10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(11):1399-1410. doi:10.7150/ijbs.27446 This issue Cite
Research Paper
Zeb1 Regulates the Symmetric Division of Mouse Lewis Lung Carcinoma Stem Cells through Numb mediated by miR-31
1. Laboratory of Translational Cancer Stem Cell Research, Institute of Life Sciences, Chongqing Medical University, Chongqing, China
2. State Key Laboratory of Ultrasound Engineering in Medicine Co-Founded by Chongqing and the Ministry of Science and Technology, Chongqing, China
3. Department of Pathology, The affiliated Hospital of Southwest medical university
Received 2018-5-24; Accepted 2018-7-15; Published 2018-7-28
Abstract
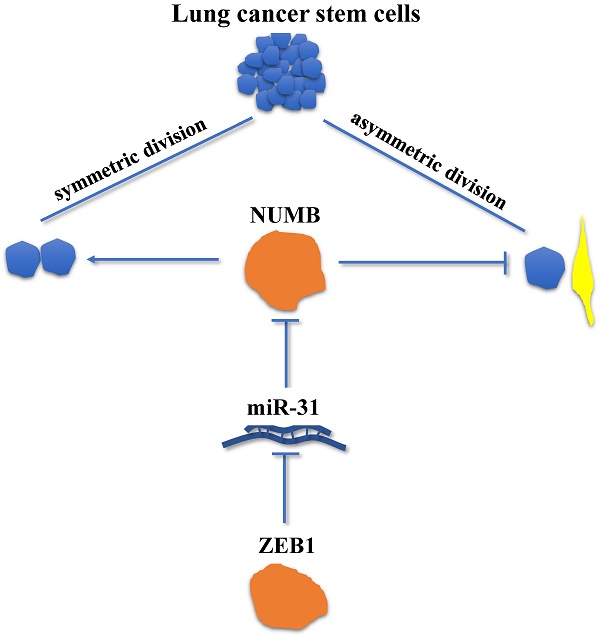
Symmetric cell division (SD) and asymmetric cell division (ASD) were the unique characteristics of stem cells and the mechanisms underlying stem cell renewal. While recent studies have identified the presence of SD and ASD in lung cancer stem cells (CSCs), the mechanisms regulating SD and ASD in cancer state have not been elucidated, mostly due to the lack of stable cellular models of SD and ASD in CSC research. In this study, the interaction between Zeb1, an Epithelial-Mesenchymal Transition (EMT) factor shown to regulate CSCs self-renew, and Numb, which regulates SD and ASD in the normal neural stem cell was investigated using the stable mouse Lewis lung adenocarcinoma SD (LLC-SD) and ASD (LLC-ASD) lines established from our previous study. The most significant finding derived from this line of research is that we have identified and molecularly ordered the axis of Zeb1-miR-31-Numb that regulates the SD, a mechanism of CSC self-renewal that has not been previously described. More specifically, the expression of Zeb1 and Numb were both significantly higher in LLC-SD than LLC-ASD cells. Silencing of Zeb1 or Numb expression lead to decreased ratio of SD and weakened single-cell cloning formation, tumor growth and tumor metastasis, respectively. The rescure experiments have molecularly ordered the regulation of Numb by Zeb1, indirectly mediated by miR-31. Moreover, we also provided preliminary evidence supporting the clinical relevance of our finding. In summary, our study provides a new insight for the self-renew of lung CSCs in which SD is regulated by the axis of Zeb1-miR-31-Numb.
Keywords: cancer stem cells, symmetric division, Zeb1, Numb, miR-31
Introduction
In normal stem cells, asymmetric division (ASD) is the unique characteristic of stem cells in which two daughter cells produced will have different cell fates: one will be a stem cell and the other will assume the fate of a differentiated cell which will generate functional cells to maintain tissue homeostasis. The symmetric division (SD), on the other hand, will produce two daughter cells that have the same fate: both are stem cells to increase the stem cell pool, or both will differentiate to increase the functional cells of the tissue[1]. Recent studies have shown the presence of these two types of cell division in the cancer stem cells (CSCs) which were thought to be important for cancer initiation[2-7]. However, mechanisms regulate SD and ASD in CSCs, as well as the relative contribution of SD and ASD to cancer initiation and progression have not been elucidated.
Numb plays an important dual-role in the regulation of the ASD and SD in neural stem cell differentiation in which Numb regulated the SD in early stage of neural stem cells differentiation and ASD in the late stage of neural stem cells differentiation[8-11]. Our previous study has shown the asymmetric partitioning of Numb in the ASD type of CSCs[12]. However, whether and how Numb regulates CSC SD and ASD have been elusive. It is notable that in a recent set of published studies, regulation of SD by microRNAs in CSCs has been demonstrated, in particular miR-146a[13] and miR-34a[14, 15]. However, the relationship between microRNAs and Numb has not been clearly established[13-17].
The Epithelial-Mesenchymal Transition (EMT) has been shown to play an important role in tumor metastasis. Amounting evidence has demonstrated the association of EMT phenotype and increased stem cell properties in CSCs[18-26]. Zeb1, one of the most import EMT transcription factors (TF), was shown to promote the generation of CSCs from the non-CSCs in breast cancer[27]. In lung cancer research, elevated Zeb1 expression is associated with poorer lung cancer prognosis[22, 28, 29] and Zeb1 expression is required for the maintenance of self-renew of lung CSCs[28, 30-32]. However, mechanistic understanding is limited. Snail was the only EMT factor identified to date that may regulate the SD of colorectal CSCs[13].
In this study, the interaction between Zeb1and Numb was investigated using the stable mouse Lewis lung adenocarcinoma SD (LLC-SD) and ASD (LLC-ASD) lines established from our previous study[12]. The most significant finding derived from this line of research is that we have identified and molecularly ordered the axis of Zeb1-miR-31-Numb that regulates the SD, a mechanism of CSC self-renewal that has not been previously described. More specifically, the expression of Zeb1 and Numb were both significantly higher in LLC-SD than LLC-ASD cells. Silencing Zeb1 or Numb expression lead to decreased ratio of SD and weakened single-cell cloning formation, tumor growth and tumor metastasis, respectively. The rescure experiments have molecularly ordered the regulation of Numb by Zeb1, indirectly mediated by miR-31. In summary, our study provides a new insight for the self-renew of lung CSCs in which SD is regulated by the axis of Zeb1-miR-31-Numb. Moreover, we also provided preliminary evidence supporting the clinical relevance of our findings.
Results
Zebl regulates the self-renew of LLC-SD through symmetric division
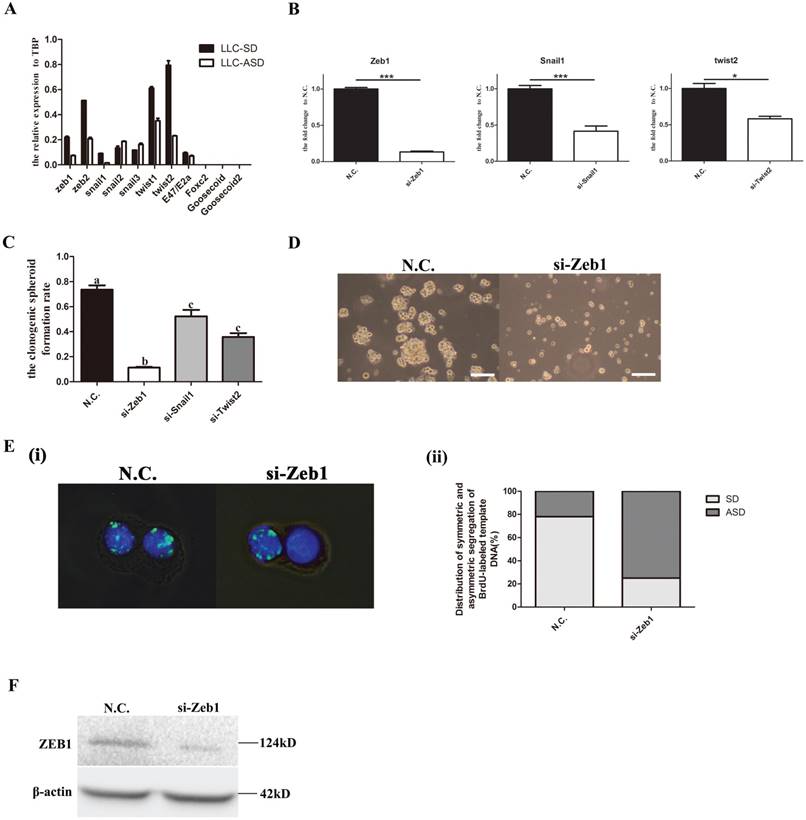
To explore the relationship between the Epithelial-Mesenchymal Transition (EMT) transcription factors (TF) and the symmetric division (SD) of lung cancer stem cells (CSCs), the expression of 11 important EMT TFs were detected by Q-PCR. And the results showed that the expression of Zeb1, Snail1 and Twist2 were significantly higher in the LLC-SD than the LLC-ASD (Figure 1A). Successful transient knock-down of Zeb1, Snail1 and Twist2 by their respective siRNAs (Figure 1B) resulted in the reduction of the single-cell cloning efficiency, and the most pronounced reduction was seen in in the siRNA-Zeb1 (11.3%) compared with siRNA-Snail1 (52.3%) and siRNA-Twist2 (35.8%) (Figure 1C). The spheroid formation was significantly inhibited upon siRNA-Zeb1 transfection in the continuous culture of LLC-SD (Figure 1D). BrdU labelling was performed to visualize and quantify SD and ASD by immunoflurescent detection of BrdU segregation pattern in the two daughter cells at the anaphase of cell division (Methods). We observed that while the ratio of SD as evident by the symmetric segregation of BrdU was decreased significantly upon siRNA-Zeb1 transfection (down from 78% vs. 25%), the ratio of asymmetric division (ASD) was increased significantly (up from 22% to 75%) (Figure 1E). The efficiency of Zeb1 knockdown was confirmed by western blot (WB) analysis (Figure 1F). These results collectively indicate that Zebl might play a role in regulating the CSC self-renew through symmetric division.
Numb, the neural stem cells SD regulator, regulates the self-renewal of LLC-SD
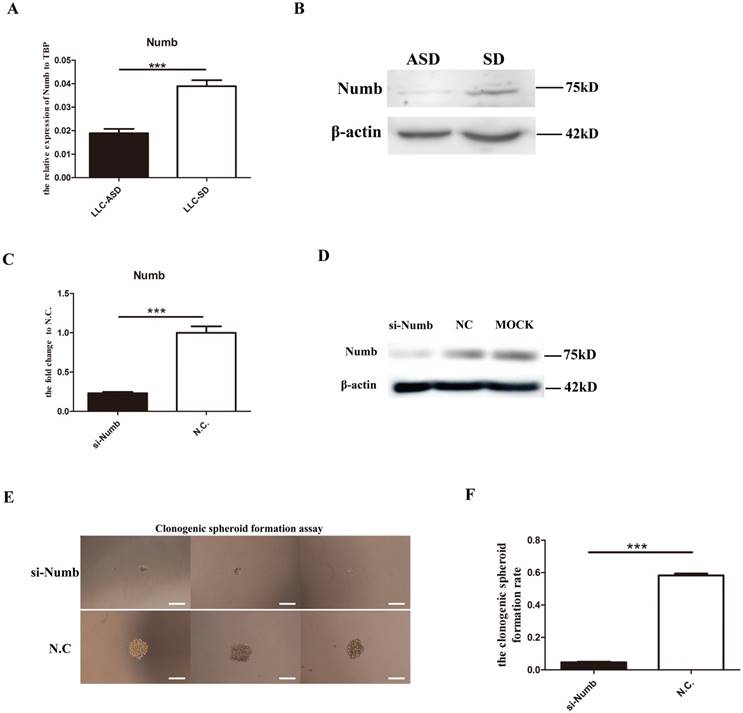
In our previous study, NUMB, an important neural stem cells SD regulator was shown to exhibit asymmetric distribution in the LLC-ASD through immunofluorescence in which Numb were segregated to the side of less differentiated daughter cell[12]. However, it was unclear whether numb regulate the self-renewal of LLC-SD. Here, the expression of Numb in LLC-SD and LLC-ASD cell lines was measured by Q-PCR and WB. LLC-SD cells had elevated Numb RNA and protein expression than the LLC-ASD cells (Figure 2A and 2B). Successful Numb silencing by siRNA was confirmed by Q-PCR and WB analysis (Figure 2C and 2D). The effect of Numb silencing on the stemness was measured by the single-cell cloning assay which showed a significant reduction of the size of the resulting colonies (Figure 2E) and the cloning efficiency (Figure 2F). These results indicate that Numb expression had positive effect on the self-renewal of the LLC-SD cells.
Zeb1 was in the upstream of the Numb and its regulation of Numb expression was mediated by miR-31
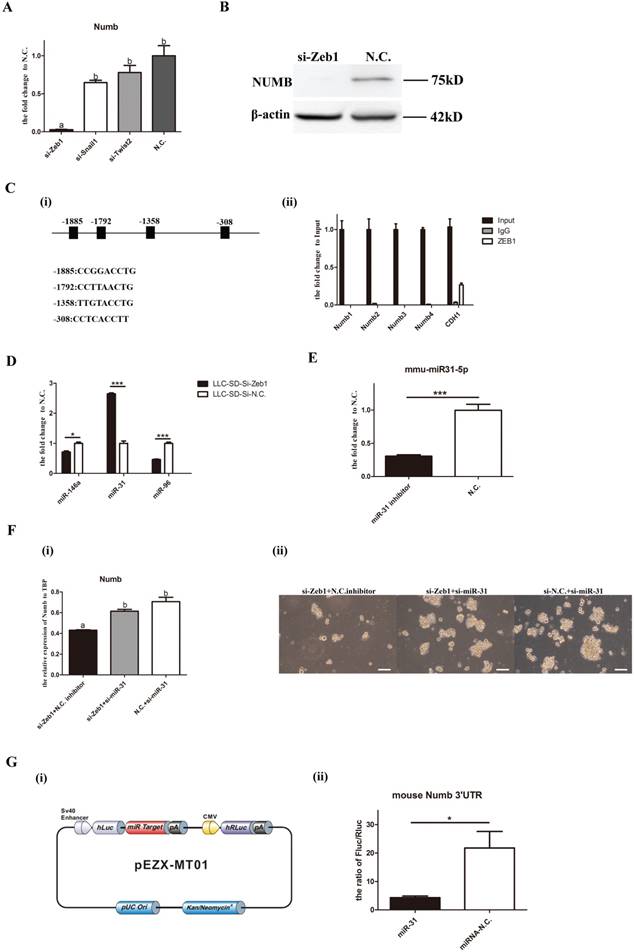
We have demonstrated thus far that both Zeb1 and Numb had positive effect on SD in LLC-SD cells (Figures 1 and 2). We next examined whether Zeb1 could directly regulate the Numb expression in LLC-SD cells. The expression of Numb in LLC-SD cells was decreased significantly upon Zeb1, Snail1 and Twist2 knockdown (Figure 3A). Down regulation of NMUB protein expression associated with Zeb-1 silencing was confirmed by WB (Figure 3B). Zeb1 can exert its positive or negative transcriptional regulation by binding to the promoter region of the target genes. To examine whether Zeb1 may regulate Numb expression directly, we analyzed the promoter region of Numb using the Jaspar tools and 4 potential Zeb1 binding sites were predicted [Figure 3C(i)]. ChiP assay was performed in which CDH1, a well-characterized target gene of ZEB1 was included as the positive control of the assay. In contrast to CDH1, no direct association between NUMB and ZEB1 was detected [Figure 3C(ii)] which ruled out the direct positive transcriptional regulation of Numb by Zeb1.
Zebl regulates the self-renewal of LLC-SD through symmetric division (SD). A. the analysis of mRNA expression of 11Epithelial-Mesenchymal Transition (EMT) factors in LLC-SD and LLC-ASD, the tata-Box binding protein gene (Tbp) was used as a reference control. B. the analysis of mRNA expression of Zeb1, Snail1 and Twsit2 after transient siRNA silencing treatment, *: p<0.05, ***: p<0.01. C. the analysis of single-cell cloning formation in which 180 wells were analyzed, different superscripts represent significant difference, p<0.05. D. morphological changes of LLC-SD upon Zeb1 knockdown, N.C. was the negative control si-RNA, bar=120um. E. (i) analysis of symmetric and asymmetric segregation of BrdU-labeled DNA during mitosis in anaphase LLC-SD cells after Zeb1 knockdown, N.C. was the negative control si-RNA, bar=120um. (ii) Quantification of SD and ASD cells in 100 dividing anaphase LLC-SD cells, si-Zeb1 and N.C. respectively. F. the analysis of protein expression of Zeb1 in LLC-SD upon Zeb1 siRNA silencing, N.C. was the negative control si-RNA, the β-actin was the reference protein.
Numb, the neural stem cells SD regulator, regulates the self-renewal of LLC-SD. A. mRNA expression of Numb in LLC-SD and LLC-ASD, ***: p<0.01. B. protein expression of Numb in LLC-SD and LLC-ASD. C. mRNA expression of Numb in LLC-SD when the Numb was knocked down, N.C. was the negative control si-RNA, ***: p<0.01. D. protein expression of Numb in LLC-SD when Numb was knocked down, N.C. was the negative control si-RNA, MOCK was blank control without si-RNA. E. analysis of single-cell cloning formation in LLC-SD upon Numb knock down, N.C. was the negative control si-RNA. F. Quantification of single-cell cloning efficiency in which 180 wells were analyzed, ***: p<0.01.
Since the direction of changes in Numb1 expression in LLC-SD cells was the same as that of Zeb1, as evident by the coordinated reduction of Numb upon silencing Zeb1expression, another possibility that could confer this pattern of gene expression regulation is the mediation of a microRNA target of Zeb1. To explore this possibility, we used TargeScan and miRTarBase and identified miR-146a, miR-31 and miR-96 as candidates. The expression of the three miRNAs were assayed upon Zeb1 knockdown. miR-31 expression were increased significantly while miR146a and miR-96 were decreased (Figure 3D), suggesting their transcriptional regulation by Zeb1. Since miRNAs act to inhibit their target gene expression, miR-31 was chosen as the potential microRNA mediator of Zeb1 for further investigation. Simultaneous knocking down of Zeb1 and miR-31 expression in LLC-SD cells was carried out (Figure 3E) and Numb expression and single-cell cloning efficiency were determined. The results showed that the expression of Numb was significantly increased in the si-Zeb1+si-miR-31 double knockout group compared to that in the si-Zeb1+N.C. inhibitor group [Figure F(i)]. In consistent with the increase in Numb expression, the stemness property as measured by the single-cell cloning efficiency, was recovered upon combinatory inhibition of miR-31 and Zeb1 [Figure F(ii)]. To confirm the direct transcriptional regulation of Numb by miR-31 and to molecularly order the Zeb1-miR-31-Numb axis, we carried out the luciferase assays in which the mouse Numb 3′UTR were inserted into the pEZX-MT01 [Figure 3G(i)]. As shown in Figure 3G(ii), miR-31 directly regulates the expression of Numb. These results collectively identified Zeb1-miR-31-Numb as a new axis of transcriptional regulation of CSC SD and self-renewal.
Molecular ordering of Zeb1-miR31-Numb axis. A mRNA expression of Numb in LLC-SD cells when Zeb1, Snail1 and Twist were silenced, different superscripts represent a significant difference, p<0.05. B. protein expression of Numb in LLC-SD cells when Zeb1 was silenced, N.C. was the negative control si-RNA. C. (i) 4 predicted binding sites of ZEB1 in the promoter of Numb. (ii) ChIP assay for determining the relationship between Zeb1 and Numb. Numb1, Numb2, Numb3, and Numb4 were 4 predicted binding sites of ZEB1 in the promoter of Numb. Input was the total DNA, IgG was the negative control, CDH1 was the positive control. D. expression of miR-146a, miR-31 and miR-96 in LLC-SD cells upon Zeb1 silencing, N.C. was the negative control si-RNA. E. decreased expression of miR-31 upon treatment with miR-31 inhibitor, the N.C. was the negative miRNA inhibitor. F. (i) mRNA expression of Numb when the Zeb1 singly silenced or co-inhibited with miR-31. (ii) the effect of Zeb1 knockdown and Zeb1/miR-31 co-knock down on the cloning formation of LLC-SD cells. G. (i) construction of plasmid made for luciferase assay, pEZX-MT01. (ii) luciferase assay for the miR-31 and Numb 3`UTR.
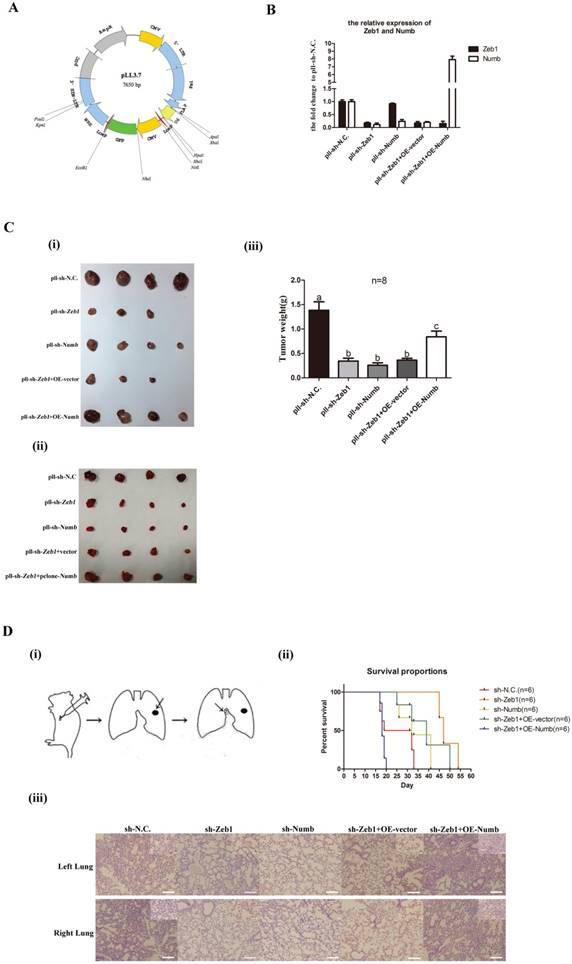
Assessment of of Zeb1-miR31-Numb axis function in vivo
In order to explore the role of Zeb1-miR31-Numb axis function in vivo, we generated LLC-SD cell line in which Zeb1 was stably inhibited while Numb was stably overexpressed using the lenti-virus expression system (Figure 4A). The following expression plasmids were constructed: pll-sh-N.C (mock Zeb-1 knockdown), pll-sh-Zeb1 (Zeb-1 knockdown only), pll-sh-Numb (Numb knockdown), pclone-vector (vector control) and pclone-Numb (Numb overexpression) (Figure 4A). LLC-SD cells were infected or transfected by the following five ways: (i) pll-sh-N.C (Zeb1 silencing vector control), pll-sh-Zeb1 (Zeb-1 knockdown), pll-sh-Numb (Numb knockdown), pll-sh-Zeb1+vector (Zeb1 knockdown mock Numb transfection vector control) and pll-sh-Zeb1+OE-Numb (Zeb1 knockdown + Numb overexpression). The expression of Zeb1 and Numb were detected by Q-PCR. Zeb1 were decreased significantly in LLC-SD cells that infected and/or transfected with pll-sh-Zeb1, pll-sh-Zeb1+vector and pll-sh-Zeb1+OE-Numb in which Zeb1 was silenced (Figure 4B). With regarding to Numb expression, while it was decreased significantly in LLC-SD cells received pll-sh-Zeb1, pll-sh-Numb, pll-sh-Zeb1+vector treatment, it was increased in the LLC-SD cells received pll-sh-Zeb1+OE-Numb. Thereafter, 105 LLC-SD of the five different experimental groups of as listed above, respectively, were injected subcutaneously into the nude mice. Tumors were taken out and weighed on day 21 after tumor cell transplantation. Inhibition of Zeb1 (pll-sh-Zeb1and pll-sh-Zeb1+vector) or Numb (pll-sh-Numb) expression in vivo resulted in lower tumor incidence and smaller tumor burden compared with the control group (pll-sh-N.C) (Figure 4C). Most importantly, overexpression of Numb in LLC-SD-pll-sh-Zeb1 cells (pll-sh-Zeb1+OE-Numb) can overcome Zeb-1-miR-31 down regulation of Numb and significantly restored LLC-SD tumor growth (Figure 4C), confirming the molecular order of Zeb1-miR-31-Numb axis and its function in vivo.
In our previous study[12], we have developed a clinically relevant syngeneic LLC orthotopic mouse model of lung cancer which allowed the evaluation of the tumor biology characteristics of LLC-SD in the C57/B6 mice [Methods and Figure 4D(i)]. 105 LLC-SD of the five experimental groups listed above respectively, were injected in the left lung of the intact C57/B6 mice. Survival assay was carried out and the time of death was recorded when it occurred. Mice died from 17th day until 33th day injected with LLC-SD in the control (pll-sh-N.C) group. Inhibition of Zeb-1 or Numb1 delayed the onset and the course of the survival assay and lead to right-shifted survival curves [Figure 4D(ii)]. In contrast, injection of Zeb-1 inhibited LLC-SD cells that overexpress Numb (pll-sh-Zeb1+OE-Numb), the survival curve was significantly left-shifted and all mouse incurred accelerated death and all died by day 20 [Figure 4D(ii)]. Histology of the left lung (the orthotopic tumor) and the right lung (metastatic foci) confirmed the presence of more severe right lung metastases in mouse received pll-sh-N.C and pll-sh-Zeb1+OE-Numb LLC-SD cells [Figure 4D(iii)]. Collectively, while the nude mice in vivo tumor growth assay confirmed the molecular order of the Zeb1-miR-31-Numb axis in vivo, the syngeneic orthotopic model of lung cancer tumorigenesis and metastasis demonstrated the importance of this axis of CSC self-renewal regulation in oncogenesis and tumor progression.
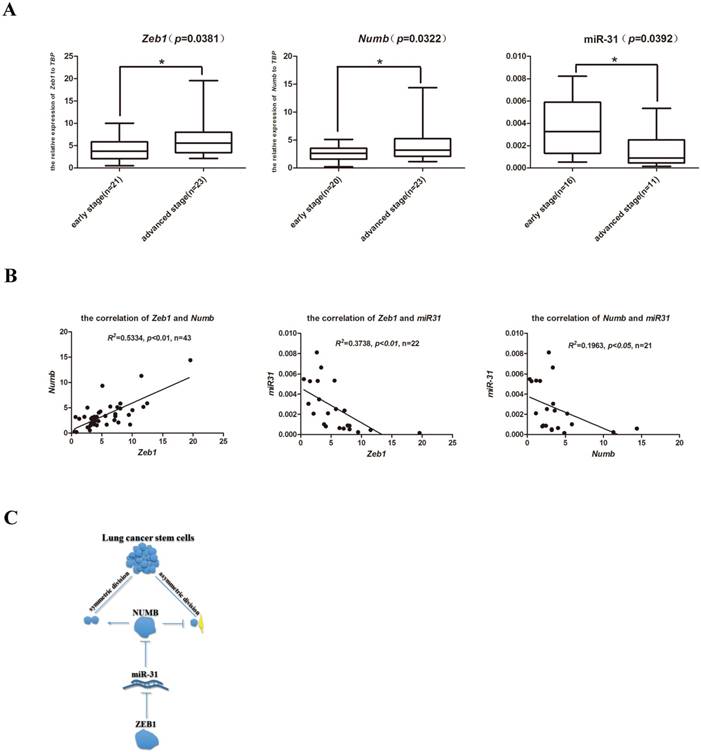
The clinical relevance of the axis of Zeb1-miR31-Numb in human lung adenocarcinoma
In order to explore the clinical relevance of the newly identified Zeb1-miR31-Numb for CSC self-renewal regulation in human lung adenocarcinoma, a total of 60 formalin fixed paraffin embedded (FFPE) clinical samples which consisted of 30 early stage (Stage I-II, no metastasis) and 30 advanced stage (Stage IV, with metastasis) lung cancer samples were identified. Total RNAs were extracted and the expression of Zeb1, Numb and miR-31 were determined by Q-PCR. While the expression of Zeb1 and Numb were both significantly elevated in the advanced-stage clinical lung cancer samples, miR-31 expression was significantly higher in the early-stage clinical lung cancer samples (Figure 5A). Statistical analysis indicated that the expression of Zeb1 was positively correlated with the expression of Numb in both early- and advanced- stage of clinical lung adenocarcinoma samples. In contrast, miR-31 expression was both negatively correlated with the expression of Zeb1 and Numb in clinical samples (Figure 5B). Thus, these preliminary observations suggest that the axis of Zeb1-miR31-Numb is of clinical relevance to human lung adenocarcinoma progression. Future investigations are required to address the prognostic importance of this newly identified axis of CSC that regulates SD, the critical step of CSC self-renewal.
In summary, we have identified a new mechanism of transcriptional regulation of stem cell self-renew regulator Numb by the key EMT regulator Zeb1 via miR-31 using the LLC-SD mouse lung cancer CSC model that we have established. We also show that Numb plays a similar role in regulating SD in CSC as in normal stem cell biology. Of important clinical relevance, we demonstrate that the complex interplay between EMT and stem cell reprograming pathway that is mediated by microRNA may also play a role in clinical lung cancer progression which merits further investigations.
The Zeb1-miR31-Numb axis functions in vivo. A. the construction of expression plasmid made for stable knock-down, pLL3.7. B. the analysis of mRNA expression of Zeb1 and Numb in the five experimental groups of LLC-SD cells: pll-sh-N.C. was the negative control for shRNA, pll-sh-Zeb1 was the knockdown of Zeb1, pll-sh-Numb was the knockdown of Numb, OE-vector was the negative control for the gene over-expression, OE-Numb was the over-expression of Numb. C. (i) and (ii) tumor formation in nude mice after injection of 05 control and experimentally modified LLC-SD cells, respectively. (iii) tumor weight measurement, different superscripts represent a significant difference, p<0.05. D. (i) the procedure of orthotopic tumor transplantation in C57BL/6 mice. (ii) the survival curve of C57/B6 mice injected with 105 control and modified LLC-SD cells, respectively. (iii) immunohistochemistry analysis of the left and right lung harvested from C57/B6 mice injected with control or modified LLC-SD cells, respectively.
The clinical relevance of the axis of Zeb1-miR31-Numb in human lung adenocarcinoma. A. analysis of mRNA expression of Zeb1, Numb and miR-31 in early-stage and advanced-stage of human lung adenocarcinoma, n was the number of human lung adenocarcinoma sample, *: p<0.05. B. the analysis of correlation between Zeb1 and Numb, Zeb1 and miR-31 and Numb and miR-31, n was the number of human lung adenocarcinoma sample. C. the mechanism of Zeb1-miR31-Numb in lung CSCs.
Discussion
While accumulated data implicating the role of lung cancer stem cells (CSCs) in lung cancer initiation and progression, the cancer biology characteristics of CSCs, including the lung cancer CSCs remain elusive due to the lack of stable CSC cellular models of human cancer. Consequently, mechanisms that regulate CSC self-renewal are poorly defined[18, 21, 33-35]. Although the Epithelial-Mesenchymal Transition pathway (EMT) was proposed as a potential regulator of CSC self-renewal, experiment evidence and clinical relevance are scant. In this study we reported here, utilizing the LLC-SD and LLC-ASD mouse lung CSC models and the syngeneic orthotopic lung cancer model that we established and characterized[12], we made a preliminary attempt to elucidate whether the EMT pathway and the stem cell self-renewal pathway have an interplay that impact stem cell characteristics as well as cancer biology features of mouse lung CSCs. Here, we reported the identification of the Zeb1-miR-31-Numb axis which represents a new mechanism of cross-talk between the EMT and stem cell pathway that is mediated by microRNAs.
In breast cancer CSCs, the promoter of Zeb1 possesses both the permissive H3K4me3 and the restrictive H3K27me3 modifications which regulates Zeb1 the switch-on and off of the TF activity of Zeb1[27], conferring CSCs plasticity. Few studies demonstrated the role of Twist2 in transcriptional regulation of breast cancer CSC properties by increasing CD24 expression[25, 26]. However, mechanisms underlying the action of EMT TFs to promote CSC properties were unclear.
In our study, the two cell lines, LLC-SD and LLC-ASD, used for exploring the mechanism of self-renewal of lung CSCs were obtained from our previous study had different stem cell characteristics[12]. Most importantly, about 80% of LLC-SD cells undergo the SD and 55% of LLC-ASD cells undergo the ASD. Using this pair of well-characterized and stable CSC cellular models of lung cancer, through genetic modification of the expression of Zeb1 and Numb, we showed a post-transcriptional regulation of Numb by Zeb1 (Figures 3A and 3B) and regulation of CSC self-renewal by influencing SD by Zeb1 (Figure 4C and 4D).
Numb has been established as the most important and dual regulator in self-renewal and differentiation of normal neural stem cells[8-11]. While Numb has a positive effect on SD in early-stage of neural stem cell differentiation, its effect on SD switched to the negative regulation mode in the late-stage of neural stem cell differentiation[10, 36]. However, only limited reports showed a correlation between Numb expression and CSC state in human cancer. In colorectal cancer CSCs, Numb could promote β-catenin degradation to direct ASD[13]. In another report, Numb had no effect on both SD and ASD in colon cancer [14, 15]. The first notable finding of our study is to provide convincing experimental evidence supporting a rather definitive role of Numb in regulating SD, CSC stemness properties in vitro (Figures 2E and 2F) as well as lung cancer oncogenesis and progression in vivo (Figures 4C and 4D) by modifying the expression of Numb, either via siRNA silencing or stable overexpression (Figures 2C-D and 4A-B).
The second notable finding of this study is that we have identified an intricate cross-talk between the EMT pathway and the stem cell reprograming pathway that is mediated by microRNA. Specifically, we identified and molecularly ordered the Zeb1-miR-31-Numb axis (Figures 3). Prior to our study, two groups reported the transcriptional regulation of Numb by miR-146a[13] in colorectal cancer and by miR-31 in breast cancer[37], consistent with our findings. Nevertheless, those studies didn't elucidate the role of these microRNAs in mediating the cross-talk between EMT and stem cell reprogramming.
The third notable observation derived from this study is the clinical relevance of the Zeb1-miR-31-Numb axis in human lung adenocarcinoma (Figure 5) which raises the possibility of exploring this axis for lung cancer prognosis. While we provided the experimental evidence that this axis is associated with lung adenocarcinoma staging, clinical cohorts that have long-term follow-up data are required to address the prognostic importance of this axis which we will pursuit in our future studies.
However, our study has raised questions that merit future investigations. First, the mechanism underlying Zeb1 transcriptional regulation of miR-31 was unclear. We observed no direct interaction between Zeb1 miR-31 by the ChIP assay (date not shown). TGF-β and Wnt are two important pathway which were downstream of EMT TFs[38-40]. Snail1 could regulate miR-146a expression in a β-catenin-dependent manner in colon CSCs[13]. Whether Wnt mediates Zeb1 regulation of miR-31 will be addressed in our future studies. Since Numb could regulate the SD and ASD of neural stem cell by inhibiting the Notch pathway[8, 9], we explored this possibility in our study but failed to detect the expression of key Notch-pathway factors, Hes1, Hes5 and Hey2, in LLC-SD and LLC-ASD cells (data not shown).
In summary, the study we presented here has provided new insights on mechanisms regulating CSC self-renewal and provided novel cellular and syngeneic orthotopic models of lung cancer for in-depth characterization of the functional importance of the mechanistic interplay both in vitro and in vivo.
Materials and Methods
Cell Culture
The mouse Lewis lung carcinoma asymmetric division cells (LLC-ASD) were maintained in DMEM high glucose supplemented with 5% FBS and passaged using 0.25% 0.25 % Trypsin-EDTA enzyme for about 1 min. The mouse Lewis lung carcinoma symmetric division cells (LLC-SD) were maintained in DMEM/F12-based normal stem-cell media (Gibico), supplemented with 20 ng/ml EGF (BD), 20 ng/ml FGF (BD), 2% B27 (BD)and 1% PS (Hyclone), and the LLC-SD were passaged by suspending to single cell without trypsinization. LLC-SD and LLC-ASD cell lines were generated as we previously reported[12].
Animals
Male athymic or female C56BLK mice, 6-8-weeks old were used in the study. Mice were obtained from the core facility of Experimental Animal Centre in Chongqing Medical University. All animal work was conducted in accordance with an approved protocol and carried out in accordance with the institutional animal welfare guidelines of the Chongqing Medical University.
Single-cell cloning assay
The single-cell cloning assay was conducted in 96-well plates as we previously described. Specifically, to set up this assay, single-cell suspension of LLC-SD cells were prepared and was plated at 1 cell/well in 96-well plates. Single-cell plating was confirmed by microscopic examination and wells containing more than one cells were marked and excluded for the analysis of single-cell cloning efficiency. After 7-10 days of growth, the colonies grown in wells that were confirmed for single cell plating and which contain more than 50 cells were counted under microscopy.
BrdU-labeling, analysis of symmetric and asymmetrical chromosome segregation
100 anaphase cells were analyzed which were labeled by 10 μM BrdU (Sigma) for 7 days followed by BrdU withdrawal and analyzed by BrdU Immunofluorescence staining. The cells were identified as in the symmetrical division were those in which each set of chromosomes in the two daughter cells were BrdU labeled. In contrast, the cells were identified as in the asymmetrical division were those in which only one daughter cell harbors BrdU labeled chromosome. Images of the BrdU-labeling analysis were taken with Olympus FSX100 Box-Type Fluorescence Imaging Device.
Immunofluorescence and immunohistochemistry staining
For immunohistochemistry, the lungs harvested from the C57BL/6 mice injected with LLC-SD cells were deparaffinized, hydrated and H&E stained. For immunofluorescence staining, cells cultured on chamber slides were fixed with 70% ethanol in 4℃ for 30 min, incubated with 2 mol/L HCL in 0.5% Triton X-100 PBS for 1 hour and followed by incubation with blocking solution (10% bovine serum albumin in PBS) for 1 hour. Thereafter, the slides were incubated with the primary antibody (anti-BrdU, Millipore) at 4°C overnight. After washing, slides were incubated with the fluorophore-labeled secondary antibody (Proteintech) for 1 hour at room temperature according to manufacturer's instructions. Prior to mounting, the slides were incubated with DAPI for nuclear staining for 10 min.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted used by TRIZOL (Invitrogen) according to the manufacturer's protocol. Standard RT-PCR was conducted using PrimeScript RT Master Mix (Takara) according to the manufacturer's instructions and miRNA RT-PCR was conducted using Mir-XTM miRNA First-Stand Syntheis Kit (Takara). And the total RNA were extracted used by RNAprep Pure FFPE Kit (TIANGEN) and miRNA were extracted used by miRNAprep Pure FFPE Kit (TIANGEN) in the formalin fixed paraffin embedded (FFPE). The sequences of PCR primers are listed in Table 1.
PCR Primer Sequence
| Gene name | Forward primers | Reverse primers |
|---|---|---|
| mouse Zeb1 | CTCCCAGTCAGCCACCTTTA | TAACTCCATCCAGCGGTAGG |
| mouse Zeb2 | TTTCCTGCCCTCTTTGTAGC | CCTTGGGTTAGCATTTGGTG |
| mouse Snail1 | CACACGCTGCCTTGTGTCT | GGTCAGCAAAAGCACGGTT |
| mouse Snail2 | TGGTCAAGAAACATTTCAACGCC | GGTGAGGATCTCTGGTTTTGGTA |
| mouse Snail3 | ACAGCGAACTGGACACACAC | GGGTAAAGGAGAGTGGAGTGG |
| mouse Twist1 | GGACAAGCTGAGCAAGATTCA | CGGAGAAGGCGTAGCTGAG |
| mouse Twist2 | GTCTCAGCTACGCCTTCTCC | AGATGTGCAGGTGGGTCCT |
| mouse E47/E2a | GCAAACGAAGGCAGAGTAGG | TGGTCAAGAGACACCTCAAGC |
| mouse Foxc2 | AACCCAACAGCAAACTTTCCC | GCGTAGCTCGATAGGGCAG |
| mouse Goosecoid | AACGCCGAGAAGTGGAACAAG | CCGAGTCCAAATCGCTTTTACC |
| mouse Goosecoid2 | CGTGGAGGTCTGGTTCAAG | CGGACAGCATCAACAACTCT |
| mouse Numb | AACCAGCCTTTGTCCCTACC | GGCGACTGATGTGGATGAG |
| ChIP Numb1 | TTTTATGGATTTTCTGTG | TTATCTTCTGGAACCTT |
| ChIP Numb2 | CAAAGTCCACTCCGGGATC | CGGGGAATAGGCAAAGC |
| ChIP Numb3 | TACCCAAATGAACGAGA | GCAGCCTAAAATAGAACC |
| ChIP Numb4 | TCTGGAAGCTGGAAAAC | TATTAAGCAGCAGTCGG |
| ChIP Cdh1 | CAGACAGGGGTGGAGGA | AGGTGAGCCCGCAGGCAC |
| mouse Tbp | AGGGATTCAGGAAGACCACA | ATGCTGCCACCTGTAACTGA |
| mmu-miR-146a-5p | TGAGAACTGAATTCCATGGGTT | |
| mmu-miR-31-5p | AGGCAAGATGCTGGCATAGCTG | |
| mmu-miR-96-5p | TTTGGCACTAGCACATTTTTGC | |
| human Zeb1 | GAGGAAGAGGAGGAGGAGGA | GCTTGACTTTCAGCCCTGTC |
| human Numb | ACTCAGCCTTCCATGTGCTT | GTGGCTGCAATTTCCTTGTT |
| human Tbp | CCACAGCTCTTCCACTCACA | CTGCGGTACAATCCCAGAAC |
| hsa-miR-31-5p | AGGCAAGATGCTGGCATAGC |
Characterization of orthotopic LLC-SD tumorigenesis and metastatic progression
| In situ | Metastasis | ||
|---|---|---|---|
| Lung (left) | Mediastinal lymph | Lung(right) | |
| sh-N.C. | 3/6 | 3/6 | 1/6 |
| sh-Zeb1 | 2/6 | 1/6 | 1/6 |
| sh-Numb | 1/6 | 0/6 | 0/6 |
| sh-Zeb1+OE-vector | 0/6 | 2/6 | 0/6 |
| sh-Zeb1+OE-Numb | 4/6 | 5/6 | 1/6 |
The siRNA design and the construction of expression plasmids
All of the siRNA were synthesized from the GenePharma and the sequence of these siRNA were listed in the Table 1. And the pLL3.7 plasmid (Plasmid #11795) and pClneo-numb (Plasmid #41712) were purchased form addgene.
Luciferase assays
Luc-PairTM luciferase Assay Kit 2.0 and pEZX-MT01(Dual luciferase vector) were obtained from GeneCopoeia. miRNA negative control and miR-31 mimic were transfected together with Numb 3' UTR (GenePharma) into 293T cells respectively for 24 hour according to the manufacturer's instructions. Expression of Firefly (FLUC) and Renilla Luciferase (RLUC) was counted using a luminometer (BioTek).
Western blot
In brief, LLC-SD were lysed with cell lysis buffer for western and IP (Beyotime). Lysates were separated in SDS-PAGE for the routine western blot assay. The primary antibodies used for incubation were ZEB1(Santa Cruz), NUMB (Abcam). The membranes were visualized by ClarityTM Western ECL Substrate (Bio-rad).
Chromatin immunoprecipitation (ChIP) assay
The ChIP assays was conducted using the Chromatin immunoprecipitation (ChIP) Assay Kit according to manufacturer's instructions (Millipore). In brief, LLC-SD cells were cross-linked with 1% formaldehyde for 10 min at room temperature and the reaction was stopped by the addition of glycine. Cells were lysed and sheared by sonication to make chromatin an average size of 500-1000bp. Chromatin was precipitated with Zeb1 antibody (Santa Cruz). IgGs purified from rabbit serum (Beyotime) were used as negative controls. The antibody-chromatin-beads complex was washed, eluted, reverse cross-linked and extracted. The chromatin-associated purified DNA was analyzed by PCR for the presence of Numb1 using Numb-specific primers. Cdh1-specifc primer which was used as positive control whose sequence were listed in the Table 1.
Orthotopic tumor transplantation studies in C57BL/6 mice
To set up the in vivo experiments, 20 μl cell suspensions containing 10 μl Matrigel Matrix (Corning) and 105 cells were injected orthotopically into the left lobe of the lungs of C57BL/6 mice. For the survival experiments, the time of death of every mouse was recorded after orthotopic tumor transplantation. For the experiments of observing the onset of tumorigenesis and progression, the mice were dissected on day 14 to determine the tumors in situ and the extent of thoracic metastasis.
Tumor transplantation studies in BALB/c nude mice
100μl suspensions containing 50μl Matrigel Matrix and LLC-SD cells were injected into the flank of nude mouse all of which were male and 6-8 weeks old. Tumor growth was measured every two days, and tumor volume was calculated as V= (length x width2)/2.
Clinical sample collection and analysis
All of the formalin fixed paraffin embedded were obtained from department of pathology in the affiliated hospital of southwest medical university which were identified the cancer progress as the TNM stage of lung cancer and received the approval of the institutional ethics committee of southwest medical university medical center.
Statistical analysis
All data were presented as mean ± standard deviation. Tumor volumes were analyzed using an ANOVA when data satisfied the Gaussian distribution assessed by the D'Agostino-Pearson normality test and the F test for equal variance. When two groups were compared, the Student's t-test with Welch's correction was used. By default, two-tailed tests were performed. KaplanMeier's method was used for survival analysis. P <0.05 was considered significant statistically and is marked with an asterisk. P < 0.01 was considered highly significant statistically and is marked with double asterisk. All statistical analyses were performed using Graphpad Prism version 5.0.
Acknowledgements
We thank the members in the Laboratory of Translational Cancer Stem Cell Research who are not listed in the authors for their encouragement. We thank the department of pathology in the affiliated hospital of southwest medical university for providing the clinical FFPE samples. This work was supported by the National Natural Science Youth Fund (Grant No.81602596) and the National Natural Science Fund (Grant No.81672908)
Competing Interests
The authors have declared that no competing interest exists.
References
1. Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068-74
2. Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N. et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer cell. 2011;20:328-40
3. Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE. et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765-8
4. Mukherjee S, Kong J, Brat DJ. Cancer stem cell division: when the rules of asymmetry are broken. Stem cells and development. 2015;24:405-16
5. Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78-81
6. Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2195-200
7. Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes & development. 2009;23:2675-99
8. Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development (Cambridge, England). 2002;129:4843-53
9. Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477-91
10. Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624-7
11. Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27-41
12. Wang J, Sun Z, Liu Y, Kong L, Zhou S, Tang J. et al. Comparison of tumor biology of two distinct cell sub-populations in lung cancer stem cells. Oncotarget. 2017;8:96852-64
13. Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW. et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nature cell biology. 2014;16:268-80
14. Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ. et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell stem cell. 2013;12:602-15
15. Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ, Xiang K. et al. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016:5
16. Vaira V, Faversani A, Martin NM, Garlick DS, Ferrero S, Nosotti M. et al. Regulation of lung cancer metastasis by Klf4-Numb-like signaling. Cancer research. 2013;73:2695-705
17. Chou CH, Tu HF, Kao SY, Chiang CF, Liu CJ, Chang KW. et al. Targeting of miR-31/96/182 to the Numb gene during head and neck oncogenesis. Head & neck. 2018;40:808-17
18. Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Seminars in cancer biology. 2012;22:396-403
19. Li P, Yang R, Gao WQ. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Molecular cancer. 2014;13:55
20. Farmakovskaya M, Khromova N, Rybko V, Dugina V, Kopnin B, Kopnin P. E-Cadherin repression increases amount of cancer stem cells in human A549 lung adenocarcinoma and stimulates tumor growth. Cell cycle. 2016;15:1084-92
21. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell stem cell. 2014;14:275-91
22. Sanchez-Tillo E, Siles L, de Barrios O, Cuatrecasas M, Vaquero EC, Castells A. et al. Expanding roles of ZEB factors in tumorigenesis and tumor progression. American journal of cancer research. 2011;1:897-912
23. Liu CW, Li CH, Peng YJ, Cheng YW, Chen HW, Liao PL. et al. Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3beta signaling pathway in non-small-cell lung cancer. Oncotarget. 2014;5:3880-94
24. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
25. Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z. et al. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene. 2011;30:4707-20
26. Liu AY, Cai Y, Mao Y, Lin Y, Zheng H, Wu T. et al. Twist2 promotes self-renewal of liver cancer stem-like cells by regulating CD24. Carcinogenesis. 2014;35:537-45
27. Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61-74
28. Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop-a motor of cellular plasticity in development and cancer? EMBO reports. 2010;11:670-7
29. Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. International journal of cancer. 2013;132:745-54
30. Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A. et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature cell biology. 2009;11:1487-95
31. Krohn A, Ahrens T, Yalcin A, Plones T, Wehrle J, Taromi S. et al. Tumor cell heterogeneity in Small Cell Lung Cancer (SCLC): phenotypical and functional differences associated with Epithelial-Mesenchymal Transition (EMT) and DNA methylation changes. PloS one. 2014;9:e100249
32. Wang H, Zhang G, Zhang H, Zhang F, Zhou B, Ning F. et al. Acquisition of epithelial-mesenchymal transition phenotype and cancer stem cell-like properties in cisplatin-resistant lung cancer cells through AKT/beta-catenin/Snail signaling pathway. European journal of pharmacology. 2014;723:156-66
33. Doherty MR, Smigiel JM, Junk DJ, Jackson MW. Cancer Stem Cell Plasticity Drives Therapeutic Resistance. Cancers. 2016:8
34. Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clinical chemistry. 2013;59:168-79
35. Sanchez-Danes A, Hannezo E, Larsimont JC, Liagre M, Youssef KK, Simons BD. et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature. 2016;536:298-303
36. Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Cheng V. et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. The Journal of cell biology. 2005;170:721-32
37. Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC. et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032-46
38. Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545:355-9
39. Hu B, Wang Q, Wang YA, Hua S, Sauve CG, Ong D. et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell. 2016;167:1281-95 e18
40. Ji W, Yu Y, Li Z, Wang G, Li F, Xia W. et al. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget. 2016;7:15118-34
Author contact
![]() Corresponding authors: H. Rosie Xing, Mailing address: 1 Yi Xue Yuan Road, Yuzhong District Chongqing, Chongqing Medical University, 400016, P.R. China. Phone and Fax number: +86-023-63738563, +86-023-68486646; Email address: xinglab310com and Jianyu Wang, Mailing address: 1 Yi Xue Yuan Road, Yuzhong District Chongqing, Chongqing Medical University, 400016, P.R. China. Phone and Fax number: +86-023-63662443, +86-023-63662443; Email address: wjy2003123com
Corresponding authors: H. Rosie Xing, Mailing address: 1 Yi Xue Yuan Road, Yuzhong District Chongqing, Chongqing Medical University, 400016, P.R. China. Phone and Fax number: +86-023-63738563, +86-023-68486646; Email address: xinglab310com and Jianyu Wang, Mailing address: 1 Yi Xue Yuan Road, Yuzhong District Chongqing, Chongqing Medical University, 400016, P.R. China. Phone and Fax number: +86-023-63662443, +86-023-63662443; Email address: wjy2003123com

 Global reach, higher impact
Global reach, higher impact