10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(12):1489-1496. doi:10.7150/ijbs.21637 This issue Cite
Review
The Molecular Mechanisms of Regulation on USP2's Alternative Splicing and the Significance of Its Products
Department of Oncology, Shanghai 9th People's Hospital, Shanghai Jiao Tong University School of Medicine, 639 Zhi Zao Ju Rd, Shanghai, China.
Received 2017-6-20; Accepted 2017-9-10; Published 2017-11-2
Abstract
Ubiquitin-specific protease 2 (USP2) has a regulatory function in cell growth or death and is involved in the pathogenesis of various diseases. USP2 gene can generate 7 splicing variants through alternative splicing, and 5 variants respectively as USP2-201, USP2-202, USP2-204, USP2-205, USP2-206 can encode proteins. The influence of circadian rhythm, nutrition and androgen on specific signaling molecules or cytokines can regulate the alternative splicing of USP2. Specifically, PKC activator, IL-1β, TNF-α, PDGF-BB, TGF-β1 are all regulatory factors for USP2's alternative splicing. USP2-201 plays a crucial role in cell cycle progression, and is also of great significance in EGFR recycling. USP2-202 can activate apoptosis signaling pathway to participate in cell apoptosis, and USP2-204 can induce cell anti-virus reaction to decrease. In general, we collect and summarize the factors involved in the alternative splicing of USP2 in this review to further understand the mechanism behind the USP2's alternative splicing.
Keywords: USP2, alternative splicing, ubiquitin, deubiquitination, regulation.
Introduction
Alternative splicing which is an important step between transcription and translation in multicellular eukaryotes affecting chromatin modification and signal transduction can be regulated by many transcriptional factors [1]. Alternative splicing allows a single gene to produce two or more similar but distinct mature mRNA variants that expand the coding capabilities of eukaryotic genomes [2-4]. Abnormal alternative splicing of some disease-related genes is also associated with pathogenesis and therapeutics, such as metabolic alterations and therapeutic tools in diabetes mellitus [5].Through alternative splicing, USP2 gene also can produce different splicing variants which have their specific roles [6, 7].
It is speculated that the different abundance of USP2 isoforms in different tissues is partly caused by splicing regulatory factors [8, 9]. These splicing regulatory factors as trans-acting factors directly or indirectly recognize and bind with cis-acting elements on the pre-mRNA in the spliceosome to play a role [10]. Currently, the cis-acting elements are usually divided into ESEs, ESSs, ISEs and ISSs according to their locations and functions.
It is of great significance to understand the regulation mechanism of the USP2's alternative splicing for the further understanding of USP2 [11-13]. Through recent studies, the alternative splicing products of USP2 have been found from primarily two splicing variants USP2-201 (USP2-69, USP-2a) and USP2-204 (USP2-45, USP-2b) to the presently seven variants [14, 15]. Therefore, it is necessary to review the molecular mechanisms involved in the regulation of USP2's alternative splicing and the significance of its products [16-18].
The alternative splicing products of USP2 gene and their distribution
The human USP2 gene containing 17 exons is located in 11q23.3 [19]. The mouse derived USP2 gene with 21 exons is localized to 9A5.1, while the location of Rat USP2 gene containing 17 exons is 8q22. USP2 gene can generate 7 splicing variants through alternative splicing, of which 5 variants can code protein. USP2-201 (USP2-a, USP2-69) mRNA has 3732 bases encoding the longest protein subtype with 605 amino acids [20]. USP2-202 (USP2-c) mRNA is composed of 2903 bases and encodes the protein with 362 amino acids. USP2-204 (USP2-b, USP2-45) mRNA including 1704 bases can encode the protein with 396 amino acids. USP2-205 and USP2-206 mRNA with 563 and 677 bases respectively were less learned than the previous three subtypes, encoding the protein composed of 137 and 167 amino acids. The USP2-203 and USP2-207 mRNA with 670 and 637 bases respectively only have introns without the ability of protein encoding [5]. These protein subtypes differ in the length and the amino acid composition of the N terminal of the protein. While all the protein subtypes have the same structure in C terminal, which contains the Cys and His residues required for the catalytic activity [21, 22]. USP2-201, USP2-202, USP2-204 contains 13, 12, 12 exons respectively. They have the same exon composition in the 3' terminal while differ in the 5' terminal (Figure 1) [23].
The mRNA expression of USP2-201 and USP2-204 in different tissues of the rat was detected by using the mRNA probes designed according to the specific region of the 5' terminal of the two isoforms [22]. USP2-204 mRNA was discovered in skeletal muscle, lung, heart, brain, liver, testis except ovary. However, USP2-201 mRNA was detected only in skeletal muscle, heart and testis. The mRNA abundance of the two subtypes was both highest in the testis. Florian Haimerl detected the expression of USP2 mRNA in mouse liver through RT-PCR and found that the proportion of the USP2-202 mRNA was up to 89%, while USP2-201 and USP2-204 mRNA occupy a litter percentage--4%、7% respectively. USP2 was also involved in the cutaneous T-cell lymphomas, the mRNA level of USP2 isoforms expressed in tumor mycosis fungoides and plaque mycosis fungoides was mearsured using RT-PCR. USP2-201 mRNA expressed in tumor mycosis fungoides was 2.3 fold lower than that in plaque mycosis fungoides, while the expression of USP2-204 and USP2-202 mRNA was not detected [24].
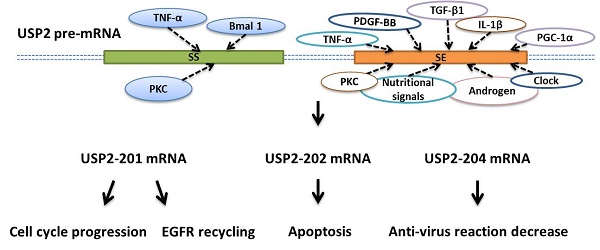
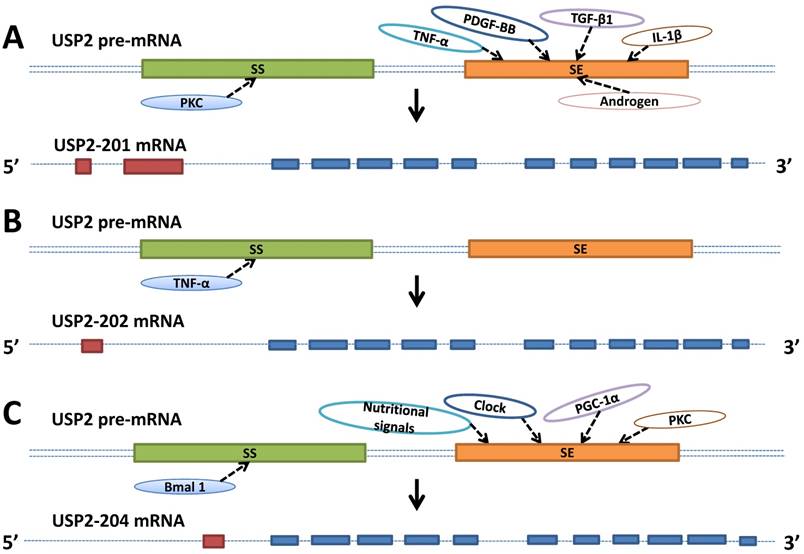
Schematic represents the factors involved in the USP2's alternative splicing. A. TNF-α, PDGF-BB, TGF-β1, IL-1β, androgen and PKC promote or inhibit USP2-201's alternative splicing by directly or indirectly influencing splicing enhancer (se, orange box) or splicing silencer (ss, green box) which are both located on the pre-mRNA (double dashed line). B. TNF-α inhibits USP2-202's alternative splicing by directly or indirectly influencing splicing silencer (ss, green box) which is located on the pre-mRNA (double dashed line). C. Nutritional signals, Clock, PGC-1α, PKC and Bmal 1 promote or inhibit USP2-204's alternative splicing by directly or indirectly influencing splicing enhancer (se, orange box) or splicing silencer (ss, green box) which are both located on the pre-mRNA (double dotted line). USP2-201, USP2-202, USP2-204 contains 13, 12, 12 exons respectively, of which 11 exons are identical (blue boxes), others are unique to each mRNA (red boxes).
To further confirm the USP2 expression at the protein level in different tissues of mouse, Florian Haimerl found that USP2-202 protein existed in mouse heart, brain, liver and kidney while USP2-204 protein was mainly expressed in kidney by using Western blot [25]. Immunofluorescence analysis of HELA cells transfected with the plasmids of USP2-201 or USP2-202 showed that USP2-201 and USP2-202 proteins were both dispersed around nucleus and adjacent to the endoplasmic reticulum while didn't coincide with mitochondria and Golgi. The similar sub-cellular location of USP2-201 and USP2-202 protein was also observed in MCF7 cells transfected with the plasmids of them [26]. Further immunofluorescence localization of GFP-USP2-201 fusion protein in HELA cells proved that GFP-USP2-201 protein localized in early endosomes involved in EGFR degradation cycle [27]. In normal fibroblasts, USP2-201, USP2-202, USP2-204 and USP2-205 protein were located in peroxisomes, cytoplasm and nucleus. The C terminals of the four subtypes above all contain a weak sequence to target the PTS1 which can interact with the corresponding peroxisome receptor--PEX5 to input USP2 protein subtypes into peroxisome matrix after translation [21].
Factors involved in the alternative splicing of USP2 gene
Circadian rhythm
In the study of the effect of USP2 gene on the maintenance of normal sodium balance and blood pressure in mouse, it was found that the expression of USP2-204 mRNA and protein in renal tissues of mice was rhythmical [28, 29]. The renal tissue lysate of mice was taken at 4, 8, 12, 16, 20, 24 o'clock of the day to detect the expression of USP2-204 protein by specific antibodies. The experiment showed that USP2-204 protein in renal tissue increased continuously in the light stage and reached the highest value at 12 o'clock which was the junction between the light and dark phases. Subsequently, the expression of USP2-204 protein decreased gradually from 12-24 o'clock which was the dark stage. Similar alterations of USP2-204 protein were also observed in the study of Daniel Pouly [30]. The expression curve of USP2-204 mRNA in renal tissue of mouse was also single peak and the highest value appeared at 8-12 o'clock. It is suggested that the alternative splicing of USP2-204 in mouse kidney may be in the pattern of circadian rhythm.
Matthew M also proved that biological clock can regulate the expression of USP2 in gluconeogenesis [31, 32]. Specific primers depending on USP2-201, USP2-204 mRNA were designed to detect the expression of two kinds of mRNA in mice liver tissue at the different time points of the day, and the results indicated that circadian rhythm could regulate USP2-204 mRNA expression but didn't have any impact on USP2-201 mRNA. The abundance of USP2-204 mRNA increased gradually in the light phase, and reached the highest value at the junction point between the light and dark phase. After entering the dark phase, the expression of USP2-204 mRNA decreased gradually. Bmal1, the key molecule involved in the biological clock in the mouse liver, was also associated with circadian rhythm: the mRNA level of Bmal1 decreased gradually in the light stage and increased entering the dark phase. Obviously, circadian rhythm regulated Bmal1 and USP2-204 in the opposite direction. The correlation between Bmal1 and USP2-204 was further validated in an experimental mouse model: the expression of USP2-204 mRNA with circadian rhythm was almost eliminated when mice liver's Bmal1 gene was knocked out. It is proved that circadian rhythm can participate in the alternative splicing of USP2-204 in mouse liver and USP2-204 is the downstream target of Bmal1 during the process (Figure 1C).
Nutrition
Matthew M further studied the influence of continuous feeding, fasting and refeeding on the expression of 47 kinds of USPs in mouse liver, and confirmed that USP2 was the only type that can be regulated among all USPs [31]. Starvation can significantly increase the mRNA level of USP2 in mouse liver, while refeeding can reverse the change. In order to determine whether the regulation of starvation on USP2 is subtype-specific, specific primers were designed according to USP2-201, USP2-204 mRNA. It was found that the level of USP2-204 mRNA in the mouse liver during fasting increased nearly 3.2 times compared with the previous feeding phase, while the expression of USP2-201 mRNA was almost not affected. And the phenomenon of starvation's regulation on USP2-204 mRNA level was liver-specific, as it was not detected in skeletal muscle and white adipose tissue. Further experiment showed that there was an interaction between USP2 and the transcription coactivator—PGC-1α which is related with various liver fasting reactions. Compared with the control group, both mouse primary hepatocytes transfected with GFP-PGC-1α and mouse liver infected with PGC-1 adenovirus were found that the level of USP2-204 mRNA nearly increased 35 times while USP2-201 mRNA wasn't affected. These results indicate that starvation can specifically promote the alternative splicing of USP2-204 in the mouse liver, but it has no effect on the alternative splicing of USP2-201 (Figure 1C).
Androgen
In the research of the expression of USP2-201 in prostate cancer, Edgard Graner treated prostate cancer cells--LNCap cells which were cultured in charcoal-stripped serum with DHT 0, 7, 24, 48 hours respectively. And he observed that USP2-201 protein gradually increased when treatment time prolonged [33]. However, when the cell treated with DHT and bicalutamide which is an anti-androgen drug simultaneously, USP2-201 protein decreased gradually when treatment time prolonged. Similarly, when the castrated rats treated with testosterone, the USP2-201 mRNA level gradually increased 0, 24, 48, 96 hours after treatment. These phenomena may suggest that androgen can promote the alternative splicing of USP2-201 (Figure 1A).
Signaling pathway molecules and cytokines involved in the alternative splicing of USP2 gene
PKC signaling pathway molecules
It was found that USP2 can change its exon usage in the process of human myeloid cell lines HL-60, THP-1, U937 differentiate into macrophage like cells induced by PMA which is a PKC activator [34]. When HL-60, THP-1, U937 cells treated with PMA for 0, 12, 24, 48, 72 hours respectively, USP2-201, USP2-204 mRNA levels were detected by RT-PCR. USP2-201 mRNA of cells treated with PMA for 12 hours was found much less than that of cells without treatment. When the processing time prlonged to 24, 48, 72 hours, the level of USP2-201 mRNA rebounded slightly but was still significantly lower than that of cells without treatment. On the other side, the expression of USP2-204 mRNA continually increased when treatment time prolonged. It may indicate that PMA could induce the decrease of USP2-201 mRNA but promote the expression of USP2-204 mRNA in HL-60, THP-1 and U937 cells. Further studies were made by performing Western blot analysis to find whether PMA also had an effect on the expression of USP2-201, USP2-204 protein. When cells' treatment time was between 0 and 8 hours, USP2-201 was the major USP2 protein subtype expressed. However, the protein expression was remarkably different when cells' treatment time prolonged to 24 hours: USP2-204 protein level was significantly improved while USP2-201 protein was almost undetectable. Further prolonging cells' treatment time to 48 hours, two subtypes maintained the same protein levels as cells' treatment time was 24 hours. All these data hint that PKC activation may regulate the exon usage of USP2 gene in the process of myeloid cells differentiating into macrophage like cells: PKC activation can promote the alternative splicing of USP2-204 while inhibit USP2-201's alternative splicing (Figure 1).
Cytokines
IL-1β, which can cause glomerular MsC proliferation, is one of the main factors of glomerulonephritis [35, 36]. Immunohistochemistry researches showed that USP2-201 was up-regulated in MsC of mesangial proliferative glomerulonephritis [36]. It was further found that there was an interaction between IL-1β and USP2-201: When rats' MsC treated with IL-1β for 12-24 hours, USP2-201 mRNA gradually increased. At the same time, the change in USP2-201 protein induced by IL-1β was detected to be the same as that of mRNA. It may suggest that IL-1β promotes the alternative splicing of USP2-201 in MsC of mesangial proliferative glomerulonephritis, in addition, it was found that IL-1β did not affect the alternative splicing of USP2-204.
Florian Haimerl et al. observed that when mice pretreated with a low dose of TNF-α by injection, the degree of liver injury induced by GalN/TNF-α can be reduced significantly, and this protective effect sustained 24 hours [25]. Reoverexpression of USP2-202 gene in the primary liver cells of mice whose USP2 gene was knocked-out can increased the degree of cell injury and apoptosis induced by Act.D/ TNF-α slightly. The mechanism behind the phenomena is that TNF-α has impact on the alternative splicing of mouse liver's main USP2 subtype-USP2-202 which can induce apoptosis of hepatocyte: TNF-α can downregulate the mRNA and protein expression of USP2-202 in mouse liver.
Xing Mao, et al. treated MsC (without other cytokine stimulation) with TGF-β1, TNF-α, PDGF-BB respectively and observed the impact of these three cytokines on USP2 alternative splicing for 36 hours [37, 38]. Compared with PDGF-BB and TNF-α, TGF-β1 had the weakest ability in promoting USP2-201's alternative splicing and worked in a time-independent manner. The ability of TNF-α to promote the alternative splicing of USP2-201 was between TGF-β1 and PDGF-BB, and can lead USP2-201 protein to increase over time. PDGF-BB was the most potent cytokine that promoted USP2-201's alternative splicing in the anti-Thy1.1 nephritis rat model, and the increase curve of USP2-201 protein showed a single peak whose highest value was found at 12 hours after stimulation (Figure 1).
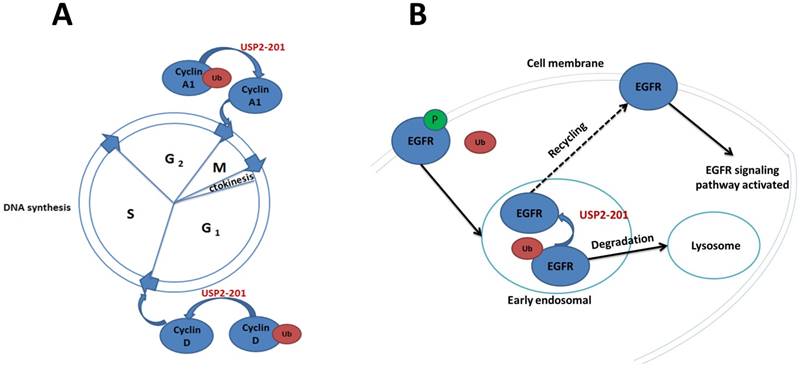
The significance of USP2-201 in cell cycle and EGFR recycling. A. USP2-201 can deubiquitinate Ub-cyclin A1 into cyclin A1 which can promote cell cycle progression from G2 to M phase. Meanwhile, USP2-201 can deubiquitinate Ub-cyclin D1 into cyclin D1 to promote cell cycle progression from G1 to S phase. B. The EGFR is ubiquitinated after its activation and can be internalized by plasma membrane to early endosome where USP2-201 exists. Then the deubiquitinated EGFR can return back to membrane, while the ubiquitinated EGFR can be transported to lysosomes for degradation.
The significance of alternative splicing products of USP2 gene
EGFR recycling
EGFR is a transmembrane tyrosine kinase receptor [39]. The activation of EGFR is involved in many signaling pathways and has effects on cell proliferation, cell differentiation and tumorigenesis [40]. As many membrane receptors, the activated EGFR can be internalized by plasma membrane to early endosomes, then it can either return back to membrane or be transported to lysosomes for degradation [41]. The process is regulated by the alternative splicing products of USP2 [27, 42]. USP2-201 located to early endosomes in HeLa cells can improve the level of membrane EGFR. The molecular mechanism is that the activated membrane EGFR phosphorylated and ubiquitinated at the same time, then the Ub-EGFR internalized to early endosomes. The Ub-EGFR can further be deubiquitinated by USP2-201 to return back to the plasma membrane, which can enhance the recycling of EGFR and activate the molecules involved in the EGFR signaling pathway. It was also found in NSCLC cells where EGFR is highly expressed, USP2-201 can prevent the degradation of both mutant and wild-type EGFR. As a matter of fact, in NSCLC tissue the expression of EGFR protein was positively correlated with USP2-201 protein (Figure 2B).
Cell cycle progression
Cyclin A1 which is essential for promoting cell cycle progression from G2 to M phase is intimately related with prostate and bladder cancer [43-45]. Jayoung et al. have found that there was also a relationship between USP2 and Cyclin A1, the alternative splicing products of USP2 promote the development of bladder cancer through regulating CyclinA1 [33, 46]. The mechanism is that USP2-201 can prevent the ubiquitination of CyclinA1 to improve the stability of CyclinA1 protein which plays a role in the development of bladder cancer. Overexpression of USP2-201 gene in T24 human bladder cancer cells can induce cell proliferation and improve cell's invasion ability. By contrast, when USP2-201 gene of T24 cell was knocked down by shRNA, cell's proliferation and invasion capabilities decreased.
CyclinD1 is an important regulatory factor during the transition from the G1 to S phase in the cell cycle [47-49]. Jing, Shan, et al. screened 76 deubiquitination enzymes' catalytic ability on Ub-CyclinD1 in vitro, and found that USP2-201 was a specific deubiquitination enzyme for CyclinD1 [50]. In 293T cells the Ub-CyclinD1 was deubiquitinated by USP2-201 thus promoting cell cycle progression from G1 to S phase. Both human breast cancer cell MCF-7 and prostate cancer cell PC3 have a high expression of CyclinD1 protein, of which the USP2-201 gene knockdown can significantly reduce the CyclinD1 protein and further result in growth inhibition. Magiera et al. demonstrated that the most potent LCA derivative, LCA hydroxyamide, can inhibit USP2-201 in cyclin D1-expressing cells, without the dependence on p53 status. Thus it can further result in the destabilization of cyclin D1, cell cycle arrest and growth inhibition [51]. Mindy I. Davis et al. also reported the discovery of a small molecule inhibitor targeting USP2-201 induced an increase in cellular cyclin D1 degradation (Figure 2A) [52].
Cell apoptosis
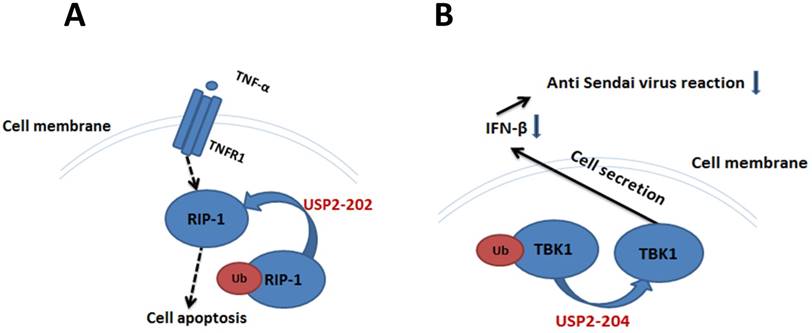
TNF-α and TNFR1 can deliver apoptosis signal, RIP1 is one of the downstream molecules in apoptosis signaling pathway [53-55]. In human breast cancer MCF7 cells, USP2-202 can elevate the level of RIP1 to promote apoptosis by deubiquitinating Ub-RIP1 [26]. The apoptosis induction effect by USP2-201 is much weaker, because the deubiquitination interaction between Ub-RIP1 and USP2-201 was inhibited by TRAF2. Overexpression of USP2-201 in TRAF2 gene knocked-out mouse embryonic fibroblast cell can significantly increase the RIP1 level and promote cell apoptosis (Figure 3A).
Cellular antiviral response
Ub-TBK1 can increase the level of IFN-β to participate in HEK293's antiviral response, and USP2-204 is a negative regulator in this process [56, 57]. When Ub-TBK1 was deubiquitinated by USP2-204 into TBK1, the ability of TBK1 to induce IFN-β was decreased, resulting in the decrease of HEK293's anti Sendai virus reaction (Figure 3B).
Conclusions and perspectives
The current researches on the alternative splicing products of USP2 mainly focus on the 3 subtypes: USP2-201, USP2-202 and USP2-204. It is found that TGF-β1, TNF-α, PDGF-BB, IL1-β and androgen are regulatory factors for USP2-201's alternative splicing to promote the expression of USP2-201 by directly or indirectly interacting with SE which is a splicing regulatory element on pre-mRNA. While the interaction between PKC and regulatory element SS inhibits the expression of USP2-201. For the alternative splicing of USP2-202, TNF-α which is a negative regulation factor for USP2-202 is the only regulatory factor found so far. Nutritional factors, biological clock, PKC, PGC-1α are USP2-204's positive alternative splicing factors which play roles in promoting the alternative splicing of USP2-204, while Bmal1 is an inhibiting alternative splicing factor for USP2-204 (Figure 1). These three alternative splicing products' functions were embodied in many ways. USP2-201 promotes cell cycle progression through the regulation on cell cycle regulatory proteins CyclinA1, CyclinD1, on the other hand, USP2-201 located in early endosomes deubiquitinates Ub-EGFR to improve the utilization rate of the EGFR by promoting EGFR to return back to the plasma membrane (Figure 2). USP2-202 deubiquitinates the Ub-RIP1 to activate apoptosis signaling pathway resulting in cell apoptosis and USP2-204 participates in cell antiviral response (Figure 3). The specific sequence of the splicing regulatory elements SE, SS on the USP2 pre-mRNA and the interaction between them and related regulatory factors are still unknown. Simultaneously, there is a need to validate the current conclusions on USP2's alternative splicing and the study on the alternative splicing of USP2 remains to be deficient. With the accumulation of data in the future, the specific process of the alternative splicing of USP2 and the molecular mechanism behind it will be further elucidated. Then it is possible to regulate USP2's alternative splicing products as needed.
The significance of USP2-202 and USP2-204 in cell apoptosis and cellular antiviral response. A. USP2-202 can elevate the level of RIP1 to induce cell apoptosis by deubiquitinating Ub-RIP1. B. USP2-204 regulate the ubiquitination degree of TBK1 to determine the level of IFN-β in cell's anti Sendai virus response.
Abbreviations
USP2: Ubiquitin-specific protease 2; ESE: exon splicing enhancer; ESS: exonic splicing silencer; ISE: intron splicing enhancer; ISS: intron splicing silencer; SR: Ser/Arg rich protein; hnRNP: heterogeneous nuclear ribonucleoprotein; PTS1: peroxisomal targeting signal of type 1; PEX5: peroxisomal targeting signal 1 receptor; Bmal1: Brain and muscle ARNT like gene; PGC-1α: peroxisome proliferator-activated receptor gamma coactivator alpha 1-α; DHT: Dihydrotestosterone; FAS: factor associated suicide; PKC: protein kinase C; IL-1β: Interleukin-1β; MsC: glomerular mesangial cell; TGF-β1: transforming growth factor-β1; TNF-α: tumor necrosis factor-α; PDGF-BB: platelet-derived growth factor BB; EGFR: Epidermal growth factor receptor; TNFR1: tumor necrosis factor receptor 1; RIP1: receptor-interacting protein 1; TRAF2: TNF receptor associated factor 2; TBK1: TANK-binding kinase 1; SS: splicing silencer; SE: splicing enhancer; IFN-β: interferon-β.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (81172322, 81302006), Shanghai Municipal Education Committee (13ZZ089), Science and Technology Committee of Shanghai (14401901500), Science and Technology Committee of Baoshan District (12-E-2), NO.3 People's Hospital affiliated to Shanghai Jiao-Tong University School of Medicine (sy2013-008).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature reviews Molecular cell biology. 2013;14:153-65
2. Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nature reviews Genetics. 2014;15:689-701
3. Li M. et al. Transcriptional landscape of human cancers. Oncotarget. 2017;8:34534-51
4. Herhaus L. Expanding the ubiquitin code through post-translational modification. EMBO reports. 2015:16
5. Dlamini Z. et al. Abnormalities in alternative splicing in diabetes: therapeutic targets. J Mol Endocrinol. 2017;59:93-107
6. Park M. et al. p53 signaling is involved in leptin-induced growth of hepatic and breast cancer cells. Korean J Physiol Pharmacol. 2016;20:487-98
7. Gopinath P. et al. Understanding and predicting the potency of ROS-based enzyme inhibitors, exemplified by naphthoquinones and ubiquitin specific protease-2. Chemical Science. 2016;7:7079-86
8. Matthew M. et al. Peroxisomal Localization and Circadian Regulation of Ubiquitin-Specific Protease. PloS one. 2012;7:1-10
9. Ryu KY. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. ANALYTICAL BIOCHEMISTRY. 2006;353:153-5
10. Pagliarini V. et al. Splicing Regulation: A Molecular Device to Enhance Cancer Cell Adaptation. BioMed research international. 2015;2015:543067
11. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773-86
12. Shin YCC. et al. The molecular determinants for distinguishing between ubiquitin and NEDD8 by USP2. Sci Rep. 2017;7:2304
13. Harrigan J. Monitoring Target Engagement of Deubiquitylating Enzymes Using Activity Probes: Past, Present, and Future. Methods Mol Biol. 2016;1449:395-410
14. Gousseva N1 BR. Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp2-45 and Usp2-69. Gene Expr. 2003;11:163-79
15. Wang CL, Wang JY, Liu ZY, Ma XM, Wang XW, Jin H. et al. Ubiquitin-specific protease 2a stabilizes MDM4 and facilitates the p53-mediated intrinsic apoptotic pathway in glioblastoma. Carcinogenesis. 2014;35:1500-9
16. ROHAN T. BAKER AMC, YAMUNA KARUNASEKARA, TATIANA A. SOBOLEVA, ROBERT SHARWOOD, SPENCER WHITNEY, and PHILIP G. BOARD. Using Deubiquitylating Enzymes as Research Tools. METHODS IN ENZYMOLOGY. 2005;398:540-54
17. Shahnawaz M. et al. Stable activity of a deubiquitylating enzyme (Usp2-cc) in the presence of high concentrations of urea and its application to purify aggregation-prone peptides. Biochemical and biophysical research communications. 2007;359:801-5
18. Ohtake F. et al. Ubiquitin acetylation inhibits polyubiquitin chain elongation. Scientific Reports. 2015;16:192-201
19. Pubmed USP2 ubiquitin specific peptidase 2 [ Homo sapiens (human) ]. https://www.ncbi.nlm.nih.gov/gene/9099
20. Benassi B, Marani M, Loda M, Blandino G. USP2a alters chemotherapeutic response by modulating redox. Cell death & disease. 2013;4:e812
21. Reglinski K, Keil M, Altendorf S, Waithe D, Eggeling C, Schliebs W. et al. Peroxisomal Import Reduces the Proapoptotic Activity of Deubiquitinating Enzyme USP2. PloS one. 2015;10:1-20
22. Park KC, Kim JH, Choi EJ, Min SW, Rhee S, Baek SH. et al. Antagonistic regulation of myogenesis by two deubiquitinating enzymes, UBP45 and UBP69. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9733-8
23. Pubmed Homo sapiens ubiquitin specific peptidase 2 (USP2), transcript variant 1, mRNA. https://www.ncbi.nlm.nih.gov/nuccore/NM_004205
24. Tianling Wei EB, Lise Mette Rahbek Gjerdrum, Omid Niazi, Niels Ødum, Robert Gniadecki. Ubiquitin-specific protease 2 decreases p53-dependent apoptosis in cutaneous T-cell lymphoma. Oncotarget. 2016;7:48391-400
25. Haimerl F, Erhardt A, Sass G, Tiegs G. Down-regulation of the de-ubiquitinating enzyme ubiquitin-specific protease 2 contributes to tumor necrosis factor-alpha-induced hepatocyte survival. The Journal of biological chemistry. 2009;284:495-504
26. Mahul-Mellier AL, Datler C, Pazarentzos E, Lin B, Chaisaklert W, Abuali G. et al. De-ubiquitinating proteases USP2a and USP2c cause apoptosis by stabilising RIP1. Biochimica et biophysica acta. 2012;1823:1353-65
27. Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR. et al. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. 2013;32:1660-9
28. Pouly D. et al. Mice carrying ubiquitin-specific protease 2 (Usp2) gene inactivation maintain normal sodium balance and blood pressure. Am J Physiol Renal Physiol. 2013;305:21-30
29. Pouly DC. et al. USP2-45 Is a Circadian Clock Output Effector Regulating Calcium Absorption at the Post-Translational Level. PloS one. 2016;11:e0145155
30. Pouly D. et al. USP2-45 Is a Circadian Clock Output Effector Regulating Calcium Absorption at the Post-Translational Level. PloS one. 2016;11:e0145155
31. Matthew M. et al. Ubiquitin-Specific Protease 2 Regulates Hepatic Gluconeogenesis and Diurnal Glucose Metabolism Through 11b-Hydroxysteroid Dehydrogenase 1. DIABETES. 2012;61:1025-35
32. Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. The Journal of biological chemistry. 2012;287:25280-91
33. Kim J, Kim WJ, Liu Z, Loda M, Freeman MR. The ubiquitin-specific protease USP2a enhances tumor progression by targeting cyclin A1 in bladder cancer. Cell cycle. 2012;11:1123-30
34. Kitamura H, Kimura S, Shimamoto Y, Okabe J, Ito M, Miyamoto T. et al. Ubiquitin-specific protease 2-69 in macrophages potentially modulates metainflammation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27:4940-53
35. Schreiber A. et al. Neutrophil Serine Proteases Promote IL-1β Generation and Injury in Necrotizing Crescentic Glomerulonephritis. J Am Soc Nephrol. 2012;23:470-82
36. Wang S, Wu H, Liu Y, Sun J, Zhao Z, Chen Q. et al. Expression of USP2-69 in mesangial cells in vivo and in vitro. Pathology international. 2010;60:184-92
37. Mao X, Luo W, Sun J, Yang N, Zhang LW, Zhao Z. et al. Usp2-69 overexpression slows down the progression of rat anti-Thy1.1 nephritis. Experimental and molecular pathology. 2016;101:249-58
38. Li Y KB, Qian S, Letterio JJ, Fung JJ, Lu L, Lin F. Hepatic stellate cells inhibit T cells through active TGFβ1 from a cell-surface-bound latent TGFβ1/GARP complex. Journal of immunology. 2015;195:2648-56
39. Yen H-Y, Liu Y-C, Chen N-Y, Tsai C-F, Wang Y-T, Chen Y-J. et al. Effect of sialylation on EGFR phosphorylation and resistance to tyrosine kinase inhibition. Proceedings of the National Academy of Sciences. 2015;112:6955-60
40. Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34-74
41. Kim KK, Han A, Yano N, Ribeiro JR, Lokich E, Singh RK. et al. Tetrathiomolybdate mediates cisplatin-induced p38 signaling and EGFR degradation and enhances response to cisplatin therapy in gynecologic cancers. Scientific Reports. 2015;5:1-11
42. Kim J. et al. A synthetic form of frizzled 8-associated antiproliferative factor enhances p53 stability through USP2a and MDM2. PloS one. 2012;7:e50392
43. Ji P, Agrawal S, Diederichs S, Bäumer N, Becker A, Cauvet T. et al. Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell cycle progression in somatic cells. Oncogene. 2004;24:2739-44
44. Dalton S. Linking the Cell Cycle to Cell Fate Decisions. Trends Cell Biol. 2015;25:592-600
45. Wei H LY, Zhao C, Jiang X, Chen H, Lang MF, Sun J. Cyclin A1 is expressed in mouse ovary. Int J Med Sci. 2014;11:754-7
46. Park CW. Cellular ubiquitin pool dynamics and homeostasis. BMB Reports. 2014;47:475-82
47. Nepal S, Shrestha A, Park PH. Ubiquitin specific protease 2 acts as a key modulator for the regulation of cell cycle by adiponectin and leptin in cancer cells. Molecular and cellular endocrinology. 2015;412:44-55
48. Tane S, Kubota M, Okayama H, Ikenishi A, Yoshitome S, Iwamoto N. et al. Repression of Cyclin D1 Expression Is Necessary for the Maintenance of Cell Cycle Exit in Adult Mammalian Cardiomyocytes. Journal of Biological Chemistry. 2014;289:18033-44
49. Bertoli C SJ, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nature Reviews Molecular Cell Biology. 2013;14:518-28
50. Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Molecular cell. 2009;36:469-76
51. Magiera K. et al. Holak, Lukasz Skalniak. Lithocholic Acid Hydroxyamide Destabilizes CyclinD1 and Induces G0/G1 Arrest by Inhibiting Deubiquitinase USP2a. Cell Chemical Biology. 2017;24:458-70
52. Mindy I. et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. The Journal of biological chemistry. 2016;291:24628-40
53. Yaoa Z. et al. RIP1 modulates death receptor mediated apoptosis. Molecular oncology. 2015;9:806-17
54. Li J. et al. Structural basis of cell apoptosis and necrosis in TNFR signaling. Apoptosis. 2015;20:210-5
55. Mbanwi AN. et al. Costimulatory TNFR family members in control of viral infection: Outstanding questions. Seminars in Immunology. 2014;26:210-9
56. Zhang L, Zhao X, Zhang M, Zhao W, Gao C. Ubiquitin-specific protease 2b negatively regulates IFN-beta production and antiviral activity by targeting TANK-binding kinase 1. Journal of immunology. 2014;193:2230-7
57. Li J LK, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, Zhou X, Yuan Z. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nature Immunology. 2013;14:793-803
Author contact
![]() Corresponding author: Feng-Hou Gao, Email: fenghougaocom
Corresponding author: Feng-Hou Gao, Email: fenghougaocom

 Global reach, higher impact
Global reach, higher impact