Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(4):427-436. doi:10.7150/ijbs.23890 This issue Cite
Research Paper
Generation of Cashmere Goats Carrying an EDAR Gene Mutant Using CRISPR-Cas9-Mediated Genome Editing
1. State Key Laboratory of Reproductive Regulation & Breeding of Grassland Livestock, Inner Mongolia University, Hohhot, 010070, P. R. China.
2. Wulanchabu Academy of Agricultural and Animal Husbandry Sciences, Wulanchabu, 012000, P. R. China.
Received 2017-11-16; Accepted 2018-2-25; Published 2018-3-11
Abstract
In recent years, while the use of the clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated protein 9 (Cas9) (CRISPR-Cas9) system for targeted genome editing has become a research hotspot, it has, to date, not proved adequate for genome editing in large mammals, such as goats. In this study, two opposite single-guide RNAs (sgRNAs) were designed for complete EDAR gene targeting in Cashmere goats, and co-transfected with a plasmid encoding Cas9 into goat fibroblasts. Among the 89 cell lines obtained through the cultivation of clonal cell lines, 62 were positive for EDAR gene targeting. Nine types of mutations were identified by sequencing analysis, and the mutation efficiency was 69.7%. Using one of these cell lines, EDAR gene-targeted Cashmere goat embryos were prepared by somatic cell cloning. Developed embryos were transferred to 79 Cashmere goat recipients, and, after a gestation period of five months six male EDAR gene-targeted Cashmere goats were born. Although only two of these goats survived, they had abnormal primary hair follicles and no hair on the top of their heads, which are the distinctive features of the EDAR gene-targeted Cashmere goats. Thus, this study provides a valuable animal model for future studies on EDAR gene-related phenotypes and hair follicle growth and development and shows that the CRISPR-Cas9 system can be used to edit genes in large mammals.
Keywords: CRISPR-Cas9, EDAR, goat, hair follicle, SCNT
Introduction
The morphogenesis and growth cycle of hair follicles (HFs), which are ectodermal accessory structures on mammalian skin, are controlled by many related signaling molecules. Along with hair growth, three processes occur during hair production: growth, cessation, and rest. In Cashmere goats, which have been used as model animals for studying the growth cycle of HFs [1-3], these structures are divided into primary and secondary HFs based on their temporal activities and structural characteristics. Primary HFs are responsible for the development of wool, and secondary HFs promote the development of cashmere [4, 5], which has been referred to as "soft gold." Therefore, understanding the molecular mechanisms controlling the growth and development of HFs might help in improving cashmere quality and yield.
Several findings have shown that the growth and development of primary and secondary HFs are regulated by different signaling pathways [6, 7]. Researchers have found that mice with spontaneous mutations in the ectodysplasin receptor (EDAR) gene lacked primary HFs; however, their secondary HFs developed normally [8, 9]. This finding indicated that the growth and development of primary HFs depend on the signaling pathway comprising the tumor necrosis factor (TNF) receptor homolog, EDAR [10, 11]. The EDAR gene is a member of the TNF receptor superfamily [12]. It consists of 12 exons and encodes the EDA-A1 receptor protein, which is a type-I transmembrane protein with an extracellular cysteine-rich domain and a potential intracellular death domain. This death domain initiates intracellular signaling by interacting with adapter proteins [13]. In mouse and human studies, the absence of EDAR gene function can lead to symptoms of hypohidrotic ectodermal dysplasia (HED) [14-16], including dysplasia of ectodermal appendages, such as hair, teeth, and exocrine glands [17]. No reports have described the effects of the EDAR gene on HF growth in Cashmere goats. Therefore, we investigated whether the EDAR gene can affect the growth and development of HFs, thus affecting cashmere yield, using EDAR-targeted Cashmere goats.
Currently, the most common gene-editing techniques used involve the zinc finger nucleases, transcription activator-like effector nucleases, and the most recently developed clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR-Cas9) system [18]. These gene-editing techniques involve engineering nucleases for cleaving a target site in a gene of interest, which, in turn, stimulates DNA repair mechanisms to complete gene modification [19]. In the CRISPR-Cas9 system, the target gene is recognized by single-guide RNAs (sgRNAs) that match the target gene and recognize the target in complex with the Cas9 protein [20]. Owing to its efficiency and advantages in terms of specificity, the CRISPR-Cas9 system has been widely used to edit genes in several species and examine gene functions in model organisms [21-26]. In this study, the CRISPR-Cas9 system and somatic cell nuclear transfer (SCNT) were used to obtain Cashmere goats with an EDAR gene mutation, providing an animal model and experimental material for studying the relationship between EDAR and HF growth and development. The efficiency and types of EDAR gene mutations in goat fibroblasts were also examined, and the comprehensive evidence of the efficiency and reliability of CRISPR-Cas9 and SCNT to generate targeted gene-modified livestock is shown.
Materials and methods
Ethics statement
All experiments performed followed the National Research Council Guide for the Care and Use of Laboratory Animals. All protocols were approved by the Institutional Animal Care and Use Committee of Inner Mongolia University. All animals were maintained at the Inner Mongolia YiWei White Cashmere Goat Limited Liability Company.
Production of sgRNAs
Using the free tool sgRNAcas9 [27] to analyze the coding sequence of EDAR, two sgRNAs were designed for targeting exon 6: sgRNA1 (5′-GGAGAACTTCTCCGCGGGGC-3′) and sgRNA2 (5′-CGGCGCCACAAGGACTGCGA-3′). Two sets of sgRNA targeted-DNA sequences were synthesized and annealed to form double-stranded DNA, which served as templates for evaluating the sgRNAs. Using the gRNA-T2 plasmid (Viewsolid Biotech, Beijing, China), the 5′ and 3′ sequences of both sgRNAs were amplified, resulting in two fragments of 319 bp and 177 bp, respectively. These two fragments were combined with the previously annealed DNA template to obtain the complete sgRNA (455 bp) for targeting exon 6 of EDAR, which was inserted into the pMD-19T vector (TaKaRa Bio, Shiga, Japan) and sequenced. Plasmid DNA was purified using the EndoFree® Plasmid Maxi Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.
Cell culture and transfection
Inner Mongolia Cashmere goat fetal fibroblasts (GFbs) were cultured in Dulbecco's modified Eagle's medium/F12 containing 10% fetal bovine serum in 5% CO2 at 37ºC. The sgRNA and Cas9 plasmids (Viewsolid Biotech) were transfected into the GFbs by electroporation at 225 v/2.5 ms, as previously described [28].
Clonal cell line screening
Twenty-four hours after transfection, the GFbs were digested in suspension with trypsin, diluted to 1 cell/100 µl, and seeded into 96-well cell culture plates for expansion. With each clonal cell line, half of the expanded cells were used to extract genomic DNA (gDNA) to identify the EDAR gene mutation, and the other half was cultured for use in further experiments. The EDAR mutant cell lines were confirmed by comparing the target sequences of wild-type cells and the clonal cell lines. Target sequence fragments were amplified by polymerase chain reaction (PCR) using the following primers: 5′-GTGGTGGTCGTCGTGGTGATGC-3′ and 5′-CTGCTCAGCCTTCCTTATGGTC-3′. The PCR conditions used were as follows: 94°C for 10 min; 35 cycles of 94°C for 30 s, 63°C for 45 s, and 72°C for 1 min; and 72°C for 10 min.
Preparation and transplantation of cloned embryos
The preparation and transplantation of cloned embryos were performed as previously described [29]. An EDAR mutant cell line was used as a donor in the preparation of the clonal Cashmere goat embryos. Oocytes used for SCNT were isolated from goat ovaries collected at a local slaughterhouse and cultured by in vitro maturation for 18 to 20 h. A single targeted GFb cell was transplanted into individual enucleated oocytes by microscopic manipulation, and the cytoplasmic membranes of the donor and recipient cells were then fused and activated to form reconstructed embryos. Finally, these embryos were cultured in an embryonic development medium at 38.5°C for 48 to 50 h. When the embryos had reached the two-to-eight-cell stage, they were transplanted into the oviduct of recipient Cashmere does.
Surveyor nuclease mutation-detection assay
Wild-type Alba white Cashmere GFbs were obtained and used as controls. These GFbs were transfected by electroporation, either with the sgRNA1 and Cas9 plasmids, or the sgRNA2 and Cas9 plasmids. At 48 h post-transfection, the gDNAs of the modified and wild-type cells were extracted using the cell genome extraction kit (TianGen Biotech, Beijing, China). These gDNAs were then used as templates to amplify the target sequence of the EDAR gene via PCR with the following primers 5′-GTGGTGGTCGTCGTGGTGATGC-3′ (forward) and 5′-CTGCTCAGCCTTCCTTATGGTC-3′ (reverse). The PCR conditions used were as described above. The resulting amplicons were purified and mixed with 10× La PCR Buffer II (TaKaRa Bio, Shiga, Japan), and a heteroduplex was formed by gradient annealing under the following conditions: 95°C for 10 min, 95 to 85°C ramping at -2°C/s, 85 to 25°C at -0.3°C/s, and a 4°C hold. The heteroduplexes were processed using the Surveyor Mutation Detection Kit (Transgenomic, Omaha, NE, USA) and run on a 2% agarose gel to detect mutation efficiency.
Western blot analysis
Expression of the EDAR gene in the skin samples of the EDAR gene-targeted Cashmere goats was detected by western blot analysis. Total protein was extracted from skin samples obtained from the head, back, and body side of the EDAR gene-targeted goats; separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis; and transferred to a nitrocellulose membrane. This membrane was blocked with 5% skimmed milk at room temperature for 1 h and incubated overnight at 4°C using an anti-EDAR antibody (1: 1,000 dilution) and an anti-β-actin antibody (1: 10,000 dilution) as a loading control. After incubation, the membrane was rinsed sequentially with phosphate-buffered saline and phosphate-buffered saline containing 0.05% Tween-20 solution. Subsequently, the membrane was treated with a secondary goat anti-rabbit antibody. Protein bands were visualized using the Tanon-5200 image-analysis system (Tanon Science & Technology Co., Ltd).
Skin tissue sections
Skin samples from the head, back, and body side of the EDAR gene-targeted Cashmere goats were fixed in 4% paraformaldehyde for 24 h, dehydrated using an alcohol-xylene series, and embedded in paraffin wax. Sample blocks were then cut into 5-µm sections using a microtome and routinely stained with hematoxylin and eosin. The growth and development of the HFs were analyzed by observing skin cross-sections under a microscope.
Off-target analysis
Potential off-target genome-editing sites (Table S1) were predicted using the free tool sgRNAcas9 [27]. Flanking sequences at each site were extracted, and the DNA sequence of each potential off-target site was amplified using the gDNA of wild-type and EDAR gene-targeted Cashmere goats as templates and the primers listed in Table S2. Variants were identified by multiple sequence alignments.
Statistical methods
Data are presented as the mean ± standard deviation. Significant differences were evaluated by performing student's t tests, and P < 0.05 was considered as the threshold for statistical significance.
Results
Efficiency of the CRISPR-Cas9 system in GFbs
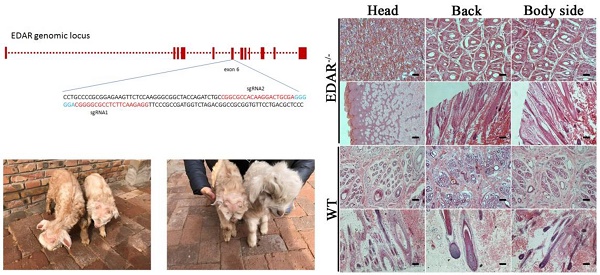
The specific target information for both opposite sgRNAs designed to remove EDAR gene function is shown in Figure 1A. To determine their cutting efficiencies, the sgRNA1, sgRNA2, and Cas9 plasmids were transfected into GFbs, and their gDNA was extracted at 48 h post-transfection. The target sequence was amplified by PCR and then analyzed using Surveyor nuclease to detect mutations. Mutations occurred in both samples in different degrees, indicating that both sgRNAs could be used in EDAR gene-targeting experiments (Figure 1B).
Mutation of the EDAR gene in clonal cell lines
Both the sgRNA plasmids and Cas9 plasmid were transfected into GFbs to establish 89 different EDAR gene-targeted cell lines, which were developed by culturing individual clones. The gDNA of each clonal cell line was extracted and the target sequence of the EDAR gene was obtained. Multiple sequence alignment revealed nine types of mutations (Figure 1D), and 62 cell lines presented mutations in the target region (Figure 2). Thus, the gene-targeting efficiency was 69.7%. The monoallelic mutation rate was 37.1% and the biallelic mutation rate was 32.6% (Table 1). Cell line 5069 grew well, showed a type-II mutation pattern (Figure 1C, 1D), and harbored a large deletion mutation in the targeted region of the EDAR gene. Therefore, this cell line was used as the donor to prepare cloned EDAR gene-mutant embryos.
EDAR gene targeting in the GFbs using the CRISPR-Cas9 system
| Monoallelic mutation | Biallelic mutation | Mutation-positive |
|---|---|---|
| 33/89 (37.1%) | 29/89 (32.6%) | 62/89 (69.7%) |
CRISPR-Cas9 system mediates EDAR gene targeting in GFbs. (a) Schematic representation of sgRNAs targeting exon 6 of the EDAR gene. The red letters represent the target sequence of each sgRNA. The blue letters indicate the PAM sequence. (b) Surveyor nuclease mutation-detection assay. Lanes 1 and 2: PCR products of the GFbs transfected with sgRNA1 + Cas9 or sgRNA2 + Cas9 plasmids, following treatment with Surveyor nuclease. Lanes 3 and 4: positive experimental group and negative experimental control group, respectively. M lane: 200-bp DNA Ladder Marker. (C) Cell line positive for a targeted EDAR gene mutation. (d) Nine different types of mutations occurred in the cell lines with EDAR gene mutations. The dotted line in the diagram represents the missing base fragment. The red box represents the inserted fragment. The red line at the top indicates the position of the sgRNA. WT, wild type
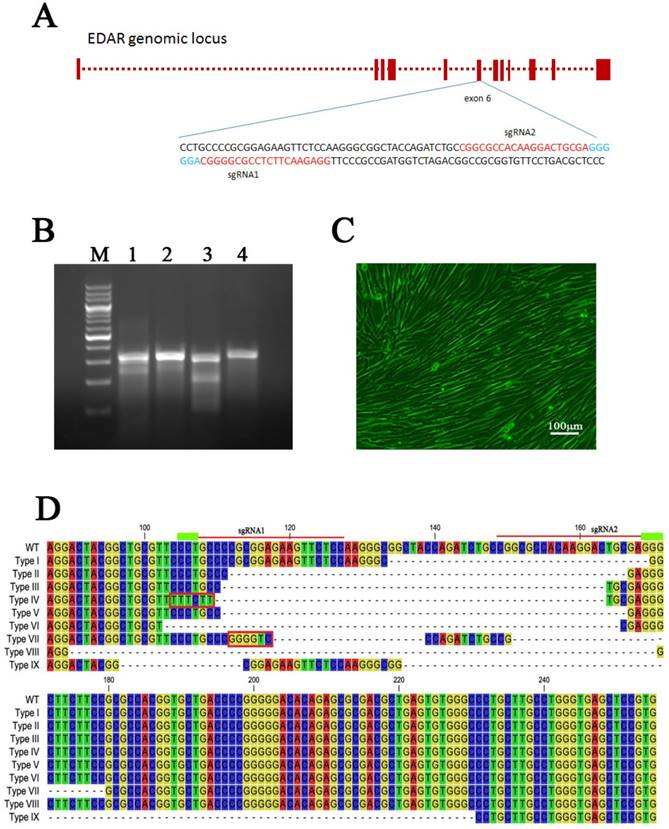
The EDAR gene-mutation genotype in the clonal cell lines. Green box: monoallelic mutation. Orange box: biallelic mutation
Cloned EDAR gene-targeted Cashmere goats
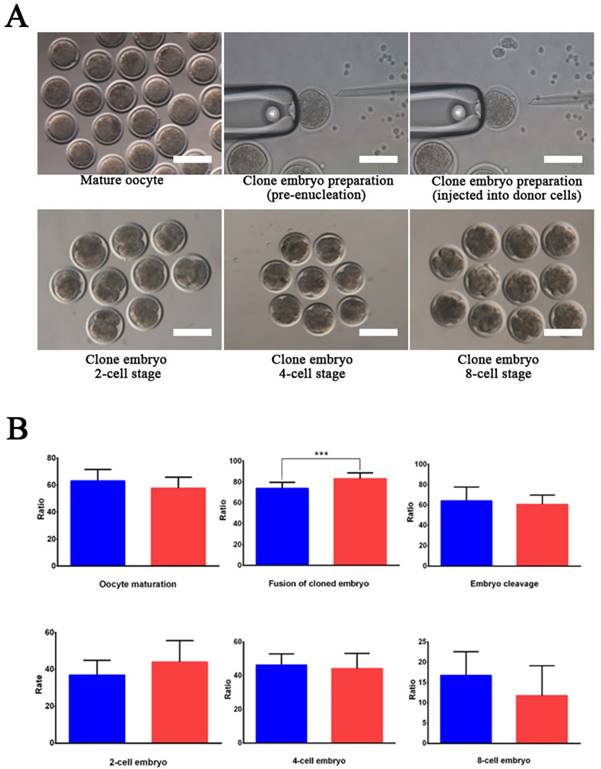
Wild-type and EDAR gene-targeted positive cells were used as nuclear donor cells. Regarding the use of wild-type cells, only 761 mature oocytes were obtained during in vitro culture of the 1,227 oocytes collected from the Cashmere goats. Among the 751 embryos that were successfully cloned, 545 embryos were fused and 542 were activated, finally resulting in 340 dividing embryos comprising 121 2-cell embryos, 160 4-cell embryos, and 59 8-cell embryos (Table 2). To construct cloned embryos from the EDAR gene-targeted positive cells, 3,339 oocytes were collected, and 1,875 mature oocytes were obtained in vitro. Among the 1,853 cloned embryos that were successfully prepared, 1,557 were fused and 1,543 were activated, finally resulting in 866 dividing embryos comprising 329 2-cell embryos, 430 4-cell embryos, and 107 8-cell embryos (Table 2). Comparison of the developmental stages of the embryos revealed that the fusion rate of the EDAR gene-targeted cloned embryos was significantly higher than that of the wild-type embryos (Figure 3B), but no significant difference was observed at other stages. Although 79 recipient goat embryo-transfer surgeries were performed (Table S3), only five resulted in pregnancy and the conception rate was 6.3%. After a gestation period of five months, six male goats were born— two stillbirths and four live goats; two of the live goats survived for only 20 h. The other two goats grew normally (Table 3), presenting the characteristic lack of hair on the top of their heads (Figure 4A).
Preparation of clonal embryos with EDAR gene mutations
| Type | Oocytes | Mature oocytes | Cloned embryos | Fused embryos | Activated embryos | Embryo type | ||
|---|---|---|---|---|---|---|---|---|
| 2-cell | 4-cell | 8-cell | ||||||
| EDAR-/- | 3,339 | 1,875 | 1,853 | 1,557 | 1,543 | 392 | 430 | 107 |
| WT | 1,227 | 761 | 751 | 545 | 542 | 121 | 160 | 59 |
The birth of EDAR-mutant Cashmere goats
| Recipient goat number | Date of lambing | EDAR mutant goat number | Lamb sex | Birth weight of lamb | Survival status | Phenotype |
|---|---|---|---|---|---|---|
| 404338 | 9-Mar-17 | 1701 | ♂ | 2.82 kg | Survived 24 h | No hair on the head |
| 916051 | 11-Mar-17 | 1702 | ♂ | 2.69 kg | Survived 20 h | No hair on the head |
| 916051 | 11-Mar-17 | 1703 | ♂ | 3.8 kg | Died | No hair on the head |
| 913006 | 14-Mar-17 | 1704 | ♂ | 5.2 kg | Died | No hair on the head |
| 922009 | 17-Mar-17 | 1705 | ♂ | 3.3 kg | Currently alive | No hair on the head |
| 118140 | 19-Mar-17 | 1706 | ♂ | 5.35 kg | Currently alive | No hair on the head |
EDAR gene-targeted cloned embryos. (A) The EDAR gene-targeted cloned embryos at various developmental stages. Scale bar = 100 μm. (B) Ratio of the cloned embryos at various developmental stages. Red indicates the EDAR gene-targeting cloned embryos and blue indicates the wild-type cloned embryos. (*** p <0.001)
Characterization of EDAR gene-targeted Cashmere goat
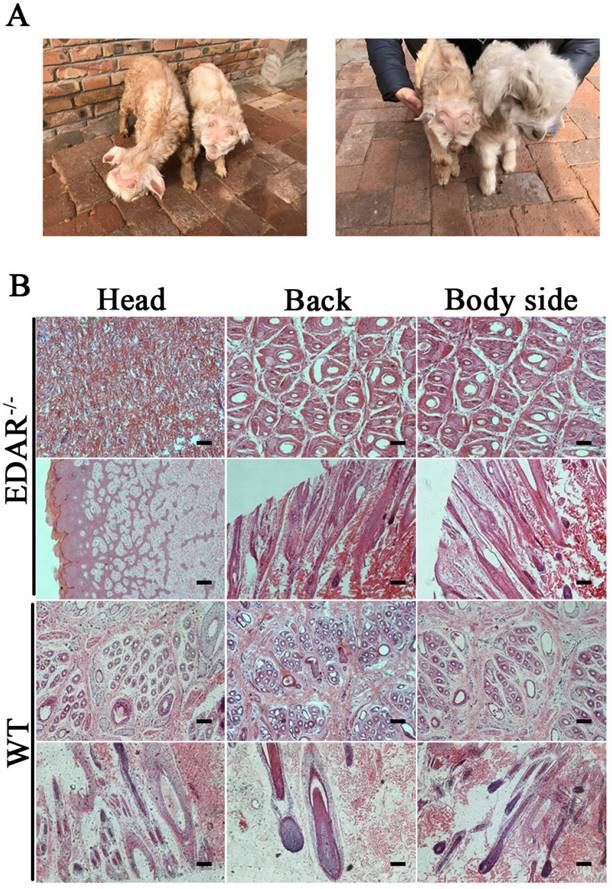
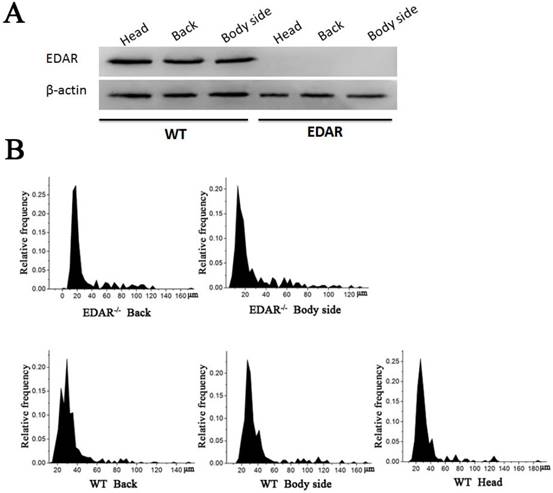
None of the newborn EDAR gene-targeted Cashmere goats had hairs on the top of their heads (Figure 4A). Histological analysis revealed that no HFs were present in the skin of the head, but back and body side skin samples had primary HFs of differing diameters randomly distributed near secondary HFs. This pattern differed from that of the wild-type samples, where three primary HFs were distributed along the side of the secondary HF clusters. However, the arrangement pattern of the secondary HFs was not significantly different between the wild-type and EDAR gene-targeted samples (Figure 4B). In addition, most HFs in the skin samples of the wild-type goats had a diameter of approximately 30 μm, whereas, most HFs in the skin samples of the EDAR gene-targeted goats had a diameter of approximately 20 μm. HFs larger than 50 μm were more frequent in the EDAR gene-targeted Cashmere goats than in the wild-type goats (Figure 5B). We could not detect EDAR protein expression by western blot analysis in the head, body, and back skin samples of the EDAR gene-targeted Cashmere goats (Figure 5A), indicating that EDAR was correctly silenced in these individuals.
EDAR gene-targeted Cashmere goats and skin tissue sections. (A) EDAR gene-targeted Cashmere goats. The left photo shows two surviving EDAR gene-targeted Cashmere goats, which displayed the characteristic hairlessness on their heads. The right panel shows a comparison of the EDAR gene-targeted Cashmere goats with the wild-type Cashmere goats. (B) Cross-sectional and longitudinal-section images of cutaneous tissues from different body parts of the EDAR gene-targeted Cashmere goats and wild-type Cashmere goats. Scale bar = 100 μm.
Off-target analysis
Although the CRISPR-Cas9 system shows high efficiency in gene editing, off-target mutations can occur. To detect off-target mutations in the EDAR gene-targeted goats, gDNA sequences compatible with sgRNA1 and sgRNA2 were examined, and 43 potential off-target sites were selected, amplified, sequenced, and analyzed. Although no off-target mutations were found, it is uncertain whether they occurred at untested sites.
Discussion
The high gene-editing efficiency of the CRISPR-Cas9 system has been confirmed in numerous studies. In this study, the gene-targeting efficiency of the CRISPR-Cas9 system in the GFbs was 69.7%, with a single-allelic mutation rate of 37.1% and a double-allelic mutation rate of 32.6%. To ensure the successful establishment of the EDAR gene-targeted Cashmere goats, two opposite sgRNAs were first designed to improve the targeting efficiency and promote a large fragment deletion. This improvement was confirmed experimentally. SCNT was also key to the success of the experiments performed in this study, as we decided not to inject the Cas9 mRNA and sgRNAs into fertilized eggs. Instead, we selected gene-targeted positive cells as nuclear donor cells and prepared the EDAR gene-targeted Cashmere goats by SCNT. This approach resulted in a low birth rate, but enabled a very high degree of gene editing that avoided uncertainty in the cloned Cashmere goat genotypes and provided a solid basis for subsequent experiments.
Western blotting and HF distribution. (A) No EDAR gene expression was detected in the head, back, and body of the EDAR gene-targeted Cashmere goats. (B) Distribution of HFs in the skin of the EDAR gene-targeted Cashmere goats.
By comparing the developmental stages of embryos, we found that the fusion rate of the EDAR gene-targeted cloned embryos was significantly higher than that of the wild-type embryos (Figure 3B), but no significant difference was observed at other stages. Because the EDAR gene-targeted positive cells have a larger diameter than the wild-type cells, these donor cell membranes have a larger area of contact with the oocyte's cellular membrane during somatic cell-cloning procedures, thus helping to reconstitute the cell membranes of embryos (i.e., fusion), thereby improving the fusion rate of the cloned EDAR gene-targeted embryos.
The EDAR gene-targeted Cashmere goats produced through CRISPR-Cas9-based gene editing followed by SCNT had no hair on the top of their heads (Figure 4A), and EDAR protein expression was not detected in their head skin tissues (Figure 5A). In addition, no HFs were found in the head skin samples of these goats, although the back and body skin samples presented primary HFs of different diameters randomly distributed around secondary HFs. This distribution was distinct from that found in the wild-type goats, where three primary HFs were distributed along the side of the secondary HF clusters. However, secondary HFs did not differ significantly between the EDAR gene-targeted and wild-type goats (Figure 4B). These findings agree with the results of a mouse EDAR gene-mutation study, which revealed that the EDAR gene affected the growth of primary HFs, but had no effect on secondary HFs [30]. Although the EDAR gene-targeted Cashmere goats provide a valuable animal model for future studies on the EDAR gene-related phenotypes, and HF growth and development, this model has some limitations, including the presence of sparse hair and dry skin, and the lack of sweat glands. Thus, the model faces an adaptability challenge. Because HFs first appear during the embryonic period [31, 32], it is necessary to study their growth and development from this stage, and the animal model presented here enables such evaluation.
Although the experiments of the present study established the founders of EDAR gene-targeted Cashmere goats, the animals have not reached sexual maturity or produced an F1 generation. Providing a suitable environment for these founders is particularly important for maintaining their health. Previous data have shown that abnormal epigenetic modifications might affect the phenotype of clonal animals produced by somatic cloning techniques [33], but such effects of epigenetic modifications are presently unknown for the EDAR gene-targeted Cashmere goats reported here. Epigenetic modifications can only be confirmed when the F1 generation is born.
Although no off-target mutations were found among the 43 potential off-target sites in the EDAR gene-targeted Cashmere goats, this finding does not confirm the lack of off-target mutations. The goat genome is not yet complete, and this limitation affected our search for potential off-targets in the genome. In addition, some clonal cell lines did not grow normally and could not be selected as donor cells for SCNT, which might reflect off-target mutations occurring in those cell lines.
In conclusion, the CRISPR-Cas9 system and SCNT can be used for gene editing in Cashmere goats. The EDAR gene-targeted Cashmere goats have abnormal primary HFs and no hair on the top of their heads. The findings of the present study provide a basis for studying gene editing in large mammals, as well as an animal model for studying EDAR gene functions in terms of HF growth and development.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by a National Transgenic Project of Genetically Modified Breeding grant (2014ZX08008-002). We are grateful to the Inner Mongolia YiWei White Cashmere Goat limited liability company for their support with the embryo-transfer experiments.
Abbreviations
EDAR: Ectodysplasin receptor; TNF: tumor necrosis factor; HED: hypohidrotic ectodermal dysplasia; HFs: hair follicles; CRISPR: clustered regularly interspaced short palindromic repeats; Cas9: CRISPR-associated protein 9; sgRNAs: single-guide RNAs; SCNT: somatic cell nuclear transfer; GFbs: goat fetal fibroblasts; gDNA: genomic DNA; PCR: polymerase chain reaction.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Faisal SM, Yan F, Chen TT, Useh NM, Guo S, Yan W. et al. Evaluation of a Salmonella vectored vaccine expressing Mycobacterium avium subsp. paratuberculosis antigens against challenge in a goat model. PloS One. 2013;8:e70171
2. Gao Y, Wang X, Yan H, Zeng J, Ma S, Niu Y. et al. Comparative transcriptome analysis of fetal skin reveals key genes related to hair follicle morphogenesis in Cashmere goats. PloS one. 2016;11:e0151118
3. Kon E, Filardo G, Robinson D, Eisman JA, Levy A, Zaslav K. et al. Osteochondral regeneration using a novel aragonite-hyaluronate bi-phasic scaffold in a goat model. Knee surgery, Sports Traumatology, Arthroscopy. 2014;22:1452-64
4. Qiao X, Wu JH, Wu RB, Su R, Li C, Zhang YJ. et al. Discovery of differentially expressed genes in cashmere goat (Capra hircus) hair follicles by RNA sequencing. Genetics and Molecular Research. 2016:15 doi: 10.4238/gmr.15038589
5. Shi B, Ding Q, He X, Zhu H, Niu Y, Cai B. et al. Tβ4-overexpression based on the piggyBac transposon system in cashmere goats alters hair fiber characteristics. Transgenic Research. 2017;26:77-85
6. Millar SE. Molecular mechanisms regulating hair follicle development. The Journal of Investigative Dermatology. 2002;118:216-25
7. Cui CY, Kunisada M, Piao Y, Childress V, Ko MS, Schlessinger D. Dkk4 and Eda regulate distinctive developmental mechanisms for subtypes of mouse hair. PloS One. 2010;5:e10009
8. Koppinen P, Pispa J, Laurikkala J, Thesleff I, Mikkola ML. Signaling and subcellular localization of the TNF receptor Edar. Experimental Cell Research. 2001;269:180-92
9. Laurikkala J, Pispa J, Jung HS, Nieminen P, Mikkola M, Wang X. et al. Regulation of hair follicle development by the TNF signal ectodysplasin and its receptor Edar. Development (Cambridge, England). 2002;129:2541-53
10. Hehlgans T, Pfeffer K. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology. 2005;115:1-20
11. Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B. et al. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Developmental Cell. 2009;17:49-61
12. Headon DJ, Overbeek PA. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nature Genetics. 1999;22:370-4
13. Fliniaux I, Mikkola ML, Lefebvre S, Thesleff I. Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Developmental Biology. 2008;320:60-71
14. Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, Ferguson B. et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nature Genetics. 1996;13:409-16
15. Kere J, Elomaa O. Healing a natural knockout of epithelial organogenesis. Trends in Molecular Medicine. 2002;8:197-200
16. van der Hout AH, Oudesluijs GG, Venema A, Verheij JB, Mol BG, Rump P. et al. Mutation screening of the Ectodysplasin-A receptor gene EDAR in hypohidrotic ectodermal dysplasia. European Journal of Human Genetics. 2008;16:673-9
17. Pozo-Molina G, Reyes-Reali J, Mendoza-Ramos MI, Villalobos-Molina R, Garrido-Guerrero E, Mendez-Cruz AR. Novel missense mutation in the EDA1 gene identified in a family with hypohidrotic ectodermal dysplasia. International Journal of Dermatology. 2015;54:790-4
18. Gaj T, Sirk SJ, Shui SL, Liu J. Genome-editing technologies: Principles and applications. Cold Spring Harbor Perspectives in Biology. 2016;8:a023754
19. Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology. 2013;31:397-405
20. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262-78
21. Albadri S, Del Bene F, Revenu C. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods. 2017;121-122:77-85
22. Guernet A, Grumolato L. CRISPR/Cas9 editing of the genome for cancer modeling. Methods. 2017;121-122:130-7
23. Li X, Zhou W, Ren Y, Tian X, Lv T, Wang Z. et al. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. Journal of Genetics and Genomics. 2017;44:175-8
24. Liu X, Xie C, Si H, Yang J. CRISPR/Cas9-mediated genome editing in plants. Methods. 2017;121-122:94-102
25. Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M. et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nature Communications. 2017;8:14716
26. Wang X, Yu H, Lei A, Zhou J, Zeng W, Zhu H. et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Scientific Reports. 2015;5:13878
27. Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS One. 2014;9:e100448
28. Niakan S, Heidari B, Akbari G, Nikousefat Z. Comparison of different electroporation parameters on transfection efficiency of sheep testicular cells. Cell Journal. 2016;18:425-37
29. Akshey YS, Malakar D, De AK, Jena MK, Pawar SK, Dutta R. et al. Effect of roscovitine treated donor cells and different activation methods on development of handmade cloned goat (Capra hircus) embryos. Theriogenology. 2011;75:1516-24
30. Pispa J, Pummila M, Barker PA, Thesleff I, Mikkola ML. Edar and Troy signalling pathways act redundantly to regulate initiation of hair follicle development. Human Molecular Genetics. 2008;17:3380-91
31. Sennett R, Rendl M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Seminars in Cell & Developmental Biology. 2012;23:917-27
32. Chen CC, Plikus MV, Tang PC, Widelitz RB, Chuong CM. The modulatable stem cell niche: Tissue interactions during hair and feather follicle regeneration. Journal of Molecular Biology. 2016;428:1423-40
33. Niemann H. Epigenetic reprogramming in mammalian species after SCNT-based cloning. Theriogenology. 2016;86:80-90
Author contact
![]() Corresponding author: Dongjun Liu (email: nmliudongjuncom)
Corresponding author: Dongjun Liu (email: nmliudongjuncom)

 Global reach, higher impact
Global reach, higher impact