Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(2):228-236. doi:10.7150/ijbs.22849 This issue Cite
Research Paper
MiR-519d suppresses breast cancer tumorigenesis and metastasis via targeting MMP3
1. Department of Oncology, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, China;
2. Department of Pharmacology (the State-Province Key Laboratories of Biomedicine-Pharmaceutics of China, Key Laboratory of Cardiovascular Research, Ministry of Education), College of Pharmacy, Harbin Medical University, Harbin 150081, China;
3. Department of Pharmacy, The First Affiliated Hospital of Harbin Medical University, Harbin 150001, China;
4. Department of Breast Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin 150086, China;
5. Institute of Metabolic Disease, Heilongjiang Academy of Medical Science, Harbin 150086, China
Received 2017-9-17; Accepted 2017-12-28; Published 2018-2-9
Abstract
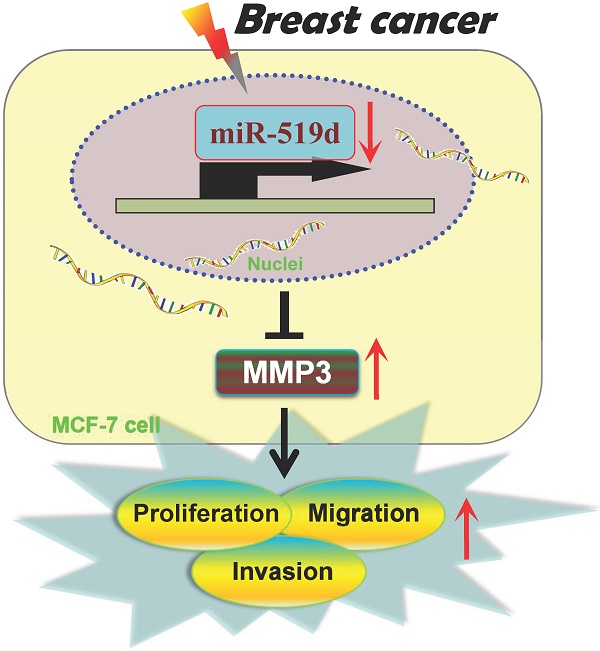
Breast cancer (BC) is the most common cause of death in women throughout the world. Although microRNAs (miRNAs) have been identified as novel regulators in carcinogenesis, there are still abundant hidden treasure needed to be excavated. In the present study, we found that miR-519d expression was remarkably decreased in both human BC tissues and MCF-7 cells. CCK8 and 5-Ethynyl-2′-deoxyuridine (EdU) assays were used to evaluate cell proliferation. Wound-healing and transwell assays were performed for detection of cell migration and invasion. The results demonstrated miR-519d overexpression dramatically suppressed MCF-7 cells proliferation, migration and invasion. While downregulation of miR-519d by miR-519d inhibitor substantially increased MCF-7 cell carcinogenesis. Further analysis identified Matrix Metalloproteinase-3 (MMP3) as a direct target of miR-519d. QRT-PCR and western blot results indicated the correlative expression of miR-519d and MMP3 in BC tissues and MCF-7 cells. In summary, our data uncovered the novel molecular interaction between miR-519d and MMP3, indicating a therapeutic strategy of miR-519d for BC.
Keywords: breast cancer, metastasis, miR-519d, MMP3
Introduction
Breast cancer (BC) is the most common cause of cancer death in women. The incidence of breast cancer worldwide has been increasing in a stable rate since 1987 [1]. It has been reported that 1 in 8 women in her life might suffer from BC in America [2]. Recent studies have focused on identifying epigenetic signatures that are associated with tumor progression and patient prognosis to improve therapy and recovery [3-5]. Although some tumor suppressors and oncogenes have been identified to play central roles in tumorigenesis and development of BC [6-8], the exact molecular mechanisms have not been totally elucidated.
MicroRNAs (miRNAs) are endogenous noncoding RNAs with 19-22 nucleotides in length that modulate variety of ceEllular processes in a post-transcriptional manner, which emerging as novel epigenetic regulators during gene expression [9]. MiRNAs are also associated with regulation of cell growth, division, apoptosis, and other physiological or pathological cellular processes within almost all cells to regulate different diseases [10, 11]. Many studies also revealed that miRNA expression signatures in tumor tissue or liquid biopsies enabled more accurate diagnosis and prognosis to be made in patients with cancer [12]. Moreover, miRNAs could even represent therapeutic targets in their own right [13, 14]. Among them, miR-519d was found highly associated with tumor progression [15], but its role in BC was not totally disclosed.
The role of matrix metalloproteinases (MMPs) in tumor metastasis has been well studied, they degrade the extracellular matrix (ECM), which is associated with tumor metastasis [16]. MMP3, a member of the family of MMPs, has been proved to promote tumor cell migration and invasion [17]. However, till now, few studies focus on the regulation of MMP3 by miRNAs in BC. In this study, we demonstrated that miR-519d play pivotal role in BC by targeting MMP3, which may provide a potential therapeutic targets for the treatment of BC.
Materials and methods
Patients and specimens
From February 2017 to June 2017, 10 pairs of human breast cancer specimens (breast tumor and adjacent non-tumor tissues) were obtained from breast cancer patients who underwent surgery at the Second Affiliated Hospital of Harbin Medical University (Harbin, China), The research were approved by the Ethics and Scientific Committees of Harbin Medical University. We have obtained the written informed consents from all the enrolled patients.
Cell culture
MCF-7, a breast cancer cell line, was cultured at the temperature of 37 °C in 5% CO2, in a culture medium of Dulbecco's modified eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). For transfection procedure, the MCF-7 cells were starved in serum-free DMEM for 24 h, then transiently transfected with MMP3 siRNA, siRNA negative control (si-NC, non-sense scramble sequence of MMP3 siRNA), miR-519d mimic, miR-519d inhibitor or negative control (NC, non-sense scramble sequence of miR-519d) (RiboBio Co., Ltd., Guangzhou, Guangdong, China) at a concentration of 100nM. X-treme RNA transfection reagent (Invitrogen., Ltd, America) was used as transfection vehicle. Cells were harvested 48 h after transfection.
Quantitative real-time PCR (QRT-PCR)
MiR-519d and MMP3 mRNA levels were determined by qRT-PCR. Total RNA from cells was extracted by TRIzol (Invitrogen, Carlsbad, USA) method. The SYBR Green Master Mix Kit (TOYOBO) was used for relative quantification of RNA levels according to manufacturer's instructions. GAPDH and U6 were chosen as an internal control.
miR-519d RT primer, 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCACTCT-3′; miR-519d forward primer, 5′-CAAAGTGCCTCCCTTT-3′; miR-519d reverse primer, 5′-CAGTGCGTGTCGTGGAGT-3′; U6 forward primer, 5′-GCTTCGGCAGCACATATACTAAAAT-3′; U6 reverse primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. MMP3 forward primer, 5′-GAGGACACCAGCATGAACCT-3′; MMP3 reverse primer, 5′-CACCTCCAGAGTGTCGGAGT-3′; GAPDH forward primer, 5′-AAGAAGGTGGTGAAGCAGGC-3′; GAPDH reverse primer, 5′ -TCCACCACCCAGTTGCTGTA-3′.
Western blotting
Homogenized specimens or cells were lysed in 300 ul of RIPA solution containing 1% protease inhibitor. Then centrifuging at 12000 g for 25 min. The protein concentration in the supernate was tested by BCA Protein Assay (Beyotime, Shanghai, China). 10% SDS-PAGE gel electrophoresis was used for fractionating total proteins, then the proteins were transferred onto nitrocellulose membranes. The membranes were blocked in the PBS solution containing 5% de-fatted milk for 2 h in room temperature following the incubation at 4℃ overnight with the primary antibodies of MMP3 (1:1000) (Abcam), cyclin D1 (1:1000) (Abcam), GAPDH (1:1000) (Wanleibio, Inc., Shenyang, China) was as an internal control. Matched secondary antibody (1:10000) (Invitrogen) was incubated for 1 h at room temperature. Protein expression was scanned and quantified by Odyssey Infrared Imaging System (LI-COR, Lincoln, NE, USA).
Cell proliferation
Cell Counting Kit-8 (CCK8) (Dojindo, Kumamoto, Japan) was used for detection of cell proliferation. MCF-7 cells were transfected with specific reagents for 24 h, then plated in a 96-well plate and cultivated for 24 h. After that, 10% CCK8 solution were added to the cell culture medium for a one-hour incubation till color change. OD value was determined by a microplate reader and proliferation rates were calculated.
EdU assay kit
MCF-7 cells were transfected as experiment design, then plated on sterile coverslips in 24-well plates. EdU kit (RiboBio) was used for detection of cell proliferation according to the manufacturer's instructions. Laser confocal microscopy (FV300, Olympus, Japan) was used to acquire the images. Nuclei that double stained with EdU and 4',6-diamidino-2-phenylindole (DAPI) (Beyotime, Shanghai, China) were considered to be positively proliferative cells.
Cell migration and Invasion
For wound healing analysis, 24 h after the transfection, the cells were plated in 6-well plates. 24 h later, the adherent cells were wounded by a 10 μl plastic pipette tip. Then rinsing the scathing cells with PBS and culturing with serum-free DMEM for 24 h. The wound closure in different groups was photographed and evaluated with the microscope. For transwell assay, 8-μm pore size chambers (Corning, NY, USA) with an insert coated Matrigel (BD Bioscience) were used. 24 h after the transfection, cells in serum-free DMEM were plated into the upper chamber, while 10% FBS DMEM was added into the lower chamber. After 24 h of incubation, the cells that invaded into the lower chamber through the membrane were fixed with 100% methanol for 15 minutes, then stained with 0.1% crystal violet for 20 minutes. Images of the cells were photographed with the microscope.
Immunofluorescence staining
For immunofluorescence staining, MCF-7 cells were fixed with 4% paraformaldehyde in PBS. Then incubating the cells with blocking solution (5% BSA and 0.1% Triton-X in PBS) for 2 h at room temperature, and subsequently blocked by 5% BSA dissolved in PBS for 2 h. Primary antibody against MMP3 (1:150) and cyclin D1 (1:150) were added overnight at 4 °C, followed by incubation with the matched secondary antibody (1:5000) (Invitrogen) for 1 h at 37 °C temperature. The nuclei were stained by DAPI (1:100) for 20 min at room temperature. Then cells were examined and analyzed by laser confocal microscopy (FV300, Olympus, Japan).
Statistical analysis
Group data was analyzed as mean ± S.E.M. Significant difference was analyzed by Student's t-test for two groups or one-way ANOVA accompanied by Bonferroni's Multiple Comparison Test for more than two groups. P< 0.05 was considered to be significant. Data were analyzed by the GraphPad Prism 5.0 software.
Results
MiR-519d is dysregulated in breast cancer
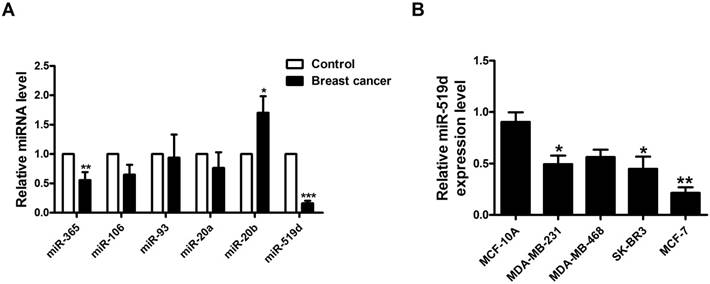
To validate the regulatory miRNAs of MMP3 in BC, we used TargetScan and microRNA.org to predict the complementary binding miRNAs with 3'UTR of MMP3. MiR-365, miR-106, miR-93, miR-20a, miR-20b, and miR-519d were tested in tumor and adjacent non-tumor tissues of BC patients by qRT-PCR. The results showed that miR-519d was the most downregulated one in BC tumor tissues compared to its relative control (Fig. 1A). We then tested the expression level of miR-519d in several breast cancer cell lines (MCF-7, SK-BR3, MDA-MB-468, MDA-MB-231). Compared with human mammary epithelial cells (MCF-10A), MCF-7 cells had a most significant downregulation of miR-519d (Fig. 1B) .
MMP3 acts as a downstream target of miR-519d
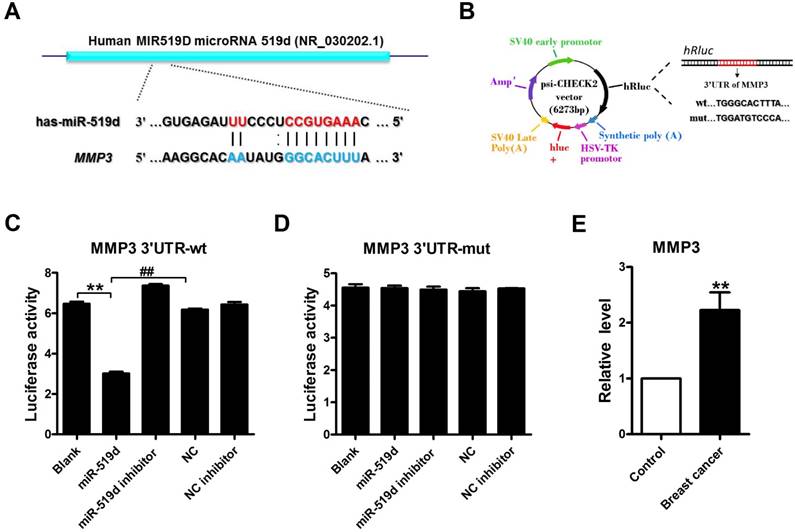
Luciferase assay was used to identify the inhibitory effect of miR-519d on MMP3 (Fig. 2A and B). The results demonstrated that miR-519d dramatically inhibited the luciferase activity of the constructed plasmid containing wild-type (WT) 3'UTR of MMP3, but not that of the mutant (Mut) 3' UTR (Fig. 2C and D). Subsequently, we found that MMP3 mRNA markedly overexpressed in tumor tissues compared to their matched non-tumor tissues (Fig. 2E). Moreover, MMP3 mRNA level was remarkable increased in MCF-7 cells (Supplementary Fig. 1). These data suggested that miR-519d was complementary base-pairing with 3'UTR of MMP3 and strongly inhibited its expression.
MiR-519d is dysregulated in breast cancer A. Expression levels of miR-365, miR-106, miR-93, miR-20a, miR-20b and miR-519d in 10 pairs of breast cancer and adjacent breast tissue specimens determined by qRT-PCR. *p< 0.05, **p< 0.01, ***p< 0.001 vs. Control. B. MiR-519d expression levels in MDA-MB-231, MDA-MB-468, SK-BR3, MCF-7 and MCF-10A cell lines, determined by qRT-PCR. The data are presented as mean ± SEM of three independent experiments. *p< 0.05, **p< 0.01 vs. MCF-10A.
MMP3 acts as a downstream target of miR-519dA. Sequence complementarity between miR-519d and MMP3 3'UTR in human. B. Luciferase reporter constructs containing 3'UTR of MMP3 or mutated 3'UTR of MMP3. (C and D). Luciferase reporter assay showed that transfected miR-519d could reduce the luciferase activity in wild-type (wt) of MMP3 but not in mutant (mut) 3'UTR of MMP3. **p<0.01, ##p< 0.01 vs. miR-519d. E. MMP3 expression levels in 15 paired breast cancer and adjacent breast tissue specimens determined by qRT-PCR. n=3 batches of cells. The data are presented as mean ± SEM. **p<0.01 vs. Control.
Overexpression of miR-519d inhibits MCF-7 cell viability or proliferation
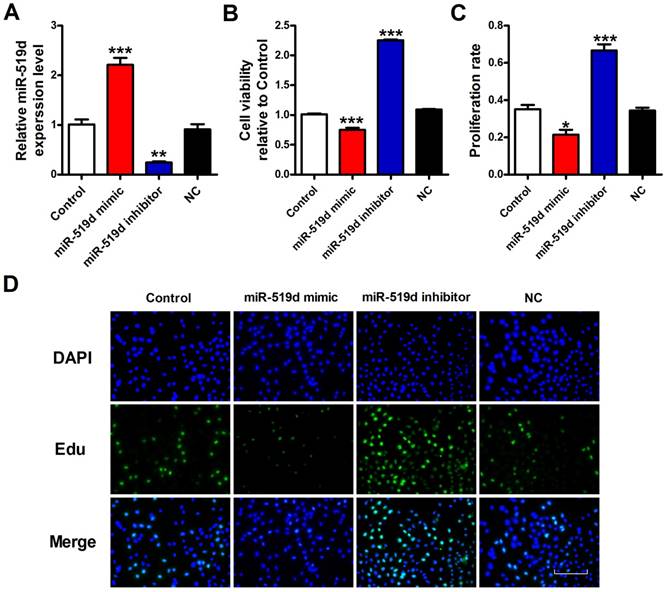
To explore the functional role of miR-519d in BC, we transfected miR-519d mimic for miR-519d overexpression, as well as miR-519d inhibitor for its suppression (Fig. 3A). CCK8 assay was used for measurement of cell viability. The results demonstrated that miR-519d overexpression remarkably inhibited MCF-7 cell viability, while miR-519d knockdown increased cell viability (Fig. 3B). Besides, EdU assay was performed to evaluate MCF-7 cell proliferation. As shown in figure 3C and 3D, less proliferation rate of MCF-7 cells were observed when treated with miR-519d mimic compared with the NC group, whereas a sharply increase was seen after transfection with miR-519d inhibitor. Consistent with Edu results, immunofluorescence analysis showed that miR-519d overexpression significantly decreased Ki67 expression in MCF-7 cells, while miR-519d inhibitor had a reverse effect (Supplementary Fig. 2). Furthermore, cell cycle analysis showed that miR-519d mimic decreased the percentage of MCF-7 cells in the S phase, miR-519d inhibitor increased the S phase (Supplementary Fig. 3). Cyclin D1 examination further demonstrated the cell cycle arrest by miR-519d mimic (Supplementary Fig. 4). These data revealed that miR-519d played a inhibitory role for BC proliferation.
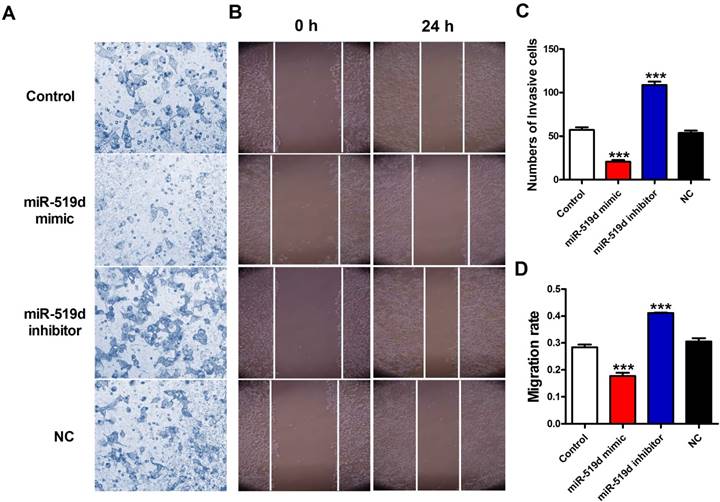
Inhibition of miR-519d promotes MCF-7 cell metastasis
Next, we detected the effects of miR-519d on migration capacities of MCF-7 cells. Transwell assay results demonstrated that the number of migratory cells were significantly reduced after miR-519d overexpression while increased after miR-519d downregulation (Figure 4A and 4C). In addition, the wound-healing assay showed that cells transfected with miR-519d mimic underwent a slower closing of scratch wounds compared with the NC group, but miR-519d inhibitor transfection accelerated the scratch closing (Figure 4B and 4D). Taken together, these results suggested that miR-519d inhibitor might act as a tumor suppressing tool through inhibiting cell proliferation, migration and invasion in BC.
Overexpression of miR-519d inhibits MCF-7 cell viability or proliferation A. MiR-519d expression levels after miR-519d mimic and inhibitor transfection in MCF-7 cells. B. MCF-7 cells viability were detected by CCK8 assay. C. The Edu positive cells were counted. D. The proliferation of MCF-7 cells was determined by EdU kit, assessed by fluorescence microscopy (200×). Representative images of Edu staining showing proliferation cells (stained in green). Nuclei were stained in blue with DAPI. Scale bar indicates 100 μm. The data are expressed as mean±SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p<0.001 vs. NC.
MMP3 is responsible for the pivotal role of miR-519d in MCF-7 cells
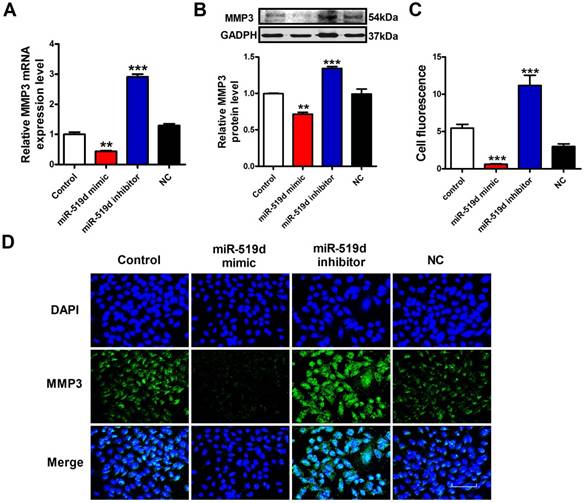
To further demonstrate whether MMP3 is the downstream regulatory target of miR-519d, we used qRT-PCR, WB and immunofluorescence staining to monitor the expression levels of MMP3 in MCF-7 cells. The results showed that the mRNA and protein levels of MMP3 were significantly decreased after miR-519d overexpression. On the contrary, inhibition of miR-519d increased MMP3 expression (Figure 5A, B and C). These data indicated that MMP3 was a direct target of miR-519d, and acted as an executor molecule for the carcinogenic effects of miR-519d in BC.
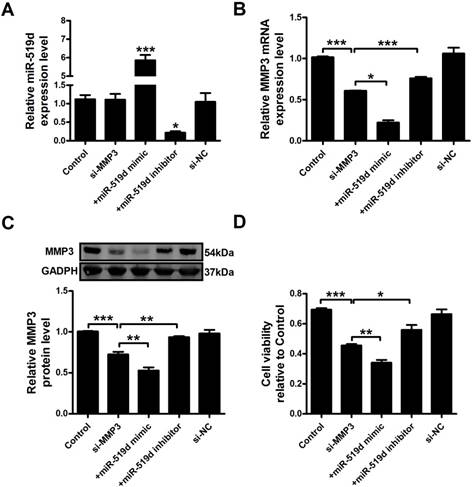
MMP3 siRNA has a synergistic effect with miR-519d mimic in MCF-7 cells
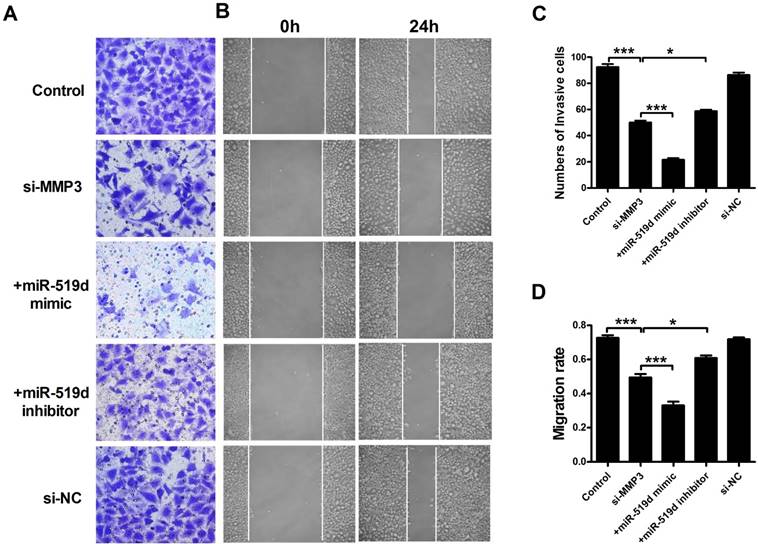
We further examined MCF-7 cell progression after inhibition of MMP3. The results showed that MMP3 siRNA remarkably inhibited its mRNA and protein expression. Co-transfection of MMP3 siRNA and miR-519d enhanced the inhibitory effect on MMP3, while miR-519d inhibitor reversed the MMP3 downregulation by siRNA (Figure 6A and B). CCK8 analysis (Figure 6C) and Ki67 staining (Supplementary Fig. 5) revealed that MMP3 inhibition resulted in a significant decrease in MCF-7 cell proliferation. MiR-519d mimic promoted the inhibitory effect of MMP3 siRNA. However, miR-519d inhibitor neutralized its promotion. Moreover, cyclin D1 detection (Supplementary Fig. 6) demonstrated the synergistic inhibitory effect of MMP3 siRNA and miR-519d mimic on MCF-7 cell proliferation. We further detected the cell metastasis by transwell healing (Figure 7A and C) and wound assays (Figure 7B and D). The results demonstrated a more severe inhibitory effects of MMP3 siRNA and miR-519d mimic on MCF-7 cell metastasis, while miR-519d inhibitor reserved this promotion effect.
Inhibition of miR-519d promotes MCF-7 cell metastasis (A and C). The effects of miR-519d mimic and miR-519d inhibitor transfection on the invasion of MCF-7 cells were assessed by transwell assay. (B and D). Wound healing assay showed that miR-519d overexpression resulted in a slower closing of scratch wounds, miR-519d inhibition caused a faster closing of scratch wounds. The data are expressed as mean±SEM of three independent experiments. ***p<0.001 vs. NC.
Discussion
Breast cancer is a clinically challenging issue that characterized by complicated variations in the genome and related signaling pathways [18]. To date, changes in epigenetic modification in cancer related genes have been the major focus. Consistently, our data suggested miR-519d was remarkably downregulated in breast cancer. Overexpression of miR-519d markedly suppressed the proliferation and metastasis capacities of MCF-7 cells, while miR-519d downregulation had the inverted effects. Moreover, we unraveled a novel regulatory mechanism of miR-519d and MMP3. These finding revealed that miR-519d might function as a tumor suppressor by directly targeting MMP3 in breast cancer.
Matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases that form a family of 24 members in mammals [19]. They facilitate cancer invasion and metastasis primarily by degradation of the extracellular matrix (ECM) surrounding tumor cells [20]. Evidence of the pathological roles of MMPs in various diseases, combined with their druggability, has made them attractive therapeutic targets [21]. Initial drug discovery efforts focused on the roles of MMPs in cancer progression, and inflammatory processes [22, 23]. Among them, MMP3 was identified as stromelysin. It can degrade numerous ECM substrates, including collagens III, IV, V, IX, X, and XI, laminins, elastin, entactin, fibronectin, fibrin, fibrillins [24]. The promotion effects of MMP3 on tumor invasive properties have been clearly uncovered [25]. Overexpressed MMP3 had been shown to drive the formation of mammary tumors in mice [26]. The hemopexin domain of MMP3 was responsible for mammary epithelial invasion [27]. In addition, MMP3 expression levels had been reported to be upregulated in BC cells and were associated with BC risk and survival [28]. In line with these published studies, we showed that MMP3 levels had a great increase in BC tumor samples compared with adjacent non-tumor samples. Moreover, inhibition of MMP3 by MMP3 siRNA remarkably decreased MCF-7 cell progression. Despite promising preclinical data supporting the blockade of matrix metalloproteinases (MMPs) as a treatment for cancer more than 10 years ago, all Phase III cancer trials failed owing to several reasons. Unwanted side effects and poor knowledge on the complexity of MMP function were the primary reasons [29]. Thereby, more studies of pharmacological mechanisms for MMPs were desiderated.
MiR-519d negatively regulates MMP3 expression A. QRT-PCR analysis of MMP3 mRNA levels after miR-519d mimic and inhibitor transfection. B. Western blot analysis of MMP3 protein levels. C.The fluorescence intensity of MMP3 were counted. D. Representative images of immunofluorescence staining for MMP3 (green) in MCF-7 cells. The nuclei were stained in blue with DAPI. Scale bar indicates 100 μm. The data are expressed as mean±SEM of three independent experiments. **p < 0.01 ***p<0.001 vs. NC.
MiR-519d inhibits MCF-7 cell proliferation by directly targeting MMP3 MiR-519d (A), MMP3 mRNA (B) levels after si-MMP3, si-MMP3 with miR-519d mimic and inhibitor transfection in MCF-7 cells. C. Western blot analysis of MMP3 protein levels. D. MCF-7 cells viability were detected by CCK8 assay. The data are expressed as mean±SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p<0.001 vs. si-MMP3.
MiR-519d inhibits MCF-7 cell migration and invasion by directly targeting MMP3 (A and C). The effects of si-MMP3, si-MMP3 with miR-519d mimic and inhibitor transfection on the invasion of MCF-7 cells were assessed by transwell assay. (B and D). Wound healing assay showed that silence the MMP3 resulted in a slower closing of scratch wounds, si-MMP3 with miR-519d inhibition caused a faster closing of scratch wounds. The data are expressed as mean±SEM of three independent experiments. *p < 0.05, ***p<0.001 vs. si-MMP3.
Plenty of studies have confirmed that miRNA dysregulation is causal in many cancers, with miRNAs acting as tumor suppressors or oncogenes, and miRNA mimics and inhibitors have shown promise in preclinical studies [30]. MiR-519d was identified as an important tumor related gene. But the regulatory role of miR-519d in diverse cancers was controversial. Hou et al. reported that miR-519d suppressed hepatocellular carcinoma (HCC) growth by targeting MKi67 [31]. On the contrary, Fornari el al. suggested miR-519d as an oncogenic gene [32]. In addition, the regulatory roles of miR-519d in gastric cancer [33], nasopharyngeal carcinoma [34], and cervical cancer [35] were also proved. There are some studies reporting the interactions between miR-519d and MMPs. Tsai et al reported the inhibitory effects of miR-519d on MMP2 in human chondrosarcoma cells [36]. Ding et al demonstrated miR-519d suppressed invasion and migration of trophoblast cells via targeting MMP2 [37]. In our study, we firstly identified MMP3 as a direct target of miR-519d using luciferase report assay. MiR-519d mimic induced the downregulation of MMP3 expression. Additionally, our data showed that the number of migrated and invasive cells were remarkably increased when miR-519d was upregulated. Co-transfection of miR-519d mimic and MMP3 siRNA enhanced MCF-7 cell proliferation and metastasis. While miR-519d inhibitor neutralized the cancer promotive effects of MMP3, which in reverse demonstrated the negatively regulatory effects of miR-519d on MMP3, as well as its effects on tumor cell progression. These data provided an evidence that the cell fate decision of miR-519d might depend upon its inhibitory effect of MMP3.
Taken together, our results demonstrated that miR-519d is downregulated in breast cancer, overexpression of miR-519d remarkably inhibits MCF-7 cell progression by targeting MMP3. However, it is undeniable that the effect of miR-519d on breast cancer was mediated by a variety of mechanisms besides our discovery. Moreover, it is hardly to identify the leading regulatory pathway among them. In summary, the present study revealed a new mechanism underlying the anti-tumor effects of miR-519d, that might serve as potential therapeutics for BC treatment.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (81773735, 81570399, 81270042) and the Program for New Century Excellent Talents In Heilongjiang Provincial University (1254-NCET-012).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
2. Tartari F, Santoni M, Pistelli M, Berardi R. Healthcare cost of HER2-positive and negative breast tumors in the United States (2012-2035). Cancer Treat Rev. 2017;60:12-17
3. Li Y, Jia R, Ge S. Role of Epigenetics in Uveal Melanoma. Int J Biol Sci. 2017;13:426-33
4. Gascard P, Bilenky M, Sigaroudinia M, Zhao J, Li L, Carles A, et a1. Epigenetic and transcriptional determinants of the human breast. Nat Commun. 2015;6:6351
5. Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34-49
6. Gilam A, Conde J, Weissglas-Volkov D, Oliva N, Friedman E, Artzi N, et a1. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat Commun. 2016;7:12868
7. Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, et a1. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci. 2017;13:1387-97
8. Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan KG, et a1. MiRNA Transcriptome Profiling of Spheroid-Enriched Cells with Cancer Stem Cell Properties in Human Breast MCF-7 Cell Line. Int J Biol Sci. 2016;12:427-45
9. Wang Z, Tan Y, Yu W, Zheng S, Zhang S, Sun L, et a1. Small role with big impact: miRNAs as communicators in the cross-talk between cancer-associated fibroblasts and cancer cells. Int J Biol Sci. 2017;13:339-48
10. Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222-29
11. Li X, Du N, Zhang Q, Li J, Chen X, Liu X, et a1. MicroRNA-30d regulates cardiomyocyte pyroptosis by directly targeting foxo3a in diabetic cardiomyopathy. Cell Death Dis. 2014;5:e1479
12. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
13. Zhang T, Hu Y, Ju J, Hou L, Li Z, Xiao D, et a1. Downregulation of miR-522 suppresses proliferation and metastasis of non-small cell lung cancer cells by directly targeting DENN/MADD domain containing 2D. Sci Rep. 2016;6:19346
14. Zhang Y, Liu X, Bai X, Lin Y, Li Z, Fu J, et a1. Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J Pineal Res. 2017 DOI: 10.1111/jpi.12449
15. Gennarino VA, D'angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, et a1. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163-72
16. Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52-67
17. Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9-34
18. Nielsen FC, Van Overeem Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16:599-612
19. Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287-97
20. Dufour A, Overall CM. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol Sci. 2013;34:233-42
21. Xian J, Shao H, Chen X, Zhang S, Quan J, Zou Q, et a1. Nucleophosmin Mutants Promote Adhesion, Migration and Invasion of Human Leukemia THP-1 Cells through MMPs Up-regulation via Ras/ERK MAPK Signaling. Int J Biol Sci. 2016;12:144-55
22. Rattanasinchai C, Llewellyn BJ, Conrad SE, Gallo KA. MLK3 regulates FRA-1 and MMPs to drive invasion and transendothelial migration in triple-negative breast cancer cells. Oncogenesis. 2017;6:e345
23. Shen N, Li X, Zhou T, Bilal MU, Du N, Hu Y, et a1. Shensong Yangxin Capsule prevents diabetic myocardial fibrosis by inhibiting TGF-beta1/Smad signaling. J Ethnopharmacol. 2014;157:161-70
24. Swarnakar S, Paul S, Singh LP, Reiter RJ. Matrix metalloproteinases in health and disease: regulation by melatonin. J Pineal Res. 2011;50:8-20
25. Shoshan E, Braeuer RR, Kamiya T, Mobley AK, Huang L, Vasquez ME, et a1. NFAT1 Directly Regulates IL8 and MMP3 to Promote Melanoma Tumor Growth and Metastasis. Cancer Res. 2016;76:3145-55
26. Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, et a1. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137-46
27. Correia AL, Mori H, Chen EI, Schmitt FC, Bissell MJ. The hemopexin domain of MMP3 is responsible for mammary epithelial invasion and morphogenesis through extracellular interaction with HSP90beta. Genes Dev. 2013;27:805-17
28. Aroner SA, Rosner BA, Tamimi RM, Tworoger SS, Baur N, Joos TO, et a1. Plasma matrix metalloproteinase 1, 3, and 7 levels and breast cancer risk in the Nurses' Health study. Cancer Causes Control. 2014;25:1717-23
29. Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov. 2014;13:904-27
30. Li Y, Sarkar FH. MicroRNA Targeted Therapeutic Approach for Pancreatic Cancer. Int J Biol Sci. 2016;12:326-37
31. Hou YY, Cao WW, Li L, Li SP, Liu T, Wan HY, et a1. MicroRNA-519d targets MKi67 and suppresses cell growth in the hepatocellular carcinoma cell line QGY-7703. Cancer Lett. 2011;307:182-90
32. Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, et a1. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275-85
33. Yue H, Tang B, Zhao Y, Niu Y, Yin P, Yang W, et a1. MIR-519d suppresses the gastric cancer epithelial-mesenchymal transition via Twist1 and inhibits Wnt/beta-catenin signaling pathway. Am J Transl Res. 2017;9:3654-64
34. Xu T, Xiao D. Oleuropein enhances radiation sensitivity of nasopharyngeal carcinoma by downregulating PDRG1 through HIF1alpha-repressed microRNA-519d. J Exp Clin Cancer Res. 2017;36:3
35. Zhou JY, Zheng SR, Liu J, Shi R, Yu HL, Wei M. MiR-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int. 2016;16:21
36. Tsai CH, Tsai HC, Huang HN, Hung CH, Hsu CJ, Fong YC, et a1. Resistin promotes tumor metastasis by down-regulation of miR-519d through the AMPK/p38 signaling pathway in human chondrosarcoma cells. Oncotarget. 2015;6:258-70
37. Ding J, Huang F, Wu G, Han T, Xu F, Weng D, et a1. MiR-519d-3p suppresses invasion and migration of trophoblast cells via targeting MMP-2. PLoS One. 2015;10:e0120321
Author contact
![]() Corresponding authors: Yong Zhang; E-mail: hmuzhangyongcom; Chaoqi Yan; Email: yanchaoqi2002com.
Corresponding authors: Yong Zhang; E-mail: hmuzhangyongcom; Chaoqi Yan; Email: yanchaoqi2002com.

 Global reach, higher impact
Global reach, higher impact