1-mediated gene knockout efficiency were also assessed.
Material and Methods
Plasmid vectors for Cas9 and sgRNA
In order to create a fluorescent reporter system to enrich CRISPR/Cas9-modified cells, a PCR forward primer was synthesized containing a portion of the 5' end of the Cas9-coding and a T2A skipping peptide sequences “GGAAGCGGAGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGACCT”, followed by amplifying the human codon-optimized Cas9 using PCR from pST1374-NLS-flag-linker-Cas9 plasmid (#44758, Addgene), and subcloning Cas9 into pEGFP-C1 vector (Clontech Laboratories, Inc.) using Kpn I and BamH I [14]. A glycine-serine-glycine-serine (GSGS) peptide linker was added at the C terminus after the addition of SV40 nuclear localization signal (NLS) into Cas9, aiming to allow the co-translation of Cas9 and GFP protein from the same expression vector (CMV-EGFP-hspCas9). The sgRNA expression vector was based on the pGL3-U6-sgRNA-PGK-puromycin expression plasmid (#51133, Addgene) containing Bsa I sites for inserting guide sequence into the sgRNA [14]. The sgRNA expression plasmid construction protocol was as follows: A pair of oligodeoxynucleotides (50 μM) was denatured using a thermocycler with the following program: 95 ℃ for 5 min, 65 ℃ for 60 min, hold at 4 ℃. Subsequently, the annealed oligos were ligated with Bsa I-digested pGL3-U6-sgRNA-PGK-puromycin vector, and then the ligation mixture was transformed into E. coli DH5α competent cells (Umibio, China). The correct ligation of sgRNAs was confirmed by Sanger sequencing using the specific primers. Highly pure plasmids of pGL3-U6-sgRNA-PGK-puromycin and CMV-EGFP-hspCas9 were isolated using Endo-Free Plasmid Mini Kit II (OMEGA). All the primer pairs and guide RNA sequences are listed in Supplemental Table S1.
Plasmids encoding Cpf1 and crRNAs
Plasmids encoding AsCpf1 (Acidaminococcus sp. Cpf1) and LbCpf1 (Lachnospiraceae bacterium Cpf1) proteins were human codon optimized. crRNA expression plasmid was U6 promoter-based and modified from the sgRNA expression plasmid described previously [15]. Briefly, the repeat region of the sgRNA was replaced with crRNA repeat sequences (AATTTCTACTCTTGTAGAT). crRNAs were inserted into crRNA expression plasmids which had been digested separately by enzyme Bsa I (NEB). crRNAs sequences are listed in Supplemental Table S1.
Cell culture and transfection conditions
HEK293T cell lines were maintained in DMEM supplemented with 10 % fetal bovine serum (HyClone) and 1 % penicillin/streptomycin (Life Technologies). All cell lines were maintained at 37 ℃ and 5 % CO2. At 70-80 % confluence, HEK293T cells were co-transfected with Cas9 and sgRNA plasmids (at ratio 1:1) in 6-well plates using NB polymer transfection Reagent (Umibio, UR51001) according to the manufacture's recommended protocol, including a single well of cells as the negative control (which can be non-relevant plasmid DNA). For Cpf1-mediated genome editing, HEK293T cells were seeded into 6-well plates at 70-80 % confluence, followed by transfection with Cpf1 expression plasmids (2 μg) and sgRNA plasmids (1 μg) also using NB polymer transfection Reagent. Blasticidin (10 µg/ml, Sigma, #15205) was added 24 h after transfection. Cells were collected 72 h after transfection and genomic DNA was isolated with the EasyPure Genomic DNA Kit (TransGen Biotech).
T7 endonuclease I mutation detection assay and Sanger sequencing
CRISPR/Cas9-induced lesions at the endogenous target site were quantified using the T7 endonuclease I (T7EN I) mutation detection assay to investigate the insertions/deletions (indels) mutation characteristics of nuclease-mediated non-homologous end joining (NHEJ). After transfection, cells were incubated for 48 h at 37 ℃ and genomic DNA was extracted using the TIANamp Genomic DNA Kit (Tiangen Biotech). The target locus was amplified by 32 cycles of PCR with the TaKaRa LA Taq kit (TaKaRa) using primers specific to each locus. Purified PCR product was denatured and reannealed using a thermocycler. Hybridized PCR products were digested with T7EN I (NEB, M0302L) for 15 min and separated by 2 % agarose gel. The gels were stained with Gel-Red and quantified by densitometry using the ImageLab software suite (Bio-Rad). The PCR products were cloned into the pMD18-T vector and analyzed by Sanger sequencing to verify the indels mutation. All primers are listed in Supplemental Table S1.
Fluorescence-activated cell sorting
HEK293T cell lines were each transfected with 1.5 μg of CMV-EGFP-hspCas9 and sgRNA expression vectors and incubated at 37 ℃ and 5 % CO2. At 24 h after transfection, cells were trypsinized with 0.25 % trypsin (Umibio, UR50301) and collected for fluorescence-activated cell sorting (FACS) using a FACSvantage II sorting machine (BD Biosciences, US). Viable cells were gated on size and shape using forward and side scatter. The GFP expression was measured using a 488 nm laser for excitation. GFP-positive cells were collected and expanded for analysis.
Results
Basics of CRISPR-offinder design
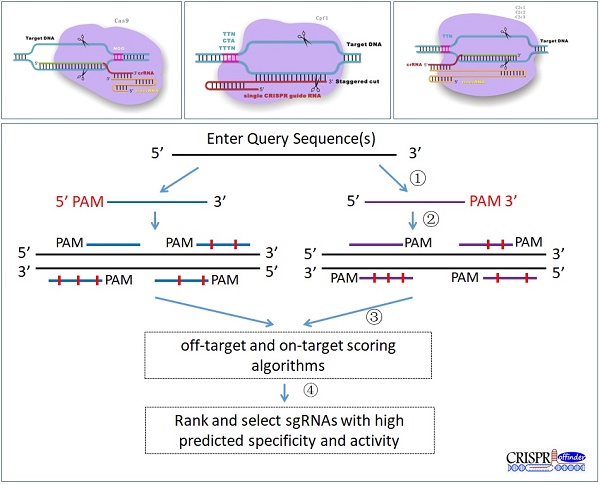
To fulfill the need of designing sgRNAs for different RNA-guided DNA endonucleases (Figure 1A, B and C), such as Cas9, Cpf1 and C2c1, CRISPR-offinder software which can design sgRNAs and evaluate off-target effects for user-defined protospacer adjacent motif (PAM) was developed. The workflow of CRISPR-offinder is shown in Figure 1D. Given a DNA query sequence, the pipeline of CRISPR-offinder program first fetches all the possible regions in the length range of 15 to 25 nts (nucleotides), with PAM either on the 5' or 3' side. These regions are then evaluated and filtered to show low/no off-target activity across the whole genome. The PAM sequences and the direction of the CRISPR target sites can be freely selected by the user. For easy use, the PAM sequences collected from different CRISPR systems have already been set as optional parameters (Table 1). PAMs show variations in size and nucleotide composition in different bacterial strains from which Cas9, Cpf1, C2c1, C2c2 and C2c3 enzymes are isolated. As presented in Table 1, all Cas9 nucleases require at least one “G” in their PAMs and must be on the 3' side, while Cpf1, C2c1, C2c2 and C2c3 nucleases require “T” in their PAMs and must be on the 5' side (Figure 1). Thus, the degeneracy, base composition and 5' or 3' side in PAM recognition by different RNA-guided endonucleases are taken into account when designing sgRNA and searching for potential off-target sites. In addition, the length, GC content and sequence features are also set as optional parameters when designing highly-efficient CRISPR sgRNAs. More importantly, the sgRNA activity and off-target sites are predicted by using sgRNA Scorer 2.0 [12], CFD_Scoring [13] and Cas-OFFinder [10] version 2.4. The output from CRISPR-offinder is parsed and only alignments including a proper PAM are listed.
CRISPR-offinder program
There are four application frameworks for the CRISPR-offinder tool: (i) length of protospacer: the length of the sgRNA target site (protospacer) ranges from 15 to 25 nt; (ii) PAM requirement: the requirements consist of sequence, orientation and location. CRISPR-offinder provides two PAM options: default PAM and user-defined PAM. With these two options, the user can define PAM for a different CRISPR system through setting the parameter using the degeneracy base, for example, R: A or G, Y: C or T, S: G or C, W: A or T, K: G or T, M: A or C, etc., and the orientation or chromosome location of PAM, such as 5' or 3' side, plus or minus DNA strand. (iii) Input parameters: CRISPR-offinder provides two options for inputting gene or genome sequence. Given an input FASTA file of the target sites to query the reference genome as well as a CRISPR system with a defined spacer length, PAM sequence and orientation, this standalone tool will identify the putative sites and assign an activity value for each sgRNA based on the support vector machine model, which will be conducted by sgRNA Scorer 2.0; (iv) off-target searching parameter: The number of nucleotide mismatches between on- and off-target is up to 9. In addition, sgRNAs with minimal off-target activity will be predicted based on the Cas-OFFinder and Off-Target Cutting Frequency Determination (CFD) program.
The off-line standalone software is developed to enable its users to edit any genome, assembled or un-assembled, with up to 9 nucleotide mismatches allowed in the standalone version for off-target evaluation, which is also suitable for designing the sgRNA library of any species. After running the CRISPR-offinder program, the information will be stored in a new file, such as “sgRNA ID”, “start”, “end”, “target sequences”, “position”, “length”, “GC%”, “sgRNA activity score”, etc. In addition, each sgRNA contains detailed information of off-target sites, which can be viewed in the table column, such as “InputSeq”, “chr (chromosome)”, “Start”, “OfftargetSeq”, “Strand”, “Mismatch”, and “specificity score”. The output results of sgRNAs can be ranked by the total number of off-targets, sgRNA activity or specificity CFD scores, enabling the user to select easily the sgRNAs with maximal on-target activity and minimal off-target effect.
Evaluation of cleavage efficiency and analysis of CRISPR/Cas9 off-target effects
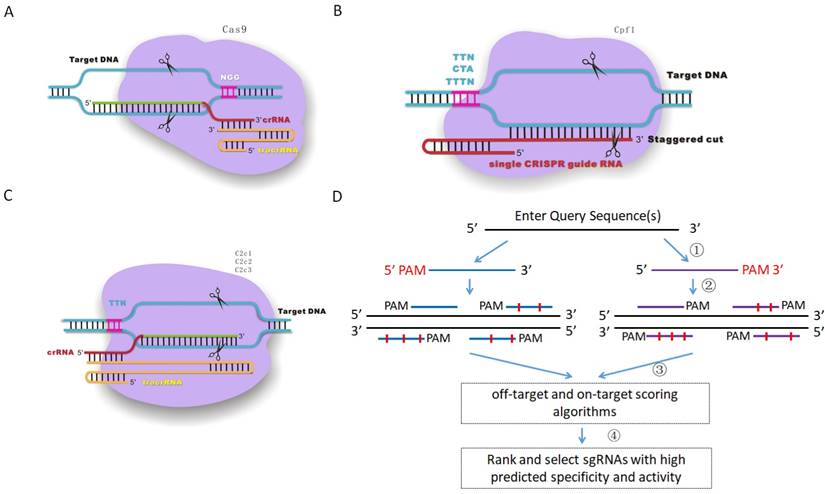
To test the functionality of CRISPR-offinder in CRISPR/Cas9-based genome editing, an androgen receptor (AR) gene was randomly selected, and four sgRNAs were designed to target the same locus of exon 1 of the AR gene, with the length of each sgRNA ranging from 17 to 20 nt. The result of CRISPR-offinder is shown in Figure 2A. The GC% content of four sgRNAs was up to 70 %. The off-target effect of each sgRNA was evaluated as previously described [16]. CRISPR/Cas9 could tolerate 1-5 nucleotide mismatches in the targets, and thus up to 5 nucleotide mismatches between the sgRNA and the off-target sites were calculated using the CRISPR-offinder. As shown in Figure 2A, the total number of predicted off-targets for over four sgRNAs ranged from 1,654 to 22,370. The result indicated that the total number of off-target sites was dramatically increased when the sgRNA was truncated, which was termed tru-sgRNA (truncated single guide RNA). When using the CMV-EGFP-hspCas9 vector system, the co-expression of Cas9 and GFP from the same mRNA created the possibility of enriching cell populations for desired genome editing outcomes via fluorescence activated cell sorting (FACS). Subsequently, sorted or unsorted cell populations were detected by T7EN I cleavage assay and DNA sequencing (Figure 2B and C). When comparing the control group to the sgRNA and Cas9 treated groups, CRISPR/Cas9-mediated indels mutation at the AR locus in HEK293T cell lines can be detected, and T7EN I-based PCR and sequencing analysis demonstrated that 55.5 to 92 % of the GFP-sorting cells were mutant cells (Figure 2D).
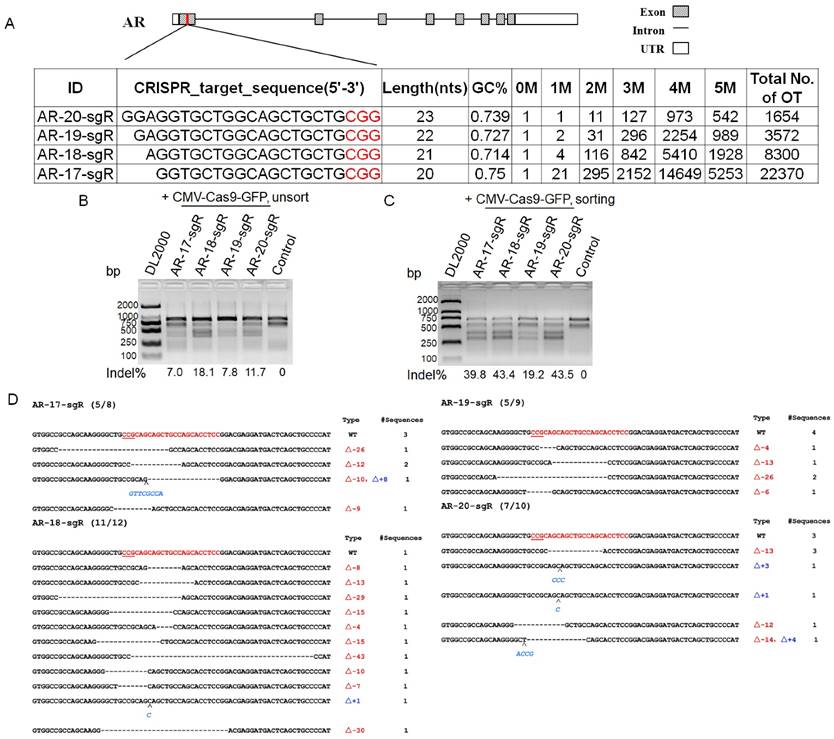
The off-target activity of sgRNAs is considered as an important indicator to study gene function. To check the specificity of each sgRNA, 19 off-target sites with 12 bp seed sequence identity to target sites were selected. In this study, TIDE (Tracking of Indels by DEcomposition), a simple assay to determine the spectrum and frequency of targeted mutations generated in a pool of cells by CRISPR/Cas9, was conducted using the on-target sites of the sgRNAs as the positive control [17], and the cleavage activity of 17-, 18-, 19-, and 20-nt sgRNAs ranged from 12.2 to 23.3 % in unsorted cells (Supplemental Figure S1). The cleavage efficiency of 19 off-target sites from different sgRNA treated groups ranged from 0.1 to 28.3 % (Figure 3A). Three potential off-targets (POTs), namely POT4, POT7 and POT12 were selected for further validation. As shown in Figure 3B, DNA cleavage of POT7 was detected in four groups by T7EN I cleavage assay, and only the activity of sgRNA with the length of 17-nt was not detected, while DNA cleavage of POT12 was only detected in the 20-nt sgRNA treated group. A comparison between the T7EN I cleavage assay and the TIDE assay results suggested that the lower bound of the TIDE method was 6% (Figure 3). Thus, there were 2, 3, 2 and 4 in the 19 off-target sites with the DNA cleavage efficiency of more than 6 % in the 17-, 18-, 19-, and 20-nt sgRNA treated groups, respectively (Figure 3A). In this study, the off-target cleavage of POT10 and POT15 was observed in the 17-nt sgRNA treated group with 3-4 rather than 1-2 nucleotide mismatches, whereas off-target cleavage of POT4 in the 20-nt sgRNA treated group with up to 3 nucleotide mismatches could induce off-target mutations (Figure 3).
RNA-guided endonucleases and protospacer adjacent motif sequences.
| RNA-guided Endonucleases (Species/variant) | PAM sequence(5'→3') | Direction(3'/5' side) |
|---|---|---|
| SpCas9 from Streptococcus pyogenes | NGG | 3' |
| SpCas9 from Streptococcus pyogenes | NRG | 3' |
| StCas9 from Streptococcus thermophilus | NNAGAAW | 3' |
| NmCas9 from Neisseria meningitidis | NNNNGATT | 3' |
| SaCas9 from Staphylococcus aureus | NNGRRT | 3' |
| SaCas9 variant (KKH SaCas9) | NNNRRT | 3' |
| SpCas9 D1135E variant | NGG (reduced NAG binding) | 3' |
| SpCas9 VRER variant | NGCG | 3' |
| SpCas9 EQR variant | NGAG | 3' |
| SpCas9 VQR variant | NGAN or NGNG | 3' |
| AsCpf1 from Acidaminococcus, LbCpf1 from Lachnospiraceae | TTTN | 5' |
| FnCpf1 from Francisella novicida strain U112 | TTN and/or CTA | 5' |
| C2c1 from four major taxa: Bacilli, Verrucomicrobia, a-proteobacteria, and d-proteobacteria | TTN | 5' |
Notes: N = A, T, C or G; R = A or G; W = A or T.
Schematic diagram of the versatile CRISPR genome editing tool and CRISPR-offinder program workflow. A. CRISPR/Cas9-based genome editing; B. CRISPR/Cpf1-based genome editing; C. CRISPR/C2c1-based genome editing; D. Workflow of CRISPR-offinder: (1) Design of CRISPR sgRNAs with user-defined PAMs; (2) Prediction of potential off-target sites; (3) Identification of putative sites and assignment of a predicted activity based on the support vector machine model, which will be conducted by sgRNA Scorer 2.0. In addition, sgRNAs with minimal off-target activity will be determined by Cas-OFFinder and Off-Target Cutting Frequency Determination (CFD); (4) Screening of the specific sgRNAs with minimal off-target sites. CRISPR stands for clustered regularly interspaced short palindromic repeat; PAM, protospacer adjacent motif; N, any of base; tracrRNA, trans-activating CRISPR RNA.
Evaluation of cleavage efficiency and analysis of CRISPR/Cpf1 off-target effects
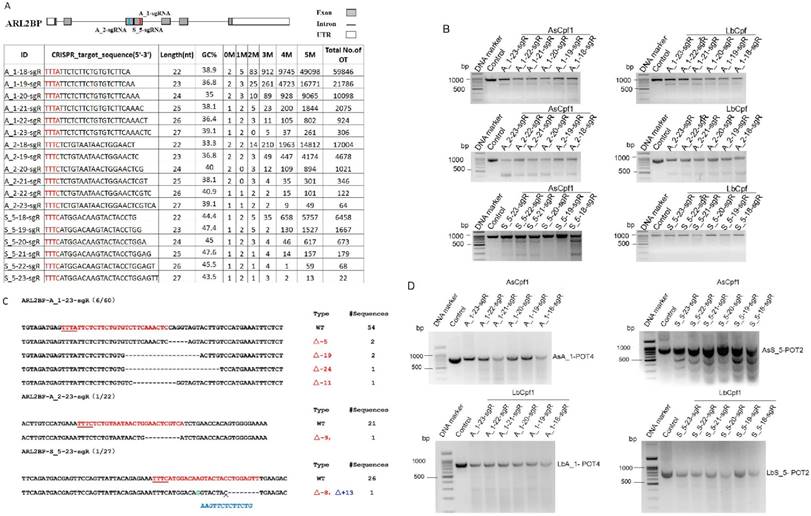
To further test the functionality of CRISPR-offinder in CRISPR/Cpf1-based genome editing, ADP ribosylation factor like GTPase 2 binding protein (ARL2BP) gene was randomly selected and 18 crRNAs were designed to target the exon 3 of the ARL2BP gene, with the length of each crRNA ranging from 18 to 23 nt. The result of CRISPR-offinder is shown in Figure 4A. The GC% content of 18 crRNAs ranged from 30 to 50 %. Up to 5 nucleotide mismatches between the crRNAs and the off-target sites were estimated, with the total number of predicted off-target sites of designed sgRNAs ranging from 22 to 59,846 (Figure 4A). The result indicated that the total number of off-target sites was also dramatically increased if crRNAs were truncated using the CRISPR/Cpf1 system. The efficiency of Cpf1-mediated genome editing in HEK293T cells was determined by co-transfection with either AsCpf1 or LbCpf1 expression plasmid. When compared with the control group, the CRISPR/Cpf1-mediated indels mutation at the ARL2BP locus could be detected by the T7EN I cleavage assay (Figure 4B and C). The off-target activity of CRISPR/Cpf1-based genome editing was also evaluated using the T7EN I cleavage assay, and 49 sites were validated using AsCpf1 or LbCpf1 (Supplemental Table S2 and Figure 4D). The results indicated that CRISPR-offinder is suitable for designing and evaluating the off-target effects of crRNAs for Cpf1.
Discussion
The CRISPR system has been adopted as an efficient genome editing tool in large animals such as pig, dog, goat, sheep and chicken [18-22]. The most important part is the selection of highly efficient sgRNAs for CRISPR/Cas9-based genome editing. However, off-target effects of the Cas nuclease activity are a recurrent concern for the CRISPR system. Thus, it is important to select sgRNAs with maximal activity and minimal off-target effects for the system. One feature of type II CRISPR-Cas systems is the requirement of a nearby PAM on the target sequence (Figure 1), and this sequence varies between different Cas9 orthologs, Cpf1 and C2c1 (Table 1). PAM functional requirements have been defined for Cas9, Cpf1 and C2c1 proteins validated for mammalian genome editing, and the PAM requirement adds a second layer of specificity for gene targeting, beyond that afforded by spacer/protospacer complementarity [23]. By developing genome-editing systems using a range of Cas9, Cpf1 and C2c1 proteins with distinct PAM requirements, the genomic regions that can be targeted by CRISPR editing will expand significantly. With the fast development of the CRISPR field, different types of CRISPR systems may still exist and need to be explored. Here, CRISPR-offinder was developed with no limitation of specific PAM types.
Targeting androgen receptor (AR) gene by CRISPR/Cas9-based genome editing. A. Scheme of sgRNAs designed to target exon 1 of the AR gene using CRISPR-offinder. B & C. Activity of 17-, 18-, 19-, or 20-nt sgRNAs targeted to the same genomic loci by cell sorting or unsorting after co-transfection with CMV-EGFP-hspCas9 plasmid. D. Indels mutation created by CRISPR/Cas9 and detected by DNA sequencing. AR, androgen receptor; M, the number of nucleotide mismatches (1M, 2M, 3M, 4M or 5M); 0M, perfect match to the on-target site, and if the number of 0M sites >1, maybe off-target sites are contained; bp, base pair; WT, wild-type; OT, off-target; GFP, green fluorescent protein; CMV (Cytomegalovirus) promoter, a constitutive mammalian promoter; Control, cells for the negative control.
Supplemental Table S3 shows the comparison results between CRISPR-offinder and other currently available tools. It can be seen that most of the tools are only suitable for the CRISPR/Cas9 system, such as sgRNAcas9, a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites [24]. In comparison with other CRISPR tools, CRISPR-offinder has the major advantage of flexibility in parameter setting for user-defined PAMs. To increase the targeting range and specificity, CRISPR-offinder provides the support for custom length sgRNAs, which is similar in function to that of CHOPCHOP v2 [25]. Moreover, the effect of sgRNA length on CRISPR/Cas9 or Cpf1-mediated gene knockout efficiency and specificity was further evaluated in this study. We found that the cleavage activity of 17-, 18-, 19- and 20-nt sgRNAs in the CRISPR/Cas9 system was increased (Figure S2 and Figure 3). As only one target site was tested, whether the activity of the tru-sgRNA is lower than that of the 20-nt full length sgRNA needs to be further validated. In addition, we found that, when using CRISPR-offinder, a larger total number of off-target sites can be predicted by the tru-sgRNA than the 20-nt sgRNA, and some predicted off-target sites in the 17-nt tru-sgRNA can be detected (Figure 2 and 4). These results suggest that the specificity of the tru-sgRNA may be equivalent to or lower than that of the 20-nt sgRNA, and the specific feature of truncated sgRNA needs to be reconsidered [26]. As only 19 off-target sites were selected based on the computer prediction method, the comparison result may be unilateral. Thus, an unbiased genome-wide method needs to be used to evaluate the specificity of sgRNAs with different lengths. In addition, we have found that the 12 bp seed sequence model is not completely predictive of SpCas9 specificity according to T7EN I cleavage assay in Figure 3 or our previous study [11], which means this new CRISPR design tool does not consider seed sequence region for evaluation of off-target effects. High-throughput screening of a CRISPR/Cas library for functional genomics is a very powerful approach [27]. To this end, CRISPR-offinder, an off-line version of software, has been developed for designing genome-wide libraries of different species.
Cpf1-mediated gene targeting can be used in generating knockout mice [28-29]. Using GUIDE-seq (genome-wide, unbiased identification of DSBs enabled by sequencing) and targeted deep sequencing analysis with both Cpf1 nucleases, Kleinstiver et al. failed to detect off-target cleavage for more than half of the 20 different crRNAs [30]. In this study, we evaluated the cleavage efficiency and CRISPR/Cpf1 off-target effects, and found that the activity of the two members of the Cpf1 family, the AsCpf1 from Acidaminococcus sp. and the LbCpf1 from Lachnospiraceae bacterium, caused lower indels frequencies than SpCas9 (Figure 2 and Figure 4).
Validation of Cas9 off-target binding sites by the TIDE and T7EN I cleavage assay. A. Evaluation of on/off target site cleavage efficiency using the TIDE method. B. Validation of the three Cas9 off-target binding sites by the T7EN I cleavage assay. POT, potential off-target sites; GFP, green fluorescent protein; CMV (Cytomegalovirus) promoter, a constitutive mammalian promoter; AR, androgen receptor; Control, cells for the negative control; bp, base pair; TIDE (Tracking of Indels by DEcomposition), a web tool for rapid assessment of genome editing by CRISPR-Cas9 of a target locus determined by a guide RNA (sgRNA), https://tide.nki.nl/.
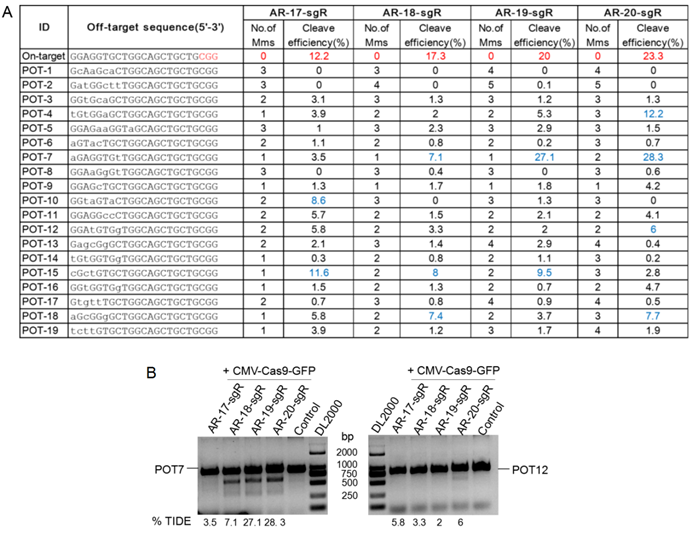
Targeting ADP ribosylation factor like GTPase 2 binding protein (ARL2BP) gene by CRISPR/Cpf1-based genome editing. A. Scheme of sgRNAs designed to target exon 3 of the ARL2BP gene using CRISPR-offinder. B. Activity of 18-, 19-, 20-, 21-, 22- or 23-nt sgRNA targeted to the different genomic loci after co-transfection with two members of the Cpf1 family, the AsCpf1 from Acidaminococcus sp. and the LbCpf1 from Lachnospiraceae bacterium. C. Indels mutation created by CRISPR/Cas9 and detected by DNA sequencing. D. Detection of the two Cpf1 off-target binding sites by the T7EN I cleavage assay. ARL2BP, ADP ribosylation factor like GTPase 2 binding protein; M, the number of nucleotide mismatches (1M, 2M, 3M, 4M or 5M); 0M, perfect match to the on-target site, and if the number of 0M sites >1, maybe off-target sites are contained; bp, base pair; OT, off-target; POT, potential off-target sites; Control, cells for the negative control.
The off-target sites can also be predicted and validated using the CRISPR-offinder. Using CRISPR-offinder, we found that the total number of predicted off-target sites was dramatically increased if sgRNAs or crRNAs were truncated. A decrease of 10-20 percentage points was observed in knock-out (KO) efficiency with 17-nt sgRNAs compared to full-length sgRNAs in HEK293T cell lines. Off-target cleavage was observed in 17-nt sgRNAs for Cas9 with 3 rather than 1-2 nucleotide mismatches, whereas 18-nt crRNAs for Cpf1 with up to 3 nucleotide mismatches could still induce off-target mutations. These results indicate the importance of balancing on-target gene cleavage potency with off-target effects: when off-target is a major concern, using bioinformatics tools to evaluate off-target effects should be the first consideration.
Recognizing and avoiding off-target effects is an important step in application of the CRISPR system, while the rules governing off-target effects are still in their infancy, especially Cpf1 and C2c1. Thus, the prediction accuracy of off-targets in the CRISPR-offinder program needs to be improved. Naturally, whether the reported genomic loci will correspond to bona fide off-targets depends on other criteria that can include the chosen span of the seed, the extent of similarity between the off-target and the sgRNA in the region immediately beyond the seed, chromatin accessibility information, methylation status, and other considerations [31]. It is worth noting that attributes such as chromatin accessibility and methylation status do not exist in all cell types, and such attributes may differ across cell types. Nonetheless, when available, this information can be easily taken into account simply by post-processing the output generated by CRISPR-offinder.
Supplementary Material
Supplementary Table S1.
Supplementary Table S2.
Supplementary Table S3.
Supplementary Figures.
Acknowledgements
The authors are grateful to Prof. Xingxu Huang for providing insightful and constructive comments on an earlier draft. We thank Hanchang Zhu for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the National High Technology Research and Development Program of China [863 Program, 2013AA102502], the National Transgenic Project of China [2016ZX08006003-004], the National Key Research and Development Program of China, Stem Cell and Translational Research [2016YFA0100203].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fu Y, Foden JA, Khayter C. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822-6
2. Hsu PD, Scott DA, Weinstein JA. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827-32
3. Hirano H, Gootenberg JS, Horii T. et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950-61
4. Kleinstiver BP, Prew MS, Tsai SQ. et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293-8
5. Kleinstiver BP, Prew MS, Tsai SQ. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481-5
6. Muller M, Lee CM, Gasiunas G. et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636-44
7. Ran FA, Cong L, Yan WX. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186-91
8. Zetsche B, Gootenberg JS, Abudayyeh OO. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759-71
9. Shmakov S, Abudayyeh OO, Makarova KS. et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385-97
10. Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473-1475
11. Changzhi Z, Yi Z, Guanglei L. et al. Development of a graphical user interface for sgRNAcas9 and its application. Yi Chuan. 2015;37:1061-72
12. Chari R, Yeo NC, Chavez A. et al. sgRNA Scorer 2.0: A Species-Independent Model to Predict CRISPR/Cas9 Activity. ACS Synth Biol. 2017;6:902-904
13. Doench JG, Fusi N, Sullender M. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184-191
14. Shen B, Zhang J, Wu H. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720-3
15. Cho SW, Kim S, Kim JM. et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230-2
16. Lin Y, Cradick TJ, Brown MT. et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473-7485
17. Brinkman EK, Chen T, Amendola M. et al. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168
18. Hai T, Teng F, Guo R. et al. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372-5
19. Zou Q, Wang X, Liu Y. et al. Generation of gene-target dogs using CRISPR/Cas9 system. Journal of Molecular Cell Biology. 2015;7:580-583
20. Ni W, Qiao J, Hu S. et al. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS One. 2014;9:e106718
21. Veitia RA, Crispo M, Mulet AP. et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. Plos One. 2015;10:e0136690
22. Veron N, Qu Z, Kipen PA. et al. CRISPR mediated somatic cell genome engineering in the chicken. Dev Biol. 2015;407:68-74
23. Hou Z, Zhang Y, Propson NE. et al. Efficient genome engineering in human pluripotent stem cellsusing Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644-9
24. Xie S, Shen B, Zhang C. et al. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448
25. Labun K, Montague TG, Gagnon JA. et al. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272-6
26. Fu Y, Sander JD, Reyon D. et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNA. Nat Biotechnol. 2014;32:279-284
27. Zhou Y, Zhu S, Cai C. et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487-91
28. Kim Y, Cheong SA, Lee JG. et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016;34:808-10
29. Hur JK, Kim K, Been KW. et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016;34:807-8
30. Kleinstiver BP, Tsai SQ, Prew MS. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869-74
31. Pliatsika V, Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biology Direct. 2015;10:4
Author contact
![]() Corresponding authors: Fei Sun (Email: sunfeiedu.cn), Shengsong Xie (Email: ssxiehzau.edu.cn) and Shuhong Zhao (Email: shzhaohzau.edu.cn).
Corresponding authors: Fei Sun (Email: sunfeiedu.cn), Shengsong Xie (Email: ssxiehzau.edu.cn) and Shuhong Zhao (Email: shzhaohzau.edu.cn).

 Global reach, higher impact
Global reach, higher impact