Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(5):549-556. doi:10.7150/ijbs.24374 This issue Cite
Research Paper
Prognostic nomogram for patients with Nasopharyngeal Carcinoma incorporating hematological biomarkers and clinical characteristics
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, China.
#Authors contributed equally to this work.
*Authors contributed equally to this work.
Received 2017-12-15; Accepted 2018-2-16; Published 2018-4-5
Abstract
Predictive models for survival prediction in individual cancer patients following the tumor, node, and metastasis (TNM) staging system are limited. The survival rates of patients who share TNM stage diseases are diversified. Therefore, we established a nomogram in which hematological biomarkers and clinical characteristics for predicting the overall survival (OS) of nasopharyngeal carcinoma (NPC) patients were incorporated. The clinicopathological and follow-up data of 690 NPC patients who were histologically diagnosed histologically at the Sun Yat-sen University Cancer Center between July 2007 and December 2011 were retrospectively reviewed. Data was randomly divided into primary (n = 460) and validation groups (n = 230). Cox regression analysis was used to identify prognostic factors for building the nomogram in primary cohorts. The predictive accuracy and discriminative ability of the nomogram were measured by the concordance index (C-index) and decision curve, and were compared with the TNM staging system, Epstein-Barr virus DNA copy numbers (EBV DNA), or TMN stage plus EBV DNA. The results were internally validated by assessment of discrimination and calibration using the validation cohorts at the same institution. Independent factors selected into the nomogram for OS included age [hazard ratio (HR): 1.765; 95% confidence interval (CI): 1.008-3.090)], TNM stage (HR: 1.899; 95% CI: 1.023-3.525), EBV DNA (HR: 1.322; 95% CI: 1.087-1.607), lactate dehydrogenase level (LDH) (HR: 1.784; 95% CI: 1.032-3.086), high sensitivity C-reactive protein (hs-CRP) (HR: 1.840; 95% CI: 1.039-3.258), high-density lipoprotein cholesterol (HDL-C) (HR: 0.503; 95% CI: 0.282-0.896), hemoglobin (HGB) (HR: 0.539; 95% CI: 0.309-0.939) and lymphocyte to lymphocyte ratio (LMR) (HR:0.531; 95% CI: 0.293-0.962). The C-index in the primary cohort and validation cohort were 0.800 and 0.831, respectively, and were statistically higher when compared to C-index values for TNM stage (0.672 and 0. 716), EBV DNA (0.668 and 0.688), and TNM stage+ EBV DNA (0. 732 and 0. 760), P < 0.001 for all. Moreover, the decision curve analyses demonstrated that the nomogram model had a higher overall net benefit compared to the TNM staging system, EBV DNA and TNM stage+ EBV DNA. Next, patients were divided into three distinct risk groups for OS based on total points (TPs) of the nomogram: a low-risk group (TPs ≤ 19.0), an intermediate risk group (19.0 < TPs ≤ 25.5) and a high risk group (TPs > 25.5), respectively. The nomogram predicting prognosis generated for NPC patients had a higher predictive power compared to the TNM staging system, EBV DNA, and TNM stage+ EBV DNA.
Keywords: Nasopharyngeal carcinoma, nomogram, prognostic
Introduction
Nasopharyngeal carcinoma (NPC) is a fairly common malignant tumor in Southeast Asia, that is closely related to Epstein-Barr virus (EBV) infection [1]. Radiotherapy is the primary treatment for early stage disease, whereas concurrent chemoradiotherapy is the standard care for advanced NPC[2]. Currently, the Union Internationale Contre le Cancer/American Joint Cancer Committee (UICC/AJCC) TNM staging system is the gold standard for predicting prognosis and guiding treatment options of NPC patients. However, significant heterogeneities of clinical outcomes in NPC patients with a similar TNM stage and receiving similar treatment strategies have been reported [3-4]. These findings indicated that the present staging system is not adequate for prognosis without considering other clinicopathological factors or serum biomarkers. Thus, it is critical to identify reliable prognostic factors to complement the TNM staging system and refine the prediction of outcomes for NPC patients.
Recently, a number of studies have demonstrated that hematological biomarkers, such as systematic inflammation indexes (high sensitivity C-reactive protein (hs-CRP), platelet to lymphocyte ratio (PLR), neutrophil to lymphocyte ratio(NLR), lymphocyte to lymphocyte ratio (LMR)), lipid metabolism indexes (high-density lipoprotein cholesterol (HDL-C), apolipoprotein A-1 (ApoA-1)), nutritional indexes (albumin (ALB), hemoglobin (HGB)), lactate dehydrogenase level (LDH), EBV DNA copy numbers, titers of IgA antibodies against EBV capsid antigen (EBV VCA-IgA, EA-IgA) were associated with poor survival for NPC patients [5-7]. However, in most of these studies, only one or two biomarkers were evaluated without considering others. Moreover, few predictive models were used including these simple and effective factors. Therefore, in this study, we established a clinically useful nomogram in which hematological biomarkers and clinicopathologic risk factors were considered to predict OS for NPC. The nomogram was validated in the validation cohort. Moreover, a test was performed to compare the predictive power of the nomogram with EBV DNA alone and current popular staging systems.
Patients and method
A retrospective observational study was performed including 690 patients with histologically diagnosed NPC, who were treated at the Sun Yat-sen University Cancer Center between July 2007 and December 2011. The inclusion criteria were as follows: histologically confirmed primary NPC and complete baseline clinical information and laboratory data. The exclusion criteria included any of the following: systemic metastasis at the time of diagnosis or the presence of other malignancies, and a history of cancer treatments. Patients were randomly divided into a primary group (460 patients, about two thirds of data) and a validation groups (230 patients, remaining one third of data). For the development of the nomogram, the primary group was used, and the generalizability of the model was evaluated using the validation cohort. The last follow-up was performed in December 2015. This study was approved by the Hospital Ethics Committee in Sun Yat-sen University Cancer Center in China. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number as RDDA2017000449.
Cut-off values of prognostic biomarkers
Of all patients, clinical data were retrieved, including age, gender, therapeutic data (radiotherapy with or without chemotherapy), EBV DNA, EBV VCA-IgA, EA-IgA, LDH, hs-CRP, ALB, HDL-C, ApoA-1, HGB, PLR, NLR, LMR, and TNM staging system. The clinical stage was classified according to the seventh edition of the AJCC/UICC TNM staging system.
Continuous variables were transformed into categorical variables. In this study, each 10-fold increase in EBV DNA levels was chosen as the cut-off level, based on a previously published method [8]. Cut-off values for other variables were calculated using the median value of variables and were as follows: age (45 years), LDH (161.6 U/L), hs-CRP (2.03 mg/L), ALB (43.40 g/L), HDL-C (1.17 mmol/L), ApoA-1 (1.28 g/L), HGB (139 g/L), PLR (141.08), NLR (2.60), LMR (3.67), VCA-IgA titer (<1:80, 1:80-1:320, ≥1:640), and EA-IgA titer (<1:10, 1:10-1:20, ≥1:40).
Statistical analyses
Statistical analyses were performed using IBM SPSS Statistical software version 19.0 (IBMCorp., Chicago, IL, USA) and R version 3.4.0 (http://www.R-project.org/). OS was defined as the time from diagnosis to the date of the patient's death or censored at the date of last follow-up. Survival curves were plotted using the Kaplan-Meier survival analysis method and compared by the log-rank test. Variables achieving significant level of P < 0.05 in the univariate analyses were subjected to multivariable Cox regression analysis. Independent prognostic factors were determined if a significant effect was observed in the Cox model (P < 0.05). To maximize the predictive ability of the model, all variables in the multivariable model were used to establish a prognostic nomogram model (by the package of rms in R). Calibration of the nomogram for 1-, 3-, and 5-year OS were performed by comparing predicted survival with observed survival. The predictive accuracy and discriminative ability of the nomogram were measured by C-index and decision curve and were compared with the TNM staging system, EBV DNA, and TMN stage plus EBV DNA. All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
Results
Patient characteristics and survival
A total of 460 patients from the primary and 230 patients from the validation cohorts were included for analyses. Patient demographic and clinical characteristics are summarized in Table 1. No differences were found between the primary and validation cohorts in terms of age, node stage, TNM stage, EBV DNA, VCA-IgA, EA-IgA, LDH, hs-CRP, ALB, HDL-C, ApoA-1, PLR, NLR, or LMR (P = 0.063-1.000). However, parameters including gender, tumor stage, treatment, and HGB were significantly different between the two cohorts (P = 0.002-0.045). For primary and validation cohorts, the median follow-up times were 46.3 months (range = 0.7 to 76.0 months) and 69.9 months (range = 2.6 to 87.7 months), respectively. In addition, the 1-, 3-, and 5-year OS were 97.2%, 87.4%, 82.7%, and 95.7%, 82.6%, 74.8% for the primary and validation cohorts, respectively.
Univariate analysis and multivariate analysis
Univariate analysis indicated that age (P < 0.001), Tumor stage (P = 0.002), Node stage (P < 0.001), TNM stage (P < 0.001), EBV DNA (P < 0.001), VCA-IgA (P = 0.013), EA-IgA (P = 0.010), LDH (P < 0.001), hs-CRP (P < 0.001), ALB (P = 0.036), HDL-C (P = 0.001), ApoA-1 (P = 0.044), HGB (P = 0.001), PLR (P = 0.035), NLR (P = 0.016) and LMR (P < 0.001) were associated with OS of NPC patients. In multivariate analysis for OS with Cox regression, the results showed that the following variables remained independently prognostic: age (P = 0.002, HR = 1.765; 95% CI: 1.008-3.090), TNM stage (P = 0.042, HR = 1.899; 95% CI: 1.023-3.525), EBV DNA (P = 0.005, HR = 1.322; 95% CI: 1.087-1.607), LDH (P = 0.038, HR = 1.784; 95% CI: 1.032-3.086), hs-CRP (P = 0.037, HR = 1.840; 95%CI: 1.039-3.258), HDL-C (P = 0.020, HR = 0.503; 95% CI: 0.282-0.896), HGB (P = 0.029, HR = 0.539; 95% CI: 0.309-0.939), and LMR (P = 0.037, HR = 0.531; 95% CI: 0.293-0.962). The detailed results of univariate and multivariate analyses are presented in Table 2.
Patient demographics and clinical characteristics
Characteristic | All patients | Primary cohort | Validation cohort | ||
|---|---|---|---|---|---|
| No(%) | No. (%) | No. (%) | P | ||
| Total | 690 | 460 | 230 | ||
| Gender | 0.045 | ||||
| Male | 505(73.2%) | 348(75.7%) | 157(68.3%) | ||
| Female | 185(26.8%) | 112(24.3%) | 73(31.7%) | ||
| Age(years) | 1.000 | ||||
| ≤45 | 345(75.7%) | 230(50.0%) | 115(50.0%) | ||
| >45 | 345(24.3%) | 230(50.0%) | 115(50.0%) | ||
| Tumor stage | 0.024 | ||||
| T1 | 41(5.9%) | 29(6.3%) | 12(5.2%) | ||
| T2 | 146(21.2%) | 82(17.8%) | 64(27.8%) | ||
| T3 | 309(44.8%) | 217(47.2%) | 92(40.0%) | ||
| T4 | 194(28.1%) | 132(28.7%) | 62(27.0%) | ||
| Node stage | 0.523 | ||||
| N0 | 101(14.6%) | 64(13.9%) | 37(16.1%) | ||
| N1 | 270(39.1%) | 186(40.4%) | 84(36.5%) | ||
| N2 | 242(35.1%) | 164(35.7%) | 79(34.3%) | ||
| N3 | 77(11.2%) | 46(10.0%) | 30(13.0%) | ||
| TNM stage | 0.090 | ||||
| I | 19(2.8%) | 13(2.8%) | 6(2.6%) | ||
| II | 83(12.0%) | 46(10.0%) | 37(16.1%) | ||
| III | 324(47.0%) | 227(49.3%) | 97(42.2%) | ||
| IV | 264(38.3%) | 174(37.8%) | 90(39.1%) | ||
| Treatment | 0.024 | ||||
| Radiotherapy | 104(15.1%) | 59(12.8%) | 45(19.6%) | ||
| Chemoradiotherapy | 586(84.9%) | 401(87.2%) | 185(80.4%) | ||
| EBVDNA, copy/mL | 0.086 | ||||
| <103 | 311(45.1%) | 213(46.3%) | 98(42.6%) | ||
| 103~9999 | 150(21.7%) | 90(19.6%) | 60(26.1%) | ||
| 10 4~99999 | 127(18.4%) | 93(20.2%) | 34(14.8%) | ||
| 105~999999 | 64(9.3%) | 43(9.3%) | 21(9.1%) | ||
| ≥ 106 | 38(5.5%) | 21(4.6%) | 17(7.4%) | ||
| VCA-IgA | 0.427 | ||||
| <1:80 | 113(16.4%) | 72(15.7%) | 41(17.8%) | ||
| 1:80-1:320 | 416(60.3%) | 286(62.2%) | 132(57.4%) | ||
| ≥ 1:640 | 159(23.0%) | 102(22.2%) | 57(24.8%) | ||
| EA-IgA | 0.196 | ||||
| <1:10 | 207(30.0%) | 135(29.3%) | 72(31.3%) | ||
| 1:10-1:20 | 226(32.8%) | 161(35.0%) | 65(28.3%) | ||
| ≥1:40 | 257(37.2%) | 164(35.7%) | 93(40.4%) | ||
| LDH, U/L | 0.572 | ||||
| ≤161.6 | 339(49.1%) | 230(50.0%) | 109(47.4%) | ||
| >161.6 | 351(50.9%) | 230(50.0%) | 121(52.6%) | ||
| hs-CRP,mg/L | 0.106 | ||||
| ≤2.03 | 362(52.5%) | 231(50.2%) | 131(57.0%) | ||
| >2.03 | 328(47.5%) | 229(59.8%) | 99(43.0%) | ||
| ALB, g/L | 0.125 | ||||
| ≤43.40 | 349(50.6%) | 223(48.5%) | 126(54.8%) | ||
| >43.40 | 341(49.4%) | 237(51.5%) | 104(45.2%) | ||
| HLD-C, mmol/L | 0.936 | ||||
| ≤1.17 | 347(50.3%) | 232(50.4%) | 115(50.0%) | ||
| >1.17 | 343(49.7%) | 228(49.6%) | 115(50.0%) | ||
| ApoA-1, g/L | 0.063 | ||||
| ≤1.28 | 329(47.7%) | 231(50.2%) | 98(42.6%) | ||
| >1.28 | 361(52.3%) | 229(59.8%) | 132(57.4%) | ||
| HGB, g/L | 0.002 | ||||
| ≤139 | 375(54.3%) | 231(50.2%) | 144(62.6%) | ||
| >139 | 315(45.7%) | 229(59.8%) | 86(37.4%) | ||
| PLR | 0.872 | ||||
| ≤141.08 | 347(50.3%) | 230(50.0%) | 117(50.9%) | ||
| >141.08 | 343(49.7%) | 230(50.0%) | 113(49.1%) | ||
| NLR | 0.258 | ||||
| ≤2.60 | 356(51.6%) | 230(50.0%) | 126(54.8%) | ||
| >2.60 | 334(48.4%) | 230(50.0%) | 104(45.2%) | ||
| LMR | 0.809 | ||||
| ≤3.67 | 349(50.6%) | 231(50.2%) | 118(51.3%) | ||
| >3.67 | 341(49.4%) | 229(59.8%) | 115(48.7%) |
Univariate and Multivariable Analysis for overall survival of the Primary Cohort
Variable | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | |||||
| Male/ Female | 0.886(0.508~1.546) | 0.670 | |||
| Age(years) | |||||
| ≤45/>45 | 2.607(1.566~4.342) | <0.001 | 1.765(1.008~3.090) | 0.047 | |
| Tumor stage | |||||
| T1/T2/T3/T4 | 1.613(1.184~2.200) | 0.002 | 0.943(0.623~1.429) | 0.783 | |
| Node stage | |||||
| N0/N1/N2/N3 | 1.858(1.399~2.468) | <0.001 | 1.176(0.849~1.628) | 0.330 | |
| TNM stage | |||||
| I/II/III/IV | 3.083(1.999~4.756) | <0.001 | 1.899(1.023~3.525) | 0.042 | |
| Treatment | |||||
| Radiotherapy/ Chemoradiotherapy | 2.090(0.842~5.188) | 0.112 | |||
| EBVDNA, copy/mL | |||||
| <103/103~9999/104~99999/105~99999/≥106 | 1.674(1.409~1.988) | <0.001 | 1.322(1.087~1.607) | 0.005 | |
| VCA-IgA | |||||
| <1:80/1:80-1:320/≥ 1:640 | 1.624(1.109~2.378) | 0.013 | 0.966(0.576~1.620) | 0.895 | |
| EA-IgA | |||||
| <1:10/1:10-1:20/≥1:40 | 1.486(1.100~2.009) | 0.010 | 1.055(0.714~1.558) | 0.788 | |
| LDH, U/L | |||||
| ≤161.6/>161.6 | 2.594(1.558~4.319) | <0.001 | 1.784(1.032~3.086) | 0.038 | |
| hs-CRP, mg/L | |||||
| ≤2.03/>2.03 | 3.451(2.019~5.899) | <0.001 | 1.840(1.039~3.258) | 0.037 | |
| ALB, g/L | |||||
| ≤43.4/>43.4 | 0.603(0.376~0.968) | 0.036 | 1.235(0.739~2.062) | 0.421 | |
| HLD-C, mmol/L | |||||
| ≤1.17/>1.17 | 0.446(0.271~0.734) | 0.001 | 0.503(0.282~0.896) | 0.020 | |
| APOPA1, g/L | |||||
| ≤1.28/>1.28 | 0.613(0.381~0.988) | 0.044 | 0.862(0.488~1.521) | 0.607 | |
| HGB, g/L | |||||
| ≤139/>139 | 0.416(0.252~0.688) | 0.001 | 0.539(0.309~0.939) | 0.029 | |
| PLR | |||||
| ≤141.08/>141.08 | 1.674(1.036~2.703) | 0.035 | 1.265(0.706~2.265) | 0.430 | |
| NLR | |||||
| ≤2.60/>2.60 | 1.805(1.114~2.926) | 0.016 | 0.839(0.455~1.547) | 0.574 | |
| LMR | |||||
| ≤3.67/>3.67 | 0.374(0.223~0.627) | <0.001 | 0.531(0.293~0.962) | 0.037 | |
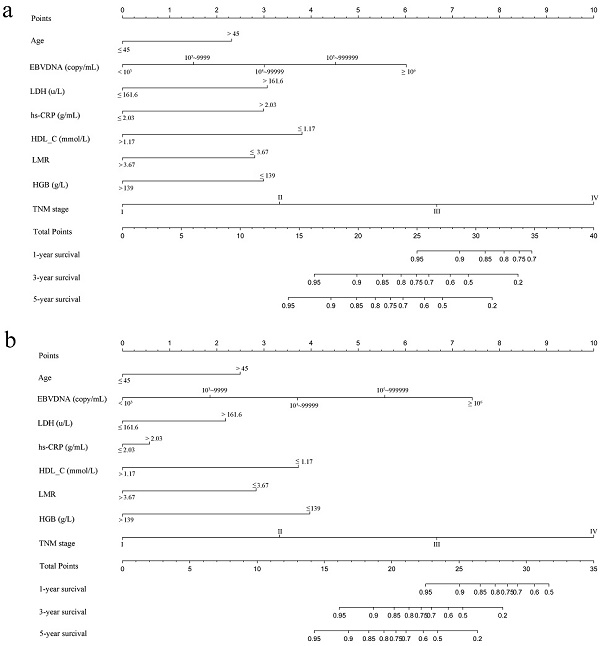
Construction of the Nomogram
Based on multivariate analysis in primary cohorts, a nomogram was constructed for OS prediction and involved all independent prognostic factors that were mentioned above in the primary cohort (Figure 1a) and validation cohort (Figure 1b). The larger points in the nomogram indicated a shorter OS. The TMN stage had the greatest impact on OS, which was subsequently followed by EBV DNA, HDL-C, LDH, hs-CRP, HGB, LMR, and age. The model demonstrated good accuracy for predicting OS rate of NPC, with a C-index of 0.800 (95% CI, 0.751-0.849). The calibration plot for the prediction of 1-year, 3-year and 5-year OS is shown in Figure 2a, 2b, and 2c. The calibration plots revealed good prediction of 1-,3- and 5-year OS.
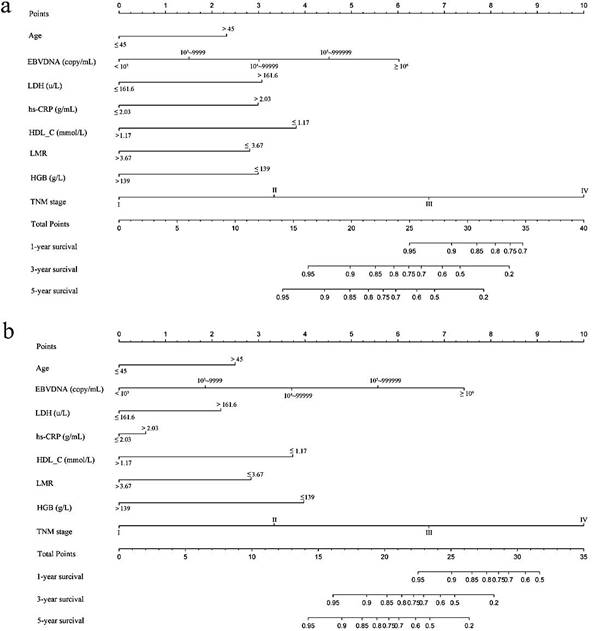
Validation of the predictive accuracy of nomograms for OS
In the validation cohorts, the C-index of the nomogram for predicting OS was 0.831 (95% CI, 0.783 ~ 0.879), and the calibration curve showed a good relation between the nomogram prediction and actual observation in the probability of 1-year, 3-year and 5-year survival (Figure 2d, 2e,and 2f).
Comparison of predictive accuracy between nomogram, EBV DNA, and TMN staging systems
In the primary cohort, the C-index of nomogram was 0.800 (95% CI = 0.751 to 0.849), which was higher than the C-index of TMN stage, EBV DNA and TMN stage + EBV DNA, with values of 0.672 (95% CI = 0.620 to 0.724, P < 0.001),0.668 (95% CI = 0.604 to 0.732, P < 0.001), and 0.732 (95% CI = 0.675 to 0.789, P < 0.001), respectively. In the validation cohort, significant differences were observed in the C-index between the nomogram and others (0.831 vs. TMN stage: 0.716; EBV DNA: 0.685; and TMN stage + EBV DNA; 0.760; P < 0.001 for all, respectively), and all C-indexes were significantly lower compared to that of the nomogram. The results are presented in Table 3.
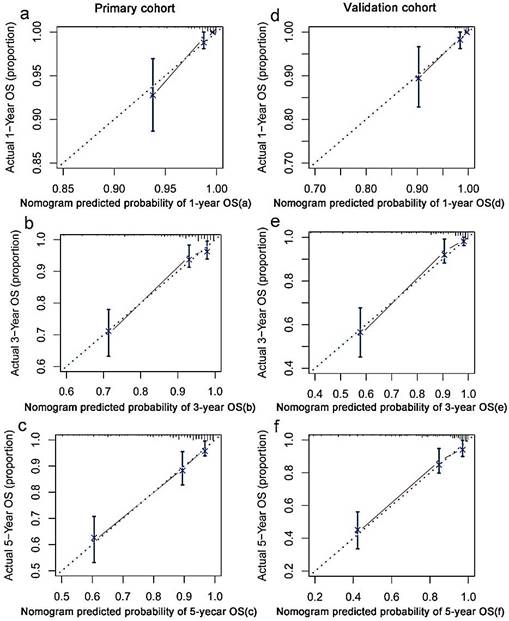
The decision curve analysis
In the decision curve analysis, the y-axis indicated the net benefit, which was calculated by summing the benefits (true positives) and subtracting the harms (false positives). The nomogram model (black dotted line) had the highest net benefit when compared with the TNM staging system (red dotted line). The straight line represented the assumption that all patients will die, and the horizontal line represented the assumption that no patients will die. Decision curve analysis showed that the nomogram had a higher net clinical benefit compared to TMN stage, EBV DNA and TMN stage + EBV DNA across a wider range of threshold probabilities for predicting OS in the primary cohort (Figure 3a) and in the validation cohort (Figure 3b), respectively.
Risk stratification of OS
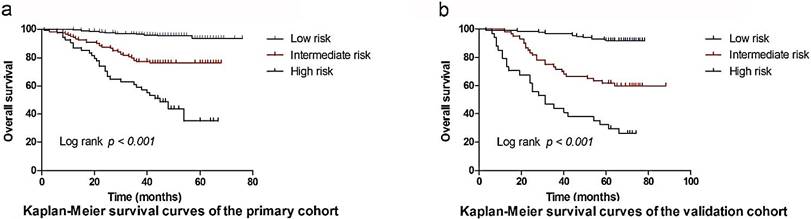
Based on the cut-off values of the total points determined by the X-tile program [9], patients were subdivided into a low-risk group (3.25-19.00), an intermediate-risk group (19.25-25.50), and a high-risk group (25.75-33.75) for OS in primary, and validation cohort. The OS rates among the 3 risk groups in the primary cohort were 93.8%, 76.2%, and 34.9% (p <0.001, Figure 4a), respectively. Likewise, significant differences were observed for OS in the validation cohort (OS: 91.9% for the low-risk group, 59.9% for the intermediate-risk group, and 26.1% for the high-risk group; P < 0.001, Figure 4b). This stratification could effectively discriminate the survival outcomes for the three proposed risk groups in both primary and validation cohorts.
Discussion
In the present study, we successfully established a nomogram model for predicting OS in patients with NPC, by incorporating hematological risk factors and clinical characteristics. Our nomogram model showed improved prognostic accuracy in NPC patients.
Compared with the current staging system, our model showed predictive accuracy and discriminative ability. Although in previous studies nomograms were established for predicting NPC survival, our study had several advantages compared to other reports. In nomogram models described by Tang, et al.[10] and Yang, et al.[11], nomogram models, they not included these common systemic inflammation parameters, such as NLR, PLR and LMR were not considered. Systemic inflammation has been shown to be involved in the initiation, development, and progression of several types of cancers [12-13]. Considering importance of inflammation in the prognosis of cancer, we investigated the prognostic factors including several systemic inflammation parameters, such as NLR, PLR and LMR. Multivariate analysis showed that only LMR was an independent risk prognostic factors in NPC patients. The addition of LMR into our model, combined with other biomarkers provided a good model to predict OS. Moreover, patients were divided into three distinct risk groups for OS based on total points of the nomogram. The Kaplan-Meier survival curves of OS separated very well both in primary cohorts and validation cohort. These findings indicated that maybe a higher total point scores were related to a higher risk of death. Therefore, different treatment strategies and active surveillance could be followed in patients in the three groups.
Nomogram model predicting 1-, 3- and 5- year OS in NPC patients. The nomogram was used summing the points identified on the points scale for each variable. The total points projected on the bottom scales indicate the probability of 1-, 3- and 5-year survival. a: The nomogram of primary cohort; b: The nomogram of validation cohort;
The C-indexes of nomograms, TNM stage ,EBV DNA, and TNM stage+ EBV DNA for prediction of OS in the primary cohort and validation cohort
Factors | Primary cohort | Prospective validation cohort | |||
|---|---|---|---|---|---|
| C-index (95% CI) | P | C-index (95% CI) | P | ||
| Nomograms | 0.800(0.751~0.849) | 0.831(0.783~0.879) | |||
| TNM stage | 0.672(0.620~0.724) | 0.716(0.653~0.779) | |||
| EBV DNA | 0.668(0.604~0.732) | 0.685(0.630~0.740) | |||
| TNM stage+ EBV DNA | 0.732(0.675~0.789) | 0.760(0.704~0.816) | |||
| Nomogram vs TNM stage | <0.001 | <0.001 | |||
| Nomogram vs EBV DNA | <0.001 | <0.001 | |||
| Nomogram vs TNM stage+ EBV DNA | <0.001 | <0.001 | |||
Nomogram: including eight risk factors (Age, TNM stage, pretreatment hs-CRP, LDH,HDL-C, HGB, LMR,and EBV DNA); C-index = concordance index; CI = confidence interval; EBV DNA = Epstein-Barr virus DNA; P values are calculated based on normal approximation using function rcorrp.cens in Hmisc package
Based on the multivariate analysis results, we identified and incorporated 8 independent clinical factors into the nomogram, including age, TNM staging system, EBV-DNA, LDH, hs-CRP, HDL-C, HGB and LMR. In line with previous studies, each of these markers were associated with prediction of outcomes in NPC patients. In our nomogram, TNM staging system and EBV DNA contributed the most to predicting OS of NPC patients. It has been widely accepted that the TNM staging system was the most important prognostic factor for NPC patients [14]. Pretreatment of EBV DNA has been shown a useful prognostic factor for the clinical management of NPC, and has been used to evaluate the disease development and treatment effectiveness [15-16]. Liaw et al.was the first to report the relationship between pretreatment with serum LDH and NPC [17]. Until now, many studies have shown that the LDH level at diagnosis is of prognostic significance in NPC patients [17-19]. Serum hs-CRP levels were associated with the acute phase of inflammation, which have been demonstrated to be correlated with a poorer prognosis in NPC patients [20]. Moreover, high-density serum lipoproteins play a fundamental role in supplying cholesterol to tumors, potentially via receptor-mediated mechanisms [21], and predicted outcome in cancer patients [22-23]. Several studies suggested that anemia correlated with a detrimental tumor oxygenation status [24-25], therefore HGB appeared to affect cancer survival. LMR reflected the diverse effects of monocytes and lymphocytes on tumor progression, which was a significant predictor of prognosis in NPC [26].
The calibration curves for predicting patient OS at (a) one year, (b) three years and (c) five years in the primary cohort and at (d) one year, (e) three years and (f) five years in the validation cohort. Nomogram model-predicted OS is plotted on the x-axis; actual OS is plotted on the y-axis. Closer alignment with the diagonal line represents a better estimation.
Many studies have shown that several molecular markers were associated with the survival of NPC patients, including epidermal growth factor receptor (EGFR) [27], long noncoding RNA [28] and microRNAs [29]. However, these prognostic factors were not included in our model, because measuring molecular markers were not routinely tested in our institution. Moreover, the high costs of molecular testing, as well as the resulting increasing patients' economic burden were reasons not to include these prognostic factors. Thus, It was not suitable for the application of basic medical institutions in China. So our nomogram was a simple, inexpensive and easily measurable tool to predict OS of NPC patients.
Decision curve analysis for 5-year survival predictions. a: The decision curve of primary cohort; b: The decision curve of validation cohort.
The nomogram showed excellent discrimination ability, which was better than the TNM staging system, EBV DNA, and TNM stage + EBV DNA. The Harrell's C-index of the nomogram was 0.800, which was higher than that of the TNM stage (0.672), EBV DNA (0.668), and TNM stage + EBV DNA (0.732) in the primary cohorts. Similar results were found in the validation cohorts. The decision curve analysis results also showed that our newly established nomogram model had a higher overall net benefit, compared to the traditional TNM staging system, EBV DNA, and TNM stage + EBV DNA across a wide range of threshold probabilities. Thus, the above results indicated that our nomogram was a reliable and precise prognostic tool to predict OS in NPC patients.
Although our nomogram represented a helpful tool for clinicians to choose and plan treatment strategies for NPC patients, our study has several limitations. First, like all retrospective study analysis, there may be a potential source for selection bias. Second, our study only evaluated the prognostic values for predicting the OS in NPC patients, and the effect of our nomogram for predicting disease-free survival (DFS) in NPC patients was not evaluated. It was better clinical application that nomogram combined with OS and DFS. Third, our sample size was relatively small and the samples were obtained from a single center. Thus, it is warranted to obtain a larger sample size from other institutes to validate the model.
In conclusion, we have established a predictive nomogram for OS in the NPC patients, which was more accurate and precise in predicting the OS compared with the traditional TNM staging system and EBV-DNA alone. The model is simple, inexpensive, and useful for clinicians in decision-making and individual patient counselling. To confirm our findings, additional studies are required using a larger, multi-center study population.
Graphs showing the results of Kaplan-Meier curves for all three groups based on the predictor from the nomogram model in the primary cohort (a) and those in the validation data set (b).
Acknowledgements
This work was supported by the Open Project Program of the State Key Laboratory of Proteomics (SKLPO201703). We thank the staff at the Director of Clinical Laboratories, Sun Yat-sen University Cancer Center for providing support on research conditions in this study.
Author contributions
All authors contributed to this manuscript, including conception and design (JPL, SLC), acquisition of data (SGP), analysis and interpretation of data (SX), material support (YJL), study supervision (XH, HC), and writing, review and revision of the manuscript (JPL, SLC, SGP, SX, XH, HC).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chinese Journal of Cancer Research. 2011;30:114-9
2. Blanchard P, Lee A, Marguet S, Leclercq J, Ng WT, Ma J. et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncology. 2015;16:645
3. Mao YP, Xie FY, Liu LZ, Sun Y, Li L, Tang LL. et al. Re-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imaging. International Journal of Radiation Oncology Biology Physics. 2009;73:1326
4. Wang HY, Sun BY, Zhu ZH, Chang ET, To KF, Hwang JS. et al. Eight-signature classifier for prediction of nasopharyngeal [corrected] carcinoma survival. Journal of clinical oncology. 2011;29:4516-25
5. Tang LQ, Li CF, Chen QY, Zhang L, Lai XP, He Y. et al. High-sensitivity C-reactive protein complements plasma Epstein-Barr virus deoxyribonucleic acid prognostication in nasopharyngeal carcinoma: a large-scale retrospective and prospective cohort study. International Journal of Radiation Oncology*Biology*Physics. 2015;91:325-36
6. Jiang R, Zou X, Hu W, Fan YY, Yan Y, Zhang MX. et al. The elevated pretreatment platelet-to-lymphocyte ratio predicts poor outcome in nasopharyngeal carcinoma patients. Tumor Biology. 2015;36:7775-87
7. Chua MLK, Tan SH, Kusumawidjaja G, Ma TTS, Cheah SL, Fong KW. et al. Neutrophil-to-lymphocyte ratio as a prognostic marker in locally advanced nasopharyngeal carcinoma: A pooled analysis of two randomised controlled trials. European Journal of Cancer. 2016;67:119-129
8. Lo YM, Chan AT, Chan LY, Leung SF, Lam CW, Huang DP. et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer research. 2000;60:6878
9. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252-9
10. Tang LQ, Li CF, Li J, Chen WH, Chen QY, Yuan LX. et al. Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma. Journal of the National Cancer Institute. 2015;108:djv291
11. Yang L, Hong S, Wang Y, Chen H, Liang S, Peng P. et al. Development and External Validation of Nomograms for Predicting Survival in Nasopharyngeal Carcinoma Patients after Definitive Radiotherapy. Scientific reports. 2015;5:15638
12. Diakos CI, Charles KA, Mcmillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncology. 2014;15:493-503
13. Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nature Reviews Clinical Oncology. 2015;12:584-96
14. Xu L, Pan J, Wu J, Pan C, Zhang Y, Lin S. et al. Factors associated with overall survival in 1706 patients with nasopharyngeal carcinoma: significance of intensive neoadjuvant chemotherapy and radiation break. Radiother Oncol. 2010;96:94-9
15. Chan KC. Plasma Epstein-Barr virus DNA as a biomarker for nasopharyngeal carcinoma. Chin J Cancer. 2014;33:598-603
16. Wang WY, Twu CW, Chen HH, Jiang RS, Wu CT, Liang KL. et al. Long-term survival analysis of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA levels. cancer. 2013;119:963-70
17. Liaw CC, Wang CH, Huang JS, Kiu MC, Chen JS, Chang HK. Serum lactate dehydrogenase level in patients with nasopharyngeal carcinoma. Acta Oncologica. 1997;36:159-64
18. Wan XB, Wei L, Li H, Dong M, Lin Q, Ma XK. et al. High pretreatment serum lactate dehydrogenase level correlates with disease relapse and predicts an inferior outcome in locally advanced nasopharyngeal carcinoma. European Journal of Cancer. 2013;49:2356-64
19. Tao CJ, Liu X, Tang LL, Mao YP, Chen L, Li WF. et al. Prognostic scoring system for locoregional control among the patients with nasopharyngeal carcinoma treated by intensity-modulated radiotherapy. Chinese Journal of Cancer Research. 2013;32:494-501
20. Xia WX, Zhang HB, Shi JL, Lu X, Wang L, Ye YF. et al. A prognostic model predicts the risk of distant metastasis and death for patients with nasopharyngeal carcinoma based on pre-treatment serum C-reactive protein and N-classification. European Journal of Cancer. 2013;49:2152-60
21. Cruz PMR, Mo H, Mcconathy WJ, Sabnis N, Lacko AG. The role of cholesterol metabolism and cholesterol transport in carcinogenesis: a review of scientific findings, relevant to future cancer therapeutics. Frontiers in Pharmacology. 2013;4:119
22. Pakzad R, Safiri S. The effect of preoperative serum triglycerides and high-density lipoprotein-cholesterol levels on the prognosis of breast cancer: Methodological issue. Breast. 2017
23. Liu YY, Lin SJ, Chen YY, Liu LN, Bao LB, Tang LQ. et al. High-density lipoprotein cholesterol as a predictor of poor survival in patients with nasopharyngeal carcinoma. Oncotarget. 2016;7:42978
24. Vaupel UP, Mayer A, Höckel M. Impact of Hemoglobin Levels on Tumor Oxygenation: the Higher, the Better? Strahlentherapie und Onkologie. 2006;182:63-71
25. Vaupel P, Mayer A. Hypoxia and anemia: effects on tumor biology and treatment resistance. Transfusion Clinique Et Biologique. 2005;12:5-10
26. Li J, Jiang R, Liu WS, Liu Q, Xu M, Feng QS. et al. A large cohort study reveals the association of elevated peripheral blood lymphocyte-to-monocyte ratio with favorable prognosis in nasopharyngeal carcinoma. PloS one. 2013;8:e83069
27. Kyzas PA, Cunha IW, Ioannidis JPA. Prognostic Significance of Vascular Endothelial Growth Factor Immunohistochemical Expression in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2005;11:1434-40
28. Zhang W, Wang L, Zheng F, Zou R, Xie C, Guo Q. et al. Long Noncoding RNA Expression Signatures of Metastatic Nasopharyngeal Carcinoma and Their Prognostic Value. Biomed Research International. 2015;2015:618924
29. Spence T, Bruce J, Yip KW, Liu FF. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:17
Author contact
![]() Corresponding author: Hao Chen, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, P. R. China (e-mail: chenhaoorg.cn). Tel./Fax: +86-20-8734-3438
Corresponding author: Hao Chen, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, P. R. China (e-mail: chenhaoorg.cn). Tel./Fax: +86-20-8734-3438

 Global reach, higher impact
Global reach, higher impact