10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(1):78-86. doi:10.7150/ijbs.22426 This issue Cite
Research Paper
Fam60al as a novel factor involved in reprogramming of somatic cell nuclear transfer in zebrafish (Danio rerio)
1. State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China;
2. University of Chinese Academy of Science, Beijing 100049, China;
3. Qingdao National Laboratory for Marine Science and Technology, Qingdao, 266237, China.
Received 2017-8-17; Accepted 2017-12-22; Published 2018-1-11
Abstract
The main reason for abnormal development of cloned animals or embryos, and inefficient animal cloning, is a poor understanding of the reprogramming mechanism. To better comprehend reprogramming and subsequent generation of pluripotent stem cells, we must investigate factors related to reprogramming of somatic cells as nuclear donors. As we know, fam60al (family with sequence similarity 60, member A, like) is a coding gene only found in zebrafish and frog (Xenopus laevis) among vertebrates. However, until now, its functions have remained unknown. Here, we generated a zebrafish fam60al-/- mutant line using transcription activator-like effector nucleases (TALENs), and found that both nanog and klf4b expression significantly decreased while myca expression significantly increased in fam60al-/- mutant embryos. Concurrently, we also uncovered that in developmentally arrested embryos of somatic cell nuclear transfer, nanog, klf4b and myca expression was down-regulated, accompanying a decrease of fam60al expression. Interestingly, we identified a long noncoding RNA (lncRNA) of fam60al, named fam60al-AS, which negatively regulated fam60al by forming double-stranded RNA (dsRNA). RNase protection assay and real-time PCR confirmed these findings. Taken together, these results suggest that fam60al is a novel factor related to the reprogramming of somatic cell nuclear transfer in zebrafish, which is regulated by its reverse lncRNA.
Keywords: Fam60al, long noncoding RNA, somatic cell nuclear transfer, reprogramming, nanog, myca.
Introduction
Reprogramming is the process that differentiated cells undergo to restore totipotency or pluripotency. Somatic cell nuclear transfer (SCNT), pluripotent or determinative factors and cell fusion in vitro, can all induce somatic cell reprogramming [1]. A study on telomerase knockout mice demonstrated that the reprogramming ability of SCNT exceeds that of induced pluripotent stem cells [2], and SCNT embryos share more similarities with embryonic stem cells, in genetic and epigenetic modifications, than with induced pluripotent stem cells [3]. Therefore, it would have attractive prospects to obtain pluripotent stem cells through reprograming of somatic cells by SCNT.
At present, somatic cell cloning has been successfully generated in fish and mammals, including the carp [4], zebrafish [5], medaka [6], sheep [7], rat [8], calve [9, 10], pig [11], horse [12], dog [13], ferret [14], camel [15], albeit, SCNT efficiency has been very low (1-3%). Moreover, animals cloned from somatic cells have developed malformations, including respiratory and circulatory problems, immune deficiency and premature aging [16]. The main cause is incomplete reprogramming of somatic cells. Complete reprogramming of somatic cell depends on whether epigenetic modification of donor cells can be restored to the state of totipotent stem cells following their transfer into recipient enucleated eggs in SCNT embryos [17-19]. Epigenetic modifications mainly include DNA methylation, histone acetylation, chromatin remodeling and the regulation of non-coding RNA, such as microRNA and long non-coding RNA (lncRNA).
LncRNAs are >200 bp transcriptional sequences that generally have no capabilities to encode protein [20]. LncRNAs are the least understood of the genomic transcription products, and previously considered as transcriptional "noise" [21]. However, with the development of transcriptional sequencing technology, a large number of lncRNAs have now been discovered [22-24]. Recent studies have shown that lncRNAs play an important role in epigenetic modification and pluripotency maintenance in somatic cell reprogramming of human pluripotent stem cells [25, 26]. LincRNA-RoR was the first reported lncRNA participating in somatic cell reprogramming, which improved somatic cell induction efficiency by blocking the degradation of miRNA to OCT4, SOX2 and NANOG [27]. Meanwhile, lincRNA-p21 has been shown to act on H3K9 methyltransferase and DNA demethylase DNMT1, to maintain CpG methylation in the promoter region of pluripotency genes, eventually inhibiting the reprogramming process [28]. Antisense transcripts are complementary to the endogenous sense transcripts and make up a substantial proportion of lncRNAs (~50-70%). Unfortunately, however, they have been ignored for many years due to their heterogeneity, low expression levels and unknown function [29].
Many aspects of zebrafish biology make them an attractive model for researchers. For example, they share high gene homology with human and other vertebrates [30], and have large fecundity and the potential for development in vitro. Moreover, because nuclear transfer technology has been well established in zebrafish, and the development of nuclear transferred embryos can be directly observed [5, 31-33], they are suitable candidate for studying somatic cell reprogramming mechanisms.
Fam60al, a coding gene only found in zebrafish and frog among vertebrates within our knowledge [34], had an unknown function till now. Here, we discovered that the expression of nanog, klf4b and myca was significantly affected in fam60al knockout zebrafish embryos. Excitingly, fam60al, nanog, klf4b and myca also exhibited differential expression in kidney cell-derived zebrafish SCNT embryos. Moreover, we identified a reverse transcriptional sequence of fam60al, named fam60al-AS. The overlapping sequence of fam60al and fam60al-AS could form dsRNA, which negatively regulated fam60al expression. These results suggest that fam60al is a novel factor related to somatic cell nuclear reprogramming in zebrafish, which is negatively regulated by its reverse transcriptional sequence.
Materials and Methods
Zebrafish strain and maintenance
In this study, AB strain zebrafish and their embryos were maintained under standard conditions at 28.5°C. Embryos development stages were strictly in accordance with previously described [35].
Establish of fam60al knockout zebrafish line with TALENs
Firstly, use the software (http://boglabx.plp.iastate.edu/TALENT/TALENT/) to predict appropriate gene knockout sites for fam60al disruption, before constructing the TALEN plasmids as previously described [36]. Two target sites were designed in the gene coding area, not overlapping with fam60al-AS, which intended to delete most of the fam60al coding sequence. Fam60al double knockout targets were as follows: Target-left 1 and Target-right 1 (5'-TCGTCAAGTTCTCGGTTC-3' and 3'-TATGAGGAGAACTTCAGA-5', respectively), and Target-left 2 and Target-right 2 (5'-TGCAAGAGCAGGACCTGGC-3' and 3'-CAAGAAAGTGAAAAACA-5', respectively). The final TALEN plasmids were linearized using NotI and transcribed using the mMESSAGE mMACHINE Sp6 Kit (Ambion, USA). TALEN mRNAs (125 ng/uL of each: Target-left 1 and Target-right 1, Target-left 2 and Target-right 2 monomer mRNAs) were microinjected into zebrafish embryos at the 1-cell stage. TALEN-injected zebrafish embryos (F0) were raised to adulthood and outcrossed with wild type zebrafish (WT) to obtain F1. The mutations in F1 were analyzed by PCR and sequencing. PCR primers are listed in Table S1. PCR amplify as follows: 3 min at 94°C, followed by 32 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C. F3 embryos (including ¼ fam60al-/-) were obtained by self-crossing of F2 with the same mutation. Figure S1 showed the flowchart of establishing fam60al-/- line.
Construct of SCNT embryos in zebrafish
The SCNT embryos were constructed based on the method described previously by our group in 2009 and 2011 [32, 33], which included preparing nuclear transfer buffer, head kidney donor cells, unfertilized zebrafish eggs and performing nuclear transfer operations. The SCNT embryos were cultured at 28.5°C.
Rapid amplification of cDNA ends (RACE)
Total RNA was extracted using TRIZOL reagent (Ambion, USA) and the first chain cDNAs were synthesized according to the SMART™ RACE cDNA Amplification Kits (Clontech, USA). 5' RACE and 3' RACE transcript fragments were amplified using nested PCR in positive and negative directions, respectively. The primers used are listed in Table S1. The corresponding PCR fragments were sequenced and spliced to obtain the full-length transcription sequences of fam60al and fam60al-AS (Figure S2, Figure S3).
Genome walking
The genome of zebrafish embryos were extracted according to the phenol chloroform method and the adapter connectors were added to the genome DNA based on the Universal GenomeWalker™ 2.0 kit (Clontech, USA). Fam60al-AS genomic information was obtained by PCR amplification, sequencing and splicing. The primers for genome walking are listed in Table S1.
Coding Potential Calculator (CPC)
CPC is an online software that assesses the protein-coding potential of transcripts [37]. It has a user-friendly web-based interface (http://cpc.cbi.pku.edu.cn/). First to paste the fasta sequence in the box, then to click run to output the analysis result.
Dot blot
Total RNA was extracted from embryos at the shield stage, 24 hours post-fertilization (hpf) and 72 hpf using TRIZOL reagent. Then 0.1-1 mg total RNA was purified using PolyATtract mRNA Isolation System IV (Promega, USA). 100 ng of purified mRNA and RNA denaturing agent were mixed for 10 min at 65°C, before placing onto nylon membranes. The membrane was then baked for 2 h at 80°C, pre-hybridized for 20 min at 42°C, hybridized overnight at 42°C using a DIG-labeled probe and washed with 2×SSC (twice for 5 min) and 0.5×SSC (twice for 5 min). The membrane was blocked for 30 min at 37°C with 5% protein powder dissolved in phosphate buffered saline (PBS), incubated for 1 h at 37°C or overnight at 4°C with 1:5000 diluted anti-Dig antibody (Roche, Switzerland) in the blocking reagent, washed in PBS with Tween20 (PBST) thrice for 10 min each time and stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate stock solution (NBT/BCIP) (Roche, Switzerland).
Whole-mount in situ hybridization (WISH)
WISH was conducted as previously described [38]. The antisense probes and sense probes were synthetized using T7 RNA polymerase and Sp6 RNA polymerase, respectively. The images were captured using a Nikon digital camera. The primers (fam60al-WISH-F/R, fam60al-AS-WISH-F/R) are shown in Table S1.
Semi-quantitative reverse transcriptase PCR (RT-PCR) and real-time quantitative PCR (RT-qPCR)
Total RNA of different developmental stage embryos was extracted using TRIZOL reagent. The samples were digested for 30 min using RNase-free DNase, and obtained cDNAs through transcription in vitro using the ReverTra Ace reverse transcriptase kit (Toyobo, Japan) and random primers. The RT-PCR conditions were as follows: 3 min at 94°C, followed by 28/35 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C. 28 cycles for amplification of β-actin and fam60al, 35 cycles for amplification of fam60al-AS. RT-qPCR was conducted on a Roche LightCycler 480 real-time PCR system, using 2xSYBR green real-time PCR mix (Toyobo, Japan). The amplified conditions were 94°C for 1 min, followed by 40 cycles of 10 s at 94°C, 10 s at 60°C, and 15 s at 72°C. The RT-PCR and RT-qPCR primers are shown in Table S1.
RNase protection assay
RNase protection assay was conducted using purified mRNAs of embryos at the sphere stage and 24 hpf, as previously described [39]. Each sample was divided into three parts: the experimental group, the positive control group and the negative control group. Firstly, the negative control group was degenerated at 95°C for 15 min. Then, three groups of samples were digested with DNase for 30 min. RNase A was added to the experimental group and the negative control group. RNaseOut was added to the control group. All samples were treated at 37°C for 1 h and retrieved using TRIZOL reagent. ReverTra Ace reverse transcriptase and random primers were used to amplify cDNAs. DsRNA in the fam60al and fam60al-AS overlapping region was detected using PCR. The PCR primers used (dsRNA-F and dsRNA-R) are shown in Table S1.
Microinjection of fam60al-AS mRNA
Fam60al-AS sequence was firstly obtained using PCR and then the PCR fragment was subcloned into pCS2+ vector. Fam60al-AS mRNA was synthesized in vitro according to the mMessage mMachine SP6 kit instructions. 50 ng/uL mRNA was microinjected into 1-cell stage zebrafish embryos. The embryos at the sphere and shield stage were collected for RT-qPCR, respectively. The fam60al-AS amplification primers (fam60al-AS-F and fam60al-AS-R) are shown in Table S1.
Data analysis
The statistical results were showed with mean±SEM. The independent sample T test was used to compare the mean value between the two groups. A P-value < 0.05 was considered statistical significance. All experiments were repeated at least three times.
Results
Identifying an antisense transcript of fam60al in zebrafish
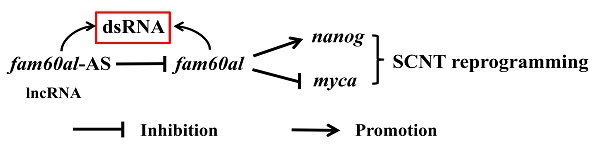
We obtained the full-length fam60al cDNA, encoding 216 amino acids, using 5' RACE and 3' RACE. Interestingly, in the process, we found an antisense sequence of fam60al with 1169bp, which we named fam60al-AS. There is a 405bp overlapping region between the antisense transcript and fam60al at the 5th and 6th exon. As the incomplete information in this region of the current zebrafish genome (GRCz11), we amplified the sequences using genome walking and found that fam60al-AS consisted of only two exons (Figure 1A). We identified fam60al-AS as an lncRNA by predicting its coding ability using CPC.
In order to further confirm the reliability of the RACE results, we synthesized the antisense and sense probes of fam60al and fam60al-AS, excluding the overlapping region, respectively. Both dot blot and WISH assays demonstrated hybridization signal of the antisense probes, whereas we observed no hybridization signal using the sense probe (Figure 1B, 1C). These results indicate that fam60al-AS, the antisense transcript of fam60al, is indeed present in zebrafish.
Expression patterns of fam60al and fam60al-AS during early embryonic development
We analyzed the expression patterns of fam60al and fam60al-AS during early embryonic developmental of zebrafish using RT-PCR (Figure 2A) and RT-qPCR (Figure 2B). Fam60al was expressed as a maternal transcript, indicating its role in the early development. As the developmental process progressed, fam60al expression gradually decreased over the period from 1-cell stage to 120hpf. Meanwhile, we only weakly detected expression of fam60al-AS before the high stage. Its expression increased over the period from sphere stage to 24hpf, and then decreased from 48hpf to 120hpf. We also found the relative expression level of fam60al was significantly higher than fam60al-AS at the same stage.
Fam60al and its antisense transcript, fam60al-AS, in zebrafish. (A) Fam60al and fam60al-AS gene structure. Red marks the antisense overlapping sequence. CPC predicted fam60al-AS as an lncRNA. (B) Dot blot of the shield stage, 24 hpf and 72 hpf embryos, using sense and antisense probes of fam60al and fam60al-AS. Antisense probes have hybridization signal, while sense probes have no hybridization signal. (C) WISH of 24hpf and 72hpf embryos using sense and antisense probes of fam60al and fam60al-AS. Antisense probes have hybridization signal, sense probes have no hybridization signal. (scale bar: 200 μm).
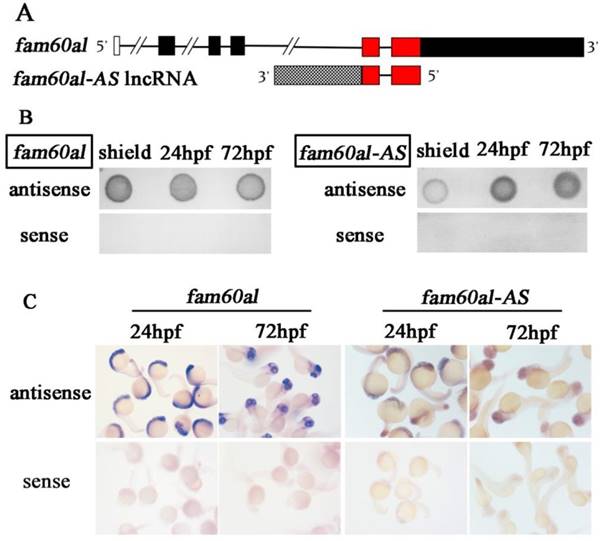
Fam60al and fam60al-AS expression at different developmental stages in zebrafish. A: RT-PCR analysis of fam60al and fam60al-AS expression during early development, normalized to β-actin. B: RT-qPCR analysis of fam60al and fam60al-AS expression. The 2-ΔΔct method was used for measuring the expression levels and normalized to β-actin. 256-cell stage fam60al-AS expression level was set to 1.
Fam60al is negatively regulated by fam60al-AS
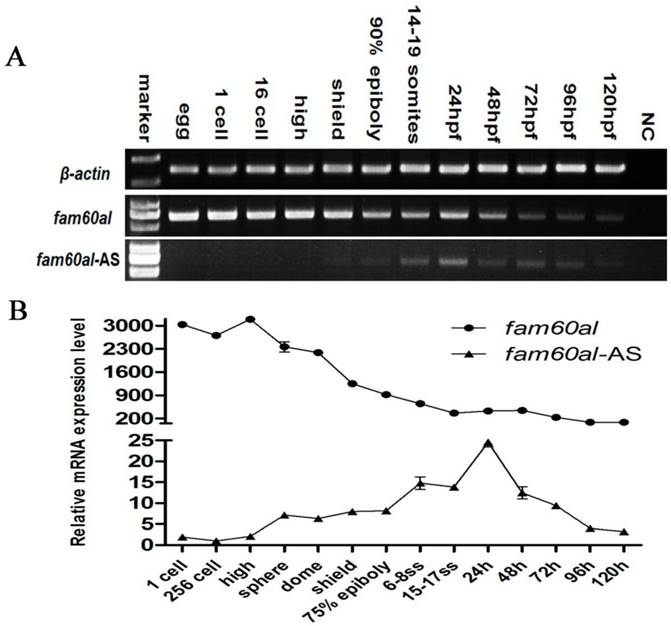
Pearson correlation analysis revealed a moderate negative correlation between fam60al and fam60al-AS (correlation coefficient: -0.604, P < 0.05), suggesting that fam60al-AS might negatively regulate fam60al. We further generated fam60al-AS overexpressed embryos by injecting fam60al-AS mRNA into zebrafish embryos at 1-cell stage, and we observed significantly down-regulated expression of fam60al at the subsequent sphere stage (Figure 3A). This experiment confirmed the negative correlation between fam60al and fam60al-AS. How did fam60al-AS influence fam60al expression in vivo? RNase protection assay showed that embryonic mRNAs at the sphere stage and 24hpf following treatment with RNase A could amplify the overlapping region of fam60al and fam60al-AS by PCR. While we firstly denatured embryonic mRNAs at 95°C and then treated with RNase A, we failed to obtain any amplified product (Figure 3B). The amplified products were confirmed by sequencing. These indicate fam60al and fam60al-AS can form dsRNA in the overlapping region. Therefore, we conclude that fam60al-AS forms dsRNA with fam60al, which negatively regulates fam60al expression.
Established a zebrafish fam60al knockout line using TALENs
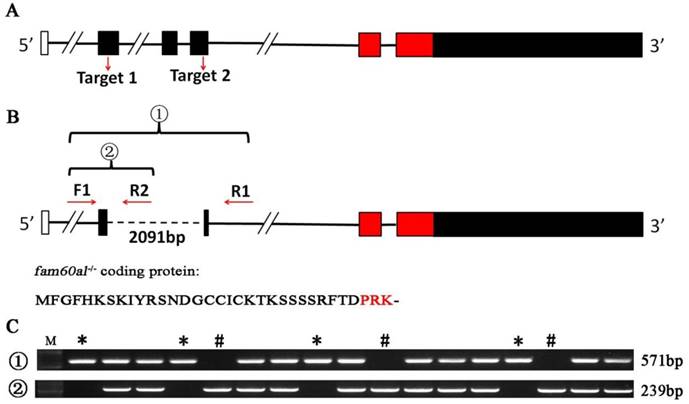
To investigate the role of fam60al during the early development of zebrafish, we designed two pairs of TALENs to target two sites in the fam60al exon region (Figure 4A). 2091bp sequence between two target sites was completely deleted in fam60al knockout zebrafish line (fam60al-/-), which confirmed by PCR and sequencing. The double sites mutant fam60al could only encode a shortened 33 amino acid peptides and avoided the production of redundant protein in single site mutant (Figure 4B). Different kinds of genetype zebrafish were detected by tri-primer-PCR (Figure 4C).
Fam60al-AS negatively regulates fam60al expression by forming dsRNA. A: RT-qPCR analysis of fam60al in fam60al-AS overexpressed embryos. Overexpression of fam60al-AS at the sphere stage, resulted in down-regulation of fam60al expression. *P < 0.05; ** P < 0.01; WT: wild type; OE: over expression. B: RNase protection assay analyzing the dsRNA formed in the overlapping region of fam60al and fam60al-AS at the sphere stage and 24 hpf. PCR bands were detected at the sphere stage and 24hpf with RNase A or RNaseOutTM digestion, while no PCR band was observed at the sphere stage and 24hpf following denaturing at 95°C and digestion with RNase A.
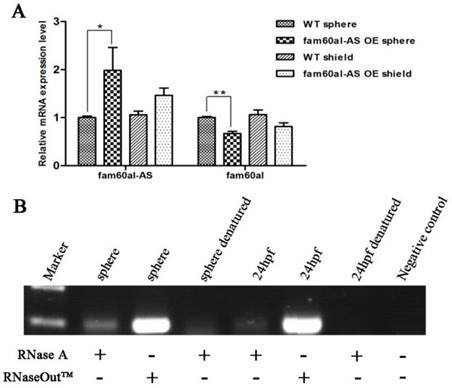
Establish of fam60al knockout line with TALENs in zebrafish. A: Schematic showing fam60al gene knockout target sites. Target 1 was on exon 2 and Target 2 on exon 4. B: Primer design sites, transcript map and protein coding sequence for fam60al in fam60al-/- line. Primers F1 and R1 were designed to amplify the fam60al gene fragment deleted 2091bp in fam60alw/- and fam60al-/-. Primers F1 and R2 were designed to amplify the wild type fam60al gene fragment in fam60alw/w and fam60alw/-. Fam60al-/- encoded a premature termination 33 amino acids peptide. C: PCR results of different genetype zebrafish. Primer set 1: amplified fragment of primers F1 and R1, Primer set 2: amplified fragment of primers F1 and R2, M: DNA marker, *: fam60al-/-, #: fam60alw/w, and the rest for fam60alw/-.
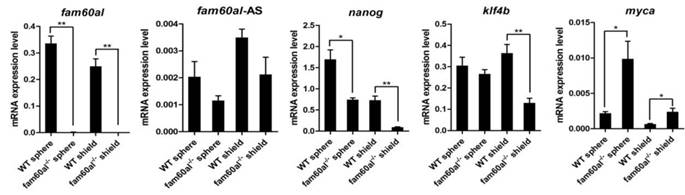
Fam60al regulates the expression of pluripotency-associated genes
In the mutant embryos without fam60al expression, we observed decreased expression of nanog and increased expression of myca at the sphere stage. We also found down-regulated expression of klf4b and nanog, up-regulated expression of myca at the shield stage. Fam60al-AS expression had no significant difference between WT and fam60al-/- mutant embryos at the sphere and shield stage as its quite lower expression (almost 100 times lower than fam60al expression) and the repeated CT values fluctuated slightly among different samples (Figure 5).
Fam60al participates in somatic cell reprogramming in zebrafish
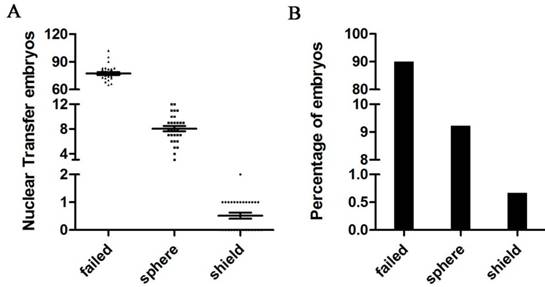
To further investigate the role of fam60al in somatic cell reprogramming, we generated SCNT embryos using zebrafish kidney cells. We conducted SCNT 29 times, with the embryo number varying from 72 to 103 in each experiment, 90.1% (2010/2231) of the transplanted eggs failed to cleave after nuclear transfer, and 9.23% (206/2231) of the SCNT embryos arrested at the sphere stage. Only 0.67% (15/2231) of the SCNT embryos successfully developed into the shield stage (Figure 6A and 6B).
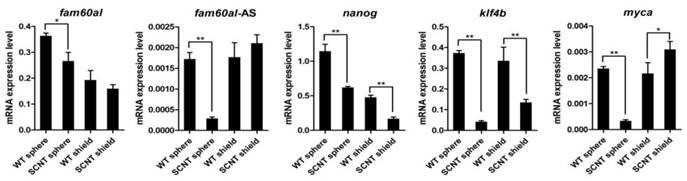
Expression of fam60al, fam60al-AS, nanog, klf4b, and myca were all down-regulated at the sphere stage in the SCNT embryos compared with WT embryos. At the shield stage, we observed decreased expression of nanog and klf4b and increased expression of myca in the SCNT embryos, while no significantly different expression levels of fam60al and fam60al-AS (Figure 7).
mRNA expression levels of fam60al, fam60al-AS, nanog, klf4b and myca in wild type embryos and fam60al mutant embryos at the sphere and shield stage. The 2-Δct method was used for measuring the expression levels. β-actin was used as the internal control. *P < 0.05, **P < 0.01.
Developmental profiles of nuclear transplants at different stages. A: The average number of different batches of kidney cell nuclear transfer embryos. N(failed) = 77.34 ± 5.65, N(sphere) = 8.03 ± 1.83, N(shield) = 0.52 ± 0.54. B: Percentage of kidney cell nuclear transfer embryos at different stages. SCNT embryos failed to cleave. SCNT embryos arrested at the sphere stage. SCNT embryos successfully developed at the shield stage.
RT-qPCR detected the expression in SCNT embryos and wild type embryos at the sphere and shield stage. The 2-Δct method was used for measuring the expression levels and β-actin was used as the internal control. *P < 0.05, **P < 0.01. WT: wild type; SCNT: somatic cell nuclear transfer.
Discussion
In this study, we provide the first evidence of fam60al as a novel factor involved in somatic cell nuclear reprogramming in zebrafish. We also identify a new antisense lncRNA that negatively regulates fam60al in zebrafish.
Nuclear reprogramming involves the process of reactivating specific genes in embryonic cells and silencing specific genes in somatic cells. Nanog, Klf4 and c-Myc, as pluripotency-associated genes, are silenced in differentiated somatic cells, but expressed in early embryonic undifferentiated stem cells [40]. Nanog prevents pluripotent cells from differentiating by inhibiting the expression of development related genes [41]. Cells without Nanog initiate the reprogramming process, but are unable to acquire the traits of developmental totipotency [42, 43]. Maternal-to-zygotic transition (MZT) represents a major reprogramming event in zebrafish. Nanog as one of the key transcription factors (TFs) in the pre-MZT transcriptome regulates the first wave of zygotic gene activation [44]. In this study, we found that nanog expression was down-regulated accompanying with the absent expression of fam60al in fam60al knockout embryos at the sphere and shield stage. Interestingly, we also observed decreased expression of fam60al and nanog in SCNT embryos at the sphere stage. These results indicate cross-talking between fam60al and nanog in SCNT reprogramming.
The balance between KLF4 and c-MYC is important for inducing mammalian pluripotent stem cells. KLF4 inhibits the programmed cell death induced by c-MYC [45, 46], and, in turn, c-MYC prevents the anti-proliferative function of KLF4 [47]. In zebrafish, we previously reported that Klf4 and c-myc could be used to evaluate the reprogramming process in SCNT embryos, and the balance between Klf4 and c-myc was important for the reprogramming process [33]. In this study, we showed that expressions of klf4b, myca, fam60al and fam60al-AS were significantly decreased in SCNT embryos at the sphere stage. The lower expression of klf4b and myca may attribute to the down-regulated of nanog which failed to trigger the first wave of zygotic gene activation [44]. Interestingly, when SCNT embryos initiated reprogramming and developed into the shield stage, we observed decreased expression of klf4b and increased expression of myca. Furthermore, klf4b was significantly down-regulated while myca was significantly up-regulated in fam60al mutant embryos at the shield stage. These results indicate cross-talking between fam60al, klf4b and myca.
Fam60al and fam60al-AS expressions were significantly decreased in SCNT embryos at the sphere stage while had no significant difference at the shield stage. Sphere stage is critical for the onset of zygotic transcription and important developmental stage for SCNT reprogramming because 93.2% (206/221) of the SCNT embryos arrested at this stage. These results provide molecular evidence that fam60al plays a vital role in the initiating reprogramming process instead of maintaining pluripotency in zebrafish. The expression differences of nanog, klf4b and myca in SCNT embryos at the shield stage may attribute to the incomplete reprogramming.
More than 1,000 lncRNAs have been reported in zebrafish, which contains 566 antisense exonic overlapping sequences of the coding gene [24]. Antisense lncRNA plays an important role in regulating the sense coding gene in transcription, splicing, mRNA processing, stability, transport and translation [48-50]. It is critical for antisense lncRNAs to form dsRNA with the sense protein coding gene in regulation [39, 51, 52]. We identified the dsRNA formed between fam60al and fam60al-AS, using the RNase protection assay. Moreover, we confirmed that fam60al-AS negatively regulates fam60al expression. While in fam60al knockout zebrafish, fam60al-AS expression just the opposite exhibited a down-regulating trend, although with no significant differences with the wild type embryos at the sphere and shield stage. We speculate that there might be some elements in fam60al deleted fragment that can regulate fam60al-AS expression, or the lower expression of fam60al-AS is regulated by other molecules.
In summary, we discovered fam60al as a novel factor participating in the initiating reprogramming process in zebrafish. We also identified, fam60al-AS, the antisense lncRNA of fam60al, which negatively regulated fam60al expression by forming dsRNA.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was financially supported by the National Natural Science Foundation (Grant No. 31325026, 31721005) and the Chinese Academy of Sciences (Grant No. XDA08010106, 2016FBZ03). We greatly appreciate Mrs. Ming Li and Mrs. Shangping Chen for their assistance with performing SCNT in zebrafish.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nature Reviews Genetics. 2013;14:427-39
2. Le R, Kou Z, Jiang Y, Li M, Huang B, Liu W. et al. Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell stem cell. 2014;14:27-39
3. Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD. et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177-83
4. Chen H, Yi Y, Chen M, Yang X. Studies on the developmental potentiality of cultured cell nuclei of fish. International Journal Of Biological Sciences. 2010;6:192-8
5. Lee KY, Huang H, Ju B, Yang Z, Lin S. Cloned zebrafish by nuclear transfer from long-term-cultured cells. Nature Biotechnology. 2002;20:795-9
6. Bubenshchikova E, Kaftanovskaya E, Hattori M, Kinoshita M, Adachi T, Hashimoto H. et al. Nuclear transplants from adult somatic cells generated by a novel method using diploidized eggs as recipients in medaka fish (Oryzias latipes). Cloning and Stem Cells. 2008;10:443-52
7. Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KHS. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810-4
8. T W, AC P, M Z, KR J, R Y. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369-74
9. Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Cathy Blackwell F. et al. Cloned Transgenic Calves Produced from Nonquiescent Fetal Fibroblasts. Science. 1998;280:1256-9
10. Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J-y, Doguchi H. et al. Eight Calves Cloned from Somatic Cells of a Single Adult. Science. 1998;282:2095-9
11. Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T. et al. Pig Cloning by Microinjection of Fetal Fibroblast Nuclei. Science. 2000;289:1188-91
12. Woods GL, White KL, Vanderwall DK, Li GP, Aston KI, Bunch TD. et al. A mule cloned from fetal cells by nuclear transfer. Science. 2003;424:635-7
13. Lee BC, Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ. et al. Dogs cloned from adult somatic cells. Nature. 2005;436:641-4
14. Li Z, Sun X, Chen J, Liu X, Wisely SM, Zhou Q. et al. Cloned ferrets produced by somatic cell nuclear transfer. Developmental biology. 2006;293:439-48
15. Wani NA, Wernery U, Hassan FA, Wernery R, Skidmore JA. Production of the first cloned camel by somatic cell nuclear transfer. Biology of reproduction. 2010;82:373-9
16. Cibelli JB, Campbell KH, Seidel GE, West MD, Lanza RP. The health profile of cloned animals. Nature Biotechnology. 2002;20:13-4
17. Rideout WM, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093-9
18. Beaujean N, Taylor J, Gardner J, Wilmut I, Meehan R, Young L. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biology of reproduction. 2004;71:185-93
19. Wakayama T. Production of Cloned Mice and ES Cells from Adult Somatic Cells by Nuclear Transfer: How to Improve Cloning Efficiency? Journal of Reproduction and Development. 2007;53:13-26
20. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs insights into functions. Nature reviews genetics. 2009;10:155-9
21. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
22. Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. Rna. 2010;16:1478-87
23. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775-89
24. Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ. et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22:577-91
25. Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell stem cell. 2014;14:752-61
26. Durruthy-Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ. et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nature genetics. 2016;48:44-52
27. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42:1113-7
28. Bao X, Wu H, Zhu X, Guo X, Hutchins AP, Luo Z. et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell research. 2015;25:80-92
29. Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z. et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927-36
30. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498-503
31. Hu W, Wang Y, Chen S, Zhu Z. Nuclear transplantation in different strains of zebrafish. Chinese Science Bulletin. 2002;47:1277-80
32. Luo D, Hu W, Chen S, Xiao Y, Sun Y, Zhu Z. Identification of differentially expressed genes between cloned and zygote-developing zebrafish (Danio rerio) embryos at the dome stage using suppression subtractive hybridization. Biology of reproduction. 2009;80:674-84
33. Luo D, Hu W, Chen S, Zhu Z. Critical developmental stages for the efficiency of somatic cell nuclear transfer in zebrafish. International Journal of Biological Sciences. 2011;7:476-86
34. Smith KT, Sardiu ME, Martin-Brown SA, Seidel C, Mushegian A, Egidy R. et al. Human family with sequence similarity 60 member A (FAM60A) protein: a new subunit of the Sin3 deacetylase complex. Molecular & Cellular Proteomics. 2012;11:1815-28
35. Kimmel CB, Schilling T, Ullmann B. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253-310
36. Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PloS one. 2013;8:1-7
37. Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35:W345-9
38. Thisse C, Thisse B. High resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59-69
39. Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Molecular cell. 2009;33:462-71
40. Guenther MG. Transcriptional control of embryonic and induced pluripotent stem cells. Epigenomics. 2011;3:323-43
41. Saunders A, Faiola F, Wang J. Concise review: pursuing self-renewal and pluripotency with the stem cell factor Nanog. Stem cells. 2013;31:1227-36
42. Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O. et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722-37
43. Serrano F, Calatayud CF, Blazquez M, Torres J, Castell JV, Bort R. Gata4 blocks somatic cell reprogramming by directly repressing Nanog. Stem cells. 2013;31:71-82
44. Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360-4
45. Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ. et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & development. 1998;12:2424-33
46. Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH. et al. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. The Journal of biological chemistry. 2000;275:18391-8
47. Seoane J, Le H-V, Massague´ J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729-34
48. Knee R, Murphy PR. Regulation of gene expression by natural antisense RNA transcripts. Neurochemistry International. 1997;31:379-92
49. Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. The Journal of biological chemistry. 2000;275:11507-13
50. Li AW, Murphy PR. Expression of alternatively spliced FGF-2 antisense RNA transcripts in the central nervous system regulation of FGF-2 mRNA translation. Molecular and cellular endocrinology. 2000;162:69-78
51. Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235-8
52. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M. et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564-6
Author contact
![]() Corresponding author: Dr. Wei Hu, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China. Tel: 86-27-68780051 Fax: 86-27-68780051 E-mail: huweiac.cn
Corresponding author: Dr. Wei Hu, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China. Tel: 86-27-68780051 Fax: 86-27-68780051 E-mail: huweiac.cn

 Global reach, higher impact
Global reach, higher impact