Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(6):608-615. doi:10.7150/ijbs.22763 This issue Cite
Research Paper
Screening of multimeric β-xylosidases from the gut microbiome of a higher termite, Globitermes brachycerastes
1. College of Plant Protection, Hunan Agricultural University, Changsha, 410128, China;
2. Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200032, China.
3. Department of Entomology, University of Kentucky, Lexington, KY, 40546-0091, USA.
* These authors contributed equally to this work.
Received 2017-9-9; Accepted 2018-1-3; Published 2018-4-26
Abstract
Termite gut microbiome is a rich reservoir for glycoside hydrolases, a suite of enzymes critical for the degradation of lignocellulosic biomass. To search for hemicellulases, we screened 12,000 clones from a fosmid gut library of a higher termite, Globitermes brachycerastes. As a common Southeastern Asian genus, Globitermes distributes predominantly in tropical rain forests and relies on the lignocellulases from themselves and bacterial symbionts to digest wood. In total, 22 positive clones with β-xylosidase activity were isolated, in which 11 representing different restriction fragment length polymorphism (RFLP) patterns were pooled and subjected to 454 pyrosequencing. As a result, eight putative β-xylosidases were cloned and heterologously expressed in Escherichia coli BL21 competent cells. After purification using Ni-NTA affinity chromatography, recombinant G. brachycerastes symbiotic β-xylosidases were characterized enzymatically, including their pH and temperature optimum. In addition to β-xylosidase activity, four of them also exhibited either β-glucosidase or α-arabinosidases activities, suggesting the existence of bifunctional hemicellulases in the gut microbiome of G. brachycerastes. In comparison to multimeric protein engineering, the involvement of naturally occurring multifunctional biocatalysts streamlines the genetic modification procedures and simplifies the overall production processes. Alternatively, these multimeric enzymes could serve as the substitutes for β-glucosidase, β-xylosidase and α-arabinosidase to facilitate a wide range of industrial applications, including food processing, animal feed, environment and waste management, and biomass conversion.
Keywords: Globitermes brachycerastes, gut microbiome, fosmid library, glycoside hydrolases, β-xylosidase, multimeric hemicellulase
Introduction
Lignocellulosic biomass is one of the most abundant carbon resources in nature. Following celluloses, hemicelluloses, the second most abundant polysaccharide in plants, can be utilized through the full-plant biomass bioconversion [1, 2]. Hemicellulose, unlike the crystalline cellulose, is composed of a random, amorphous matrix of polysaccharide structure containing various pentose and hexose sugars [3]. L-arabinoside residue exists largely in hemicellulose, and L-arabinoside residue is attached to the main chain of xylose with monomer or oligomer lateral branch [4]. The complete hydrolysis of hemicelluloses requires endo-xylanase (EC.3.2.1.8), exo-β-xylosidase (EC.3.2.1.37), a-L-arabinofuranosidase (EC 3.2.1.53), mannanase and arabic xylanase to release xylose molecules [5-7]. β-xylosidase has been reported to synergistically act with other enzymes in the degradation processing of xylan, which breaks down into multiple monosaccharides [8]. Specifically, it breaks down xylan by hydrolyzing xylobiose to xylose from the nonreducing end and by decreasing the inhibition of endo-xylanases by the end product of xylan hydrolysis [9]. The synergism among hemicellulases leads to highly efficient degradation processing of xylan, which breaks down the bonds between backbone residues and substituent groups. Therefore, it is critical to identify and characterize individual enzymes involved in this process and to utilize them in the conversion of agricultural byproducts into products with industrial applications [4, 10].
Termites, the most efficient bioreactor in nature, have a substantial ecological impact on global carbon recycling [11]. Termite guts harbor a dense assemblage of microorganisms that are essential for lignocellulose digestion [12, 13]. The major players in higher termites consist of bacteria and archaea, whereas in lower termites cellulolytic flagellates contribute greatly to degrade plant biomass [14]. However, the relative importance of endogenous enzymes in the foregut and midgut to the overall degradation process remains to be assessed [15]. Recent studies have revealed that nasute termites possess cellulase producing microbes in three gut regions: foregut, midgut and hindgut [16]. Enzymatic properties of termite hindguts have indicated that it can provide large amounts of novel enzymes for future biorefining processes [17, 18]. Their abundance of digestive enzymes provided by both the symbionts and the host make termites a model bioreactor for biotechnological applications, particularly the conversion of lignocelluloses to biofuels.
G. brachycerastes, a subterranean higher termite, belongs to the genus Globitermes and family Termitidae. As a common Southeast Asian wood-feeding genus, Globitermes has three species, and distributes predominantly in tropical rain forests. G. brachycerastes relies on the lignocellulases from themselves and bacterial symbionts to digest woody materials. The 16S rRNA analysis of G. brachycerastes gut metagenome showed that Spirochaetaceae, a group of spiral-shaped bacteria, was the most abundant [19]. Han et al (2013) cloned and characterized a bacterial xylanase, Xyl-ORF19, from G. brachyceraste gut symbionts [20].
Metagenomic screening allows us to identify genes-of-interest from unculturable microorganisms, in which the total are extracted directly from samples, collecting from different environments. DNA fragments are cloned into cosmid, fosmid or bacterial artificial chromosome (BAC) libraries [21-23]. Then, libraries are used to screen for functional clones. Metagenomics provides a powerful tool to search for lignocellulolytic enzymes for future bio-refinery processes. Some of these novel sequences are hitherto unknown from the natural environment, including the microbial communities of thermophilic methanogenic digester, termite guts, biogas slurry, yak rumen and ocean floors [17, 24-28].
Based on the preliminary research, we hypothesized that termite guts contain novel β-xylosidases, which has potential for industrial application. To examine this hypothesis, we carried out the following objectives: 1) screened β-xylosidases from an existing fosmid library of G. brachycerastes gut metagenome [29]; 2) pyrosequenced selected positive clones containing β-xylosidase activity; 3) heterologously expressed resultant β-xylosidases in E. coli BL21 competent cells; and 4) enzymatically characterized these recombinant enzymes and discussed their potential in industrial application.
Materials and Methods
Chemicals and reagents
All chemicals were purchased from Sangon Biotech (Shanghai, China). MiniBEST plasmid purification kit and restrictions enzymes were purchased from Takara (Dalian, China). The large-construction DNA kit for extracting the DNA of positive clones was obtained from Qiagen (Shanghai, China), while DNA Gel Extraction Kit was purchased from Axygen (Hangzhou, China). Clone Express®II-One Step Cloning Kit was obtained from Vazyme (Nanjing, China) and 5-Br-4-Chl-3-Indole β-D-xylopyranoside was purchased from Energy chemical (Shanghai, China). The protein concentration was determined by Modified Bradford Protein Assay Kit (Sangon Biotech, Shanghai, China). HisSep Ni-NTA Agarose Resin was purchased from Yeasen (Shanghai, China).
Termite collection and sample preparation
G. brachycerastes (family Termitidae, subfamily Amitermitinae) was collected from an underground nest in Xishuang Banna, Yunnan Province, China, in March 2008. Termites were firstly surface sterilized in 70% ethanol for 1 min, rinsed in phosphate-buffered saline (PBS), and then the entire digestive tract of the worker termites was immediately removed and transferred into a 1.5 mL sterilized Eppendorf tube containing 100 µl of PBS. The whole gut of about 2,500 G. brachycerastes workers was used for the metagenomic DNA extraction. Intestinal microbial DNA extraction was carried out according to Liu et al (2011) [30]. pCC2FOS vector was used for the fosmid library construction. β-xylosidase genes were cloned into the expression vector pET28a, and expressed in E.coli BL21 competent cells (Novagen, Shanghai, China).
Functional screening for β-xylosidases
5-Bromo-4-chloro-3-indolyl β-D-xylopyranoside was used as a chromogenic substrate to screen for β-xylosidase activity [31]. The plates were incubated at 50 °C for 30 min and the development of turquoise color indicates the presence of β-xylosidase activity. In the secondary screening, p-nitrophenyl-β-D-xylopyranoside (pNPX) and p-nitrophenyl-β-D-glucoside (pNPG) were used as the substrates to detect the corresponding β-glucosidase and arabinosidase activity, respectively. The clones with higher enzyme activity or higher thermal stability in the acid environment (pH 5.0) were subjected to 454 pyrosequencing to identify hemicellulases.
The fosmid clones were preserved in 384-well plates and the 384-pin replicator was used to inoculate the fosmid clones. The fosmid clones were inoculated in Luria-Bertani (LB) agar with 12.5 µg/mL of chloramphenicol and then incubated at 37 °C for 16-18 h. To screen for β-xylosidase activity, 5-Bromo-4-chloro-3-indolyl β-D-xylopyranoside was added into the 0.7% agarose with a final concentration of 140 µg/mL [17]. The agarose plates were incubated at 50 °C for 30 min. Positive clones were visually observed based on the colony coloration or haloes around the colony.
Sequencing of fosmid positive clones
Each positive fosmid clone was extracted by the MiniBEST plasmid purification kit and verified by RFLP using BamHI digestion. Selected fosmid positive clones (11) were mixed and extracted by the large-construction DNA kit according to the manufacturer instruction. The DNA samples were sheared and then subjected to shotgun pyrosequencing with 454 FLX sequencing system as described previously [26]. The 454 reads were assembled by Newbler software V.2.7 (http//www.454.com). Open reading frames (ORFs) were predicted by National Center for Biotechnology Information's Non-Redundant (NCBI-NR) database. For the eight GH genes, we located their nearest neighbors using BlastP (NCBI database), evolutionary distance was calculated by the maximum likehood, and the phylogenetic relationship was resolved using MEGA version 5.0 [32, 33]. The bootstrap values were obtained based on 1,000 replications. Signature domains of each protein were identified by the search against NCBI's Conserved Domain Database (CDD).
Cloning, expression and purification of β-xylosidases
To express β-xylosidases in the recombinant protein systems, ORFs encoding GH1, GH3 and GH43 enzymes were cloned from the fosmid DNA (Table S1). All the polymerase chain reaction (PCR) products were purified using DNA Gel Extraction Kit. The purified fragments were ligated into the pET28a (+) vector with BamHI/HindIII restriction site and transformed into E. coli BL21. Each clone was incubated at 37 °C overnight in LB medium containing 50 µg/mL of kanamycin. Protein expression products was subsequently induced by the addition of 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) when the OD value ranged between 0.6 and 0.8 at 600 nm. The cells were incubated at 16 °C with shaking at 110 rpm for 18-20 h, and then collected by centrifugation at 12,000 × g for 5 min at 4 °C. The pellets were washed three times using a phosphate buffer (20 mM; pH 7.4), and followed by breaking the cell under constant cell disruption systems (Constant Systems LTD, Northants, England) at 4 °C. After centrifugation at 12,000 × g at 4 °C for 15 min, the supernatant was collected for the subsequent protein purification.
With a N-Terminal 6 × His tag, all of the recombinant β-xylosidases were successfully purified by the HisSep Ni-NTA Agarose Resin (Yeasen, Shanghai, China) column. A total of 10 mL supernatant was equilibrated with equilibration buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). After binding, the column was rinsed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). The bound enzyme was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) at a flow rate of 0.5 mL/min, all the collected active fractions were concentrated to 1 mL. The supernatants were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was used to detect the expression of soluble β-xylosidase protein for each clone by using anti-His tag antibody (Yeasen, Shanghai, China). The size of each enzyme was analyzed by SDS-PAGE. Protein concentration was determined by Modified Bradford Protein Assay Kit from Sangon [34]. The resultant β-xylosidases (Xyl1-Xyl8) derived from G. brachycerastes gut metagenome have been deposited into GenBank (accession number: KY618667- KY618674).
Enzyme activity assays
β-xylosidase, β-glucosidase and arabinosidase activity was measured using pNPX, pNPG and pNPA as substrate, respectively. For pH optimum, enzyme activity was measured using the following buffer systems, including sodium acetate (100 mM; pH 3.0-6.0), sodium phosphate (100 mM; pH 7.0-8.0), and Tris-HCl buffer (100 mM; pH 9.0). To examine the thermal stability, enzyme activity was tested under a temperature gradient between 20 °C to 80 °C. The enzyme activity was calculated by the hydrolysis of 4 mM of pNPX, pNPG and pNPA, respectively, and the reaction was terminated by the addition of 100 μL of 1 M Na2CO3. The released p-nitrophenol (pNP) was measured immediately at 405 nm using a Multiskan Spectrum spectrophotometer (Thermo Scientific, Finland). One unit (U) enzyme activity was defined as the release of 1 μmol of pNP per minute under the standard assay conditions. All assays were carried out in triplicate.
Results
Screening of β-xylosidases from a termite gut metagenome
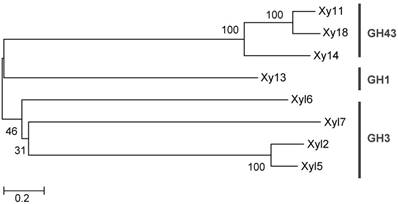
The screening process detected 22 positive clones containing β-xylosidase activity (turquoise color) with a hit rate of 1:545. The positive clones were digested by a single restriction endonuclease, BamH I, to check for RFLP. A total of 11 positive clones representing the different RFLP patterns were pooled and submitted to 454 pyrosequencing [26]. A total of 37,330 reads averaging 536 base pairs (20 Mb raw data) were obtained and assembled into 41 contigs by Newbler software V 2.7 (Table S2). Eight ORFs were predicted for β-xylosidase genes based on their DNA sequences (Table 1). Phylogenetic analysis of the GHs suggested that they belong to 3 glycoside hydrolase families, including GH1, 3 and 43 (Figure 1).
Positive clones from G. brachycerastes metagenomic fosmid library
| Gene | ORF* | Contig Number | Accession Number | Annotation | Identity (%)** |
|---|---|---|---|---|---|
| xyl1 | 32 | 1 | KY618667 | β-xylosidase /arabinosidase | 63 |
| xyl2 | 109 | 3 | KY618668 | β-xylosidase /β-glucosidase | 58 |
| xyl3 | 167 | 5 | KY618669 | β-xylosidase | 68 |
| xyl4 | 175 | 5 | KY618670 | β-xylosidase /arabinosidase | 73 |
| xyl5 | 187 | 6 | KY618671 | β-xylosidase/β-glucosidase | 70 |
| xyl6 | 230 | 7 | KY618672 | β-glucosidase/β-xylosidase | 57 |
| xyl7 | 304 | 10 | KY618673 | β-xylosidase /β-glucosidase | 60 |
| xyl8 | 436 | 26 | KY618674 | β-xylosidase /arabinosidase/α-arabinofuranosiase | 63 |
“*”: Eight β-xylosidases belong to GH1, GH3 and GH43 families, respectively. All the recombinant β-xylosidases derived from G. brachycerastes gut microbiome were expressed in the soluble fractions of E. coli and exhibited the predicted activity.
“**”: Identity of these positive clones was obtained using NCBI's BLAST search.
Analysis of functional domains of G. brachycerastes symbiotic β-xylosidases
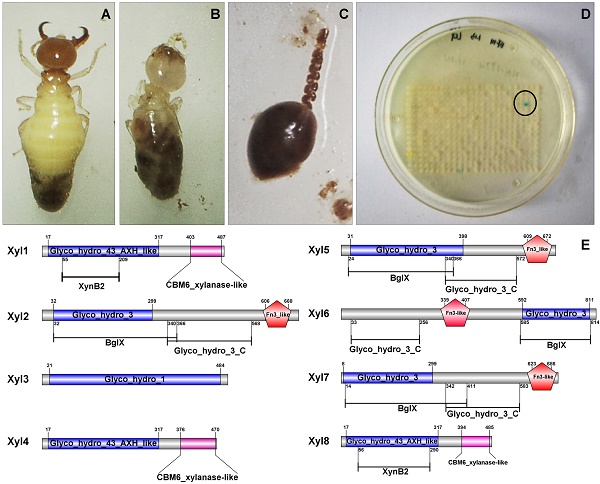
After sequencing and assembly, β-xylosidase positive clones (424,401bp contig) contained eight putative ORFs. All the ORFs begin with the putative initiation codon ATG and ends with codon TAG. Among the eight β-xylosidases (Xyl1 to Xyl8), no coding sequence for an apparent signal peptide was identified. Non-catalytic carbohydrate-binding modules (CBMs) are carbohydrate-binding modules, which bind to the surface of insoluble carbohydrates (Figure 2). The CBM6 xylanase-like domain of Xyl1, Xyl4, Xyl8 enhanced the enzyme binding to substrate surface at C-terminal [35]. The Fn3-like domain at C-terminal of Xyl2, Xyl5, Xyl6, Xyl7 has no defined function, but is probably associated with the thermal stability of the enzyme. While BglX of Xyl2, Xyl5, Xyl6, Xyl7 represents a typical β-glucosidase-β-xylosidase domain, which related to the function in the periplasmic region in Gram-negative bacteria [36] (Figure 2).
Phylogenetic analysis of the predicted hemicelluases from G. brachycerastes gut microbiome. These genes belong to three glycoside hydrolase families, GH1, 3, and 43, and their protein sequences were aligned using clustalX. This unrooted phylogenetic tree was established with MEGA 5.0 by the neighbor-joining method. Bootstrap values were derived from 1,000 replications.
Expression and purification of G. brachycerastes symbiotic β-xylosidases in E. coli
All the eight genes were cloned into pET-28a (+) expression vector and successfully expressed in E.coli BL21. The molecular mass of eight purified proteins were determined between 53-90 kDa by comparing standard protein marker in SDS-PAGE, when the gel was stained with Coomassie Brilliant Blue R-250 (Figure S1). The concentration of purified proteins was 4.68 ± 1.63 mg/mL, 0.63 ± 0.03 mg/mL, 0.82 ± 0.012 mg/mL, 0.64 ± 43 mg/mL, 1.10 ± 0.070 mg/mL, 0.81 ± 0.013 mg/mL, 1.12 ± 0.079 mg/mL, 3.03 ± 0.98 mg/mL, and 2.33 ± 0.053 mg/mL for Xyl1, Xyl2, Xyl3, Xyl4, Xyl5, Xyl6, Xyl7 and Xyl8, respectively. Theoretical pI value of the most of the purified proteins was around 5, while the pI value of Xyl6 shows 7.04 (Table S3).
Characterization of G. brachycerastes symbiotic β-xylosidases
The substrate specificity of the purified proteins was examined by incubating the enzyme with different substrates (including pNPX, pNPA and pNPG for β-xylosidase, β-glucosidase and α-arabinosidase activity, respectively) under a range of pH (3.0-9.0). As expected, all eight recombinant proteins exhibited β-xylosidase activity. The optimum pH for Xyl2 and Xyl4 were 5.0, while other enzymes were 6.0 using pNPG and pNPA as substrates (Figure 3; Table 2). The optimum temperature ranged between 30-50 °C using pNPG and pNPA as substrates (Figure 4; Table 2). Xyl1 activity against pNPX and pNPA was 4.52 ± 0.41 U/mg and 0.56 ± 0.04 U/mg of protein, respectively, while Xyl8 exhibited activities of 11.98 ± 1.21 U/mg and 1.05 ± 0.07 U/mg of protein. Xyl6 showed specific activity of 25.27 ± 1.09 U/mg and 5.98 ± 0.45 U/mg of protein using pNPG and pNPX as substrate, respectively. These combined results from enzymatic assays suggest the existence of multimeric β-xylosidases in the gut microbiome of G. brachycerastes.
Predicted functional domains of β-xylosidases from G. brachycerastes gut microbiome. Besides the signature motifs for GH1, GH3, and GH43 families, respectively, four of the recombinant G. brachycerastes symbiotic β-xylosidases, Xy12, Xy15, Xy16, and Xy17, possess a Fn3-like domain, which mostly exists at the C-terminus. BgIX, an E. coli gene encoding a β-D-glucosidase (EC 3.2.1.21), is a shared feature among Xy12, Xy15, and Xy17.
Enzymatic properties of the recombinant β-xylosidases from G. brachycerastes gut microbiome
| β-xylosidase | Substrate | pH | Temperature (°C) | Specific Activity (U/mg) |
|---|---|---|---|---|
| Xyl1 | pNPX/pNPA | 6.0 | 50 | 4.52 ± 0.49 /0.56±0.04 |
| Xyl2 | pNPX | 5.0 | 40 | 11.51±1.03 |
| Xyl3 | pNPX | 5.5 | 50 | 0.537±0.004 |
| Xyl4 | pNPX | 5.5 | 40 | 2.59±0.53 |
| Xyl5 | pNPX | 6.0 | 50 | 8.78±0.98 |
| Xyl6 | pNPX/pNPG | 6.0 | 40/50 | 5.98±0.42/32.27±2.11 |
| Xyl7 | pNPX/pNPG | 5.0 | 40 | 14.47±1.24/0.033±0.024 |
| Xyl8 | pNPX/pNPA | 6.5 | 40/50 | 11.98 ±1.21/1.05±0.012 |
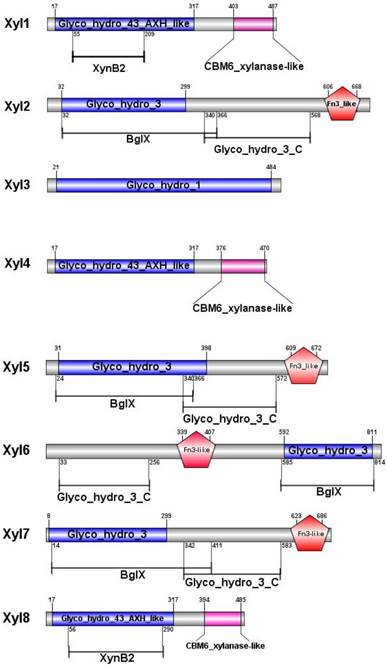
Effects of pH and temperature on enzyme activity
Recombinant β-xylosidases from termite guts exhibited activity across a broad range of pH (3.0-9.0) under optimal temperatures (Figure 3). β-xylosidases activity of Xyl1, Xyl6 and Xyl8 increased linearly from pH 2.0 to 6.5. It retained more than 80% activity in the pH range of 5.0-7.0. At pH 8.0, they retained about 40% of relative activity. Xyl2 and Xyl7 activities decreased mildly when pH was increased from 5.5 to 9.0. But their activities changed substantially when pH was increased from 3.0 to 5.0. Similarly, Xyl3, Xyl4 and Xyl5 were sensitive to pH changes. Therefore, Xyl1, Xyl6 and Xyl8 showed potential for industrial application.
Optimal pH for recombinant β-xylosidases from G. brachycerastes gut microbiome. Experiments were conducted at their optimal temperature using 4 mM pNPX as the substrate. Enzyme activity was measured in 100 mM sodium acetate buffer (pH 3.0~6.0), 100 mM sodium phosphate buffer (pH 7.0~8.0), and 100 mM Tris-HCl buffer (pH 9.0), respectively. Error bar represents the standard deviation between three independent measurements.
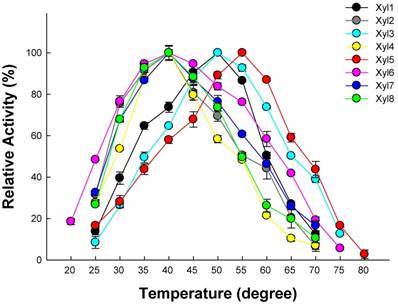
Effect of different temperatures on the eight β-xylosidases activity was shown in Figure 4. The activities of Xyl1, Xyl3, Xyl6 and Xyl8 were active across a broad temperature range (20-75 °C) using their optimal pH. Their β-xylosidases activity increased linearly between 20 to 50 °C. While the other four β-xylosidases, Xyl2, Xyl4, Xyl5 and Xyl7 were only active within a narrow temperature range, which limit their potential in industrial applications.
Optimal temperature for recombinant β-xylosidases from G. brachycerastes gut microbiome. Experiments were conducted at their optimal pH using 4 mM pNPX as the substrate. Enzyme activity was tested under a temperature gradient ranging from 20 to 80°C. Error bar represents the standard deviation between three independent measurements.
Discussion
Metagenomic analysis is a powerful tool to investigate microbial diversity at any given environment, and to search for novel biocatalyst for industrial applications [17, 27, 28, 37]. Termite guts harbor a dense and diverse microbiota that is essential for lignocellulose digestion. Recent progress in symbiotic digestion was driven by the introduction of high-throughput screening and sequencing techniques [38]. They provide sufficient resolution and sampling depth to explain the distribution patterns of microbial lineages across the wide range of host species. Considered as a rich reservoir for lignocellulosic enzymes, 0.26% of the genes identified in the fungus-growing termite Pseudacanthotermes militaris gut metagenome are hemicellulases [17], which is comparable to the rate in this study (0.18%). A slightly lower hit rate was probably due to the screening conditions in this study, in which enzymes were incubated at 50 °C for 30 min, while the temperature optimum for G. brachycerastes is about 20-35 °C. In a previous study, several xylanase genes were discovered in the microbiome of G. brachycerastes [30]. Here, we screened β-xylosidases from the same metagenomic library. As a result, we identified, cloned, and expressed eight β-xylosidases with molecular weights ranging from 53 to 90 kDa.
The optimal pH of these β-xylosidases ranged between 5.0 and 6.0, which is comparable to a model industrial strain, Trichoderma reesei [39]. For T. reesei, however, the low hemicellulase activity is one of the major barriers for its extensive application [40, 41]. Although the activity level of these newly identified β-xylosidases from G. brachycerastes bacterial symbionts is modest, Xyl1, Xyl6 and Xyl8 are relatively thermostable and pH tolerant, and have the potential for industrial application.
In general, plant cell wall degrading enzymes are modular proteins containing catalytic domains linked to one or more CBMs. CBMs are critical components of glycoside hydrolases (GHs) that degrade insoluble polysaccharides. Up to date, 81 CBM families have been cataloged in the CAZy database (http://www.cazy.org/Carbohydrate-Binding-Modules.html). CBM6 is responsible for binding to the insoluble carbohydrate surfaces. In this study, Xyl1, Xyl4 and Xyl8 contain CBM6_Xylanase-like domain at the C-terminus, which can enhance catalytic efficiency of β-xylosidases binding to the xylanase surface, and eventually can disrupt the polysaccharide structure [35]. β-xylosidases are distributed throughout GH families, including 1, 3, 30, 39, 43, 51, 52, 54, 116, and 120. Enzymes from GH43 and GH3 families exhibited bifunctional or multifunctional activities [27, 42]. In this study, Xyl1, Xyl6, Xyl7 and Xyl8, which belong to GH3 and GH43, demonstrated bifunctional activities. A thermostable β-xylosidase cloned from the ethanologenic thermophilic anaerobe Thermoanaerobacter ethanolicus type strain JW200 also exhibited α-arabinosidase activity and this bifunctional xylosidase-arabinosidase (xarB) showed great potential in industrial applications [43]. Justo et al (2015) reported that xynB5, a gene derived from a Gram-negative, oligotrophic bacterium, Caulobacter crescentus, encoded a multifunctional β-xylosidase [44]. With BglX domain at its N-terminus, xynB5 exhibited β-glucosidase, β-xylosidase, and α-arabinosidase activities. The multifunctional enzymes usually contain several distinct catalytic domains within the same polypeptide structure. The multimodularity can facilitate the degradation of substrate mixtures [45]. Commercial cellulase complexes are typically supplemented with β-gulcosidase [46] and hemicelluase to stimulate lignocellulosic biomass hydrolysis [40]. In comparison to multimeric protein engineering, the involvement of naturally occurring multifunctional biocatalysts streamlines the genetic modification procedures and simplifies the overall synthesis processes. Due to the clean and efficient catalytic performance, more and more chemical catalysts are gradually replaced by the sustainable biocatalysts with high stereo-specificity. Alternatively, these multimeric enzymes could serve as the substitutes for β-glucosidase, β-xylosidase and α-arabinosidase to facilitate a wide range of industrial applications, including food processing, animal feed, environment and waste management, and biomass conversion [2, 47].
Abbreviations
BAC: bacterial artificial chromosome; CBM: carbohydrate binding module; CDD: Conserved Domain Database; E. coli: Escherichia coli; GH: glycoside hydrolases; LB: Luria-Bertani; IPTG: isopropyl-β-D-thiogalactopyranoside; NCBI: National Center for Biotechnology Information; NR: Non-Redundant; PBS: phosphate-buffered saline; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; ORF: open reading frame; pNP: p-nitrophenol; pNPG: p-nitrophenyl-β-D-glucoside; pNPA: p-nitrophenyl-α-L-arabinofuranoside; pNPX: p-nitrophenyl-β-D-xylopyranoside; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the High-tech Research and Development Program of China (863:2013AA102806) to X.Y., and the Shen-Nong Visiting Scholar Funding Program of Hunan Agricultural University and a Hatch fund (Accession Number: 1004654; Project Number: KY008071) from the USDA National Institute of Food and Agriculture to X.Z. The information reported in this paper (No. 18-08-034) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director. These agencies had no role in study design, data collection/analysis, manuscript preparation, or the decision to publish.
Author Contributions
X.Y, C.L designed the experiments; C.L, G.Z carried out the research; X.Y, X.Z contributed reagents /materials/analysis tools; C.L, X.Z analyzed the data; C.L, X.Z wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Aristidou A, Penttilä M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187-198
2. Bajpai P. Microbial xylanolytic enzyme system: properties and applications. Adv. Appl Microbiol. 1997;43:141-194
3. McMillan JD. Pretreatment of lignocellulosic biomass. In: (ed.) Himmel ME, Baker JO, Overend RP. Enzymatic conversion of biomass for fuels production. Washington: American Chemical Society. 1994:292-324
4. Saha BC. Hemicellulose bioconversion. J Indust Microbiol Biotechnol. 2003;30:279-291
5. Subramaniyan S, Prema P. Biotechnology of microbial xylanases: enzymology, molecular biology and application. Crit Rev Biotechnol. 2002;22:33-64
6. Biely P. Microbial xylanolytic systems. Trends Biotechnol. 1985;3:286-290
7. De Vries R P, Kester HC, Poulsen CH. et al. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr Res. 2000;327:401-410
8. Wong KK, Tan LU, Saddler JN. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988;52:305-317
9. Sunna A, Antranikian G. Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol. 1997;17:39-67
10. Polizeli ML. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;67:577-591
11. Watanabe H, Noda H, Tokuda G. et al. A cellulase gene of termite origin. Nature. 1998;394:330-331
12. Breznak JA, Brune A. Role of microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol. 1994;39:453-487
13. Brune A. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 1998;16:16-21
14. Ohkuma M. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl Microbiol Biot. 2003;61:1-9
15. Brune A, Ohkuma M. Role of the termite gut microbiota in symbiotic digestion. In: (ed.) Bignell D, Roisin Y, Lo N. Biology of Termites: a Modern Synthesis. Dordrecht, Springer. 2010:439-475
16. Brune A, Dietrich C. Dietrich. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol. 2015;69:145-166
17. Bastien G, Arnal G, Bozonnet S. et al. Mining for hemicellulases in the fungus-growing termite Pseudacanthotermes militaris using functional metagenomics. Biotechnol Biofuels. 2013;6:78
18. Scharf ME. Termites as targets and models for biotechnology. Annu Rev Entomol. 2015;60:77-102
19. Long Y H, Hui X, Lei X. et al. Intra- and interspecific analysis of genetic diversity and phylogeny of termites (Isoptera) in East China detected by ISSR and COII markers. Sociobiology. 2009;53:411-430
20. Han Q, Liu N, Robinson H. et al. Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain. Biotechnol Bioeng. 2013;110:3093-3103
21. Schloss PD, Handelsman J. Biotechnological prospects from metagenomics. Curr Opin Biotechnol. 2003;14:303-310
22. Streit W R, Schmitz RA. Metagenomics-the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492-498
23. Helen LS, Streit WR. Metagenomics: Advances in ecology and biotechnology. FEMS Microbiol Lett. 2005;247:105-111
24. Wang M, Lai G L, Nie Y. et al. Synergistic function of four novel thermostable glycoside hydrolases from a long-term enriched thermophilic methanogenic digester. Front Microbiol. 2015;6:509
25. Zhang M, Liu N, Qian C. et al. Phylogenetic and functional analysis of gut microbiota of a fungus-growing higher termite: bacteroidetes from higher termites are a Rrich source of beta-glucosidase genes. Microb Ecol. 2014;68:416-425
26. Yan X, Geng A, Zhang J. et al. Discovery of (hemi-) cellulase genes in a metagenomic library from a biogas digester using 454 pyrosequencing. Appl Microbiol Biotechnol. 2013;97:8173-8182
27. Chang L, Ding M Z, Bao L. et al. Characterization of a bifunctional xylanase/endoglucanase from yak rumen microorganisms. Appl Microbiol Biotechnol. 2011;90:1933-1942
28. MatsuzawaT Kimura N, Suenaga H et al. Screening, identification and characterization of alpha-xylosidase from a soil metagenome. J Biosci Bioeng. 2016;122:393-399
29. Wang Q, Qian C, Zhang X Z. et al. Characterization of a novel thermostable β-glucosidase from a metagenomic library of termite gut. Enzyme Microb Technol. 2012;51:319-324
30. Liu N, Yan X, Zhang M. et al. Microbiome of fungus-growing termites: a new reservoir for lignocellulase genes. Appl Environ Microbiol. 2011;77:48-56
31. Dougherty M J, Patrik D, Hazen T C. et al. Glycoside hydrolases from a targeted compost metagenome, activity-screening and functional characterization. BMC Biotechnol. 2012;12:38
32. Tamura K, Daniel P, Nicholas P. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9
33. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-425
34. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254
35. Varnai A, Makela MR, Djajadi DT. et al. Carbohydrate-binding modules of fungal cellulases: occurrence in nature, function, and relevance in industrial biomass conversion. Adv Appl Microbiol. 2014;88:103-165
36. Yang M, Luoh S M, Goddard A. et al. The bglX gene located at 47.8 min on the Escherichia coli chromosome encodes a periplasmic β-glucosidase. Microbiology. 1996;7:1659-1665
37. Smith WS, Hale JR, Neylon C. Applying neutral drift to the directed molecular evolution of a beta-glucuronidase into a beta-galactosidase: Two different evolutionary pathways lead to the same variant. BMC Res Notes. 2011;4:1-10
38. Mewis K, Taupp M, Hallam SJ. A high throughput screen for biomining cellulase activity from metagenomic libraries. J Vis Exp. 2011;1:489-495
39. Hari Krishna S, Chowdary GV. Optimization of simultaneous saccharification and fermentation for the production of ethanol from lignocellulosic biomass. J. Agric. Food Chem. 2000;48:1971-1976
40. Hu J, Arantes V, Saddler JN. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol. Biofuels. 2011;4:1-14
41. Bischof RH, Ramoni J, Seiboth B. Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Fact. 2016;15:106
42. Gruninger RJ, Gong X, Forster RJ. et al. Biochemical and kinetic characterization of the multifunctional β-glucosidase/β-xylosidase/α-arabinosidase, Bgxa1. Appl Microbiol Biot. 2014;98:3003-3012
43. Shao W, Wiegel J. Purification and characterization of a thermostable β-xylosidase from Thermoanaerobacter ethanolicus. J Bacteriol. 1992;174:5848-5853
44. Justo PI, Correa JM, Maller A. et al. Analysis of the xynB5 gene encoding a multifunctional GH3-BglX β-glucosidase-β-xylosidase-α-arabinosidase member in Caulobacter crescentus. Antonie Van Leeuwenhoek. 2015;108:993-1007
45. Vrzheshch P V. Steady-state kinetics of bifunctional enzymes. Taking into account kinetic hierarchy of fast and slow catalytic cycles in a generalized model. Biochemistry (Mosc). 2007;72:936-943
46. Lassa M, Fidantsef A, Gorre-Clancy B. Variants of beta-glucosidases. Patent No WO2004099228A2. World Intellectual Property Organization. 2004
47. Beg QK, Kapoor M, Mahajan L. et al. Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol. 2001;56:326-338
Author contact
![]() Corresponding authors: xuguozhouedu and yanxingac.cn
Corresponding authors: xuguozhouedu and yanxingac.cn

 Global reach, higher impact
Global reach, higher impact