10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(11):1586-1598. doi:10.7150/ijbs.23401 This issue Cite
Research Paper
Dedifferentiation of human uterine polyp stem cells into embryo-like cells during inducing pluripotency
1. Department of Obstetrics and Gynecology, Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, Hualien, Taiwan
2. Institute of Medical Sciences, Tzu Chi University, Hualien, Taiwan
3. Department of Occupational Medicine, Hualien Tzu Chi Hospital; Tzu Chi University, Hualien, Taiwan
Received 2017-10-19; Accepted 2018-7-29; Published 2018-9-7
Abstract
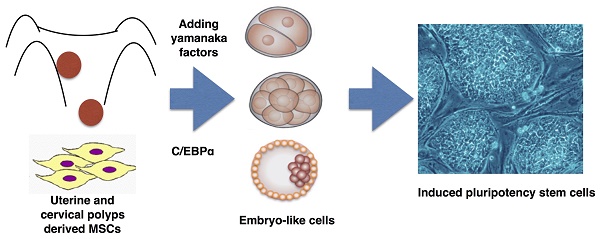
By introduction of Oct4, Sox2, Klf4 and cMyc, human adult somatic cells can be reprogrammed into embryonic stem cell capable of pluripotent differentiation. In several lines of human endometrial polyp- and cervical polyp-mesenchymal stem cells (EPMSCs and CPMSC), we showed introduction of the four transcription factors led to a dedifferentiation of these cells into early embryo-like cells in three days, ranging from one-cell, two-cell, four-cell embryos, and morula to blastocyst. These early embryo-like cells resembled human early embryo derived from in vitro fertilization (IVF) in morphology, and hatching activity. These cells also expressed hypoblast (GATA4) and trophoblast (Cdx2) markers. After culturing the embryo-like cells for one month, the induced pluripotency stem cells (iPSC) could be formed (proved by pluripotency gene expression, by in vitro and in vivo differentiation). C/EBPα expression was also increased in uterine polyps. In contrast, MSCs derived from normal endometrium could not be induced to dedifferentiation to such early embryo-like cells. We conclude that EPMSCs and CPMSCs could be dedifferentiated to early embryo-like cells by the iPSC cocktail. This suggests that polyps of the organ derived from Mullerian duct may harbor epigenetic markers making them vulnerable to reprogramming to the earliest developmental stage. This study provides a simple model to derive early human embryo-like cells by in vitro.
Keywords: induced pluripotent stem cells, embryo, dedifferentiation, endometrial polyp, cervical polyp
Introduction
Reprogramming of fibroblasts and mesenchymal stem cells (MSCs) is a promising strategy for stem cell biology and regenerative medicine. It can be induced by nuclear transfer, cell fusion, or forced transcription factors expression [1, 2]. Reprogramming factors (protein, RNAs and small molecules) enables somatic cells to reprogram [3, 4]. By introduction of Oct4, Sox2, Klf4 and cMyc (OSKM), human adult somatic cells can reprogram into embryonic stem cell (ESC) and be capable of pluripotent dedifferentiation; the reprogramming ESC is called induced pluripotency stem cell (iPSC) [5]. Nuclear reprogramming, induced by transcription factors, resets the epigenetic markers, leading to the global reversion of the somatic epigenomes to an ESC-like state [6, 7].
During the phase for inducing pluripotency, fibroblasts and MSCs undergo a mesenchymal-epithelial transition (MET) to activate endogenous pluripotency genes, paralleled by changes in histone markers, nucleosome positioning and chromatin accessibility [8]. Their promoters, in addition, become de-methylated, preceded by oxidation of methylated CpGs through the dioxygenases Tet2 and Tet1 which involve establishing pluripotency [9, 10].
In histone markers, the promoter-specific H3K4me3 is related to active transcription. H3K4me3 in ESC and iPSC is enriched in the DNMT3B promoter and is associated with DNA hypomethylation in many genomic loci [11, 12]. Therefore, H3K4me3 is an important epigenetic marker for successful reprogramming [13]. The H3K27me3 is a repressive chromatin marker in dedifferentiating somatic cells to iPSCs. The H3K27me3 domains in iPSC are smaller, interspersed, and spanning less base pairs as compared with somatic cells [14].
Recently, co-expression of CCAAT/enhancer binding protein-α (C/EBPα) with OSKM increases the reprogramming efficiency of B cells [15]. A pulse of C/EBPα followed by an over expression of OSKM permits a rapid and highly efficient reprogramming of B cells into iPSCs [16].
To date, germ-like cells can be induced from fetal stem cells [17], bone marrow stem cells [18, 19], pancreatic stem cells [20], embryonic stem cells [21, 22], and iPSCs [23-25]. However, embryo-like cells have not been induced from these stem cells and any other cells.
MSCs are multipotent stromal cells capable of differentiation into mesoderm cells, such as fat, bone, and cartilage [26]. MSCs also harbor great potential in gene therapy, regenerative therapy and immunotherapy [26]. The MSCs can be found in various tissues, such as bone [27], umbilical cord [28], and endometrial polyp [29].
In order to investigate whether embryo-like cells can be induced from MSCs, the MSCs derived from endometrial polyp and cervical polyp were used for inducing pluripotency with OSKM factors.
Materials and Methods
Tissue collection
The local Research and Ethics Committee approved this study, and informed consent was obtained from each patient prior to tissue harvesting (IRB 105-96-A). Endometrial polyp, cervical polyp and endometrial tissue samples were harvested by cutting a polyp or endometrium from hysterectomy or surgical specimens (endometrial polyp = 2, age =46 and 54; endometrium =1, age=38; cervical polyp =1, age=71). All polyps were pathologically proven benign endometrial and cervical polyps. Tissue samples were placed in Ca2+/Mg2+ - free phosphate-buffered saline (PBS, Biowest, Nuaille, France), and were immediately transferred to the laboratory.
Tissue dissociation and cell isolation
Endometrial and cervical polyp tissues, removed from the transport medium, were placed in a Petri dish, and minced into small pieces (1-2 mm3) in the presence PBS. Tissues were dissociated with 0.5% collagenase (Sigma, St Louis, MO, USA) and 0.05% type 1 deoxyribonuclease (Sigma) and incubated for 60 min at 37oC with gentle pipetting at 15-min interval.
Cell suspensions were filtered through a 40 mm sieve (Becton Dickinson, Franklin Lakes, NJ, USA) to eliminate aggregated cells, and washed with PBS. The solution containing predominantly endometrial glands was centrifuged, and the supernatant was discarded. The pellet was treated with 0.25% trypsin/0.03% ethylenediamine tetraacetic acid (EDTA, Sigma) at 37 oC for 10 min, and the reaction was stopped by adding cold Dulbecco's Modified Eagle Medium-low glucose (DMEM-LG, Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Biological Industry, Kibbutz, Israel). Cell suspensions were filtered as mentioned above. Cells were resuspended in DMEM-LG and centrifuged on a Ficoll-Paque (Pharmacia LKB, Uppsala, Sweden) for 15 min at 500 g to remove erythrocytes. Cells were collected from the interface, washed, and resuspended in DMEM with 10% FBS.
Primary endometrial and cervical polyp MSC culture
The detailed methods were described in the previous study [29]. Briefly, the isolated cells were seeded at a density of 1 x 104 cells/cm2 in DMEM-LG medium supplemented with 10% FBS, 100 mg/ml penicillin G sodium and 100 mg/ml streptomycin sulfate in culture dishes. Cultures were incubated at 37 oC under 5% CO2 and 95% humidity. Supernatant and debris were removed from the culture dish on day 2 of culturing. The resulting MSC culture was denoted as passage 0. To prevent spontaneous differentiation, cultures were maintained at subconfluent levels (< 80% confluency). We usually passaged cells at a ratio of 1:3. Passagings of MSCs cultures were performed using 2.5% trypsin/0.23 mM EDTA. Passaged cultures were defined as passage 1.
Flow cytometry
Surface molecules of endometrial polyp MSCs (EPMSCs), endometrial MSCs (EMSCs), and cervical polyp MSCs (CPMSCs) cultures of passage 3 were characterized by flow cytometry. Cells were detached with 2 mM EDTA in PBS, washed with PBS containing 2% bovine serum albumin (BSA) and 0.1% sodium azide (Sigma, St Louis, MO, USA). The detached cells were incubated with their respective antibody conjugated with fluorescein isothiocyanate (FITC) or phycoerythrin (PE), including clustering of differentiation (CD)29, CD34, CD44, CD45, CD73, CD90, CD105 and human leukocyte antigen (HLA)-ABC and HLA-DR (BD, PharMingen, Franklin Lakes, NJ, USA). Cells were analyzed using a flow cytometer (Becton Dickinson, San Jose, CA, USA).
In vitro differentiation assay
EPMSCs, CPMSCs and EMSCs were passaged and cultured to confluence. The confluence was then shifted to osteogenic medium (DMEM supplemented with 10% FBS, 0.1 mmol/L dexamethasone, 10 mmol/L β-glycerol phosphate, and 50 mmol/L ascorbate) and adipogenic medium (DMEM supplemented with 10% FBS, 1 mmol/L dexamethasone, 5 mg/mL insulin, 0.5 mmol/L isobutylmethylxanthine, and 60 mmol/L indomethacin) for 3 weeks. The above chemicals were purchased from Sigma Company. The differentiation potential for osteogenesis was assessed by mineralization of calcium accumulation by Alizarin Red S staining (Sigma, St Louis, MO, USA). For assessment of adipogenic differentiation, intracellular lipid droplets were observed microscopically and confirmed by Oil Red O (Sigma, St Louis, MO, USA) staining. For chondrogenesis, micromass cultures were established. MSCs from three origins were seeded in a total volume of 30 μl onto the bottom of a dry 15-ml test tube (BD, Franklin Lakes, NJ, USA) at a density of 25 × 106 cells/ml. The plate was placed in the humidified CO2 incubator at 37°C for 2 h and additional chondrogenic medium (0.75 mL) was added to each tube. Media were changed every 48 h. Micromass cartilages were formed and retrieved after three weeks of culturing. After being photographed, the micromass cartilages were fixed in 4% paraformaldehyde at 4°C for 24 h. They were then washed in PBS, transferred in 70% ethanol, and processed for histology. The paraffin sections (5 μm) were assessed for cartilage by type 2 collagen (1:100, GeneTex, Irvine CA, USA) immunohistochemistry staining.
RNA isolation and reversed transcription
Total RNA was purified with RNEasy reagent (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. One microgram of total RNA was used for reverse transcription reaction; complementary DNA was synthesized using a SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA, USA). Complimentary DNA was amplified by PCR using the AmpliTag Gold Kit (Applied Biosystems, Foster City, CA, USA). The PCR products were resolved on 2% agarose gels. Quantitative PCR was performed with Platinum SYBR Green qPCR Supermix UDG (Invitrogen, USA) and analyzed with the Step One Plus real-time PCR system (Applied Biosystems, USA). Primer sequences are shown in Table 1.
SiRNA transfection
All siRNA transfections were performed using Dharmafect transfection reagent (GE Healthcare, Buckinghamshire, UK) following the manufacturer's protocol. Briefly, cells were plated in 24 well plates (2x104) and the transfection mixture (containing 2.5 μl Dharmafect and siRNA with serum/antibiotics free medium) were added directly to the medium. Total RNA were isolated from cells 24hr post-transfection and were analysed using RT-qPCR. The following siRNA was use: ON-TARGETplus Human C/EBPɑ siRNA SmartPool (L-006422-00-0005), ON-TARGETplus Non-targeting Pool (D-001810-10-05), and the primers for C/EBPɑ were 5'-AAGAAGTCGGTGGACAAGAACAG-3'(sense) and 5'-TGCGCACCGCGATGT-3'(antisense).
Karyotyping of EPMSCs
The karyotypes of cells were studied in passage 3. On day 7 after passage, cells were treated with 0.1 μg/ml colcemid (Gibco) for 4h. After the cells were washed, they were treated with either 0.25% trypsin for 3-5 min or collagenase type IV for 8 min, pipetted and harvested. They were then fixed and mounted on glass slides. The metaphases were analyzed using the standard G-banding method in a certified cytogenetic laboratory in Hualien Tzu Chi Hospital.
PCR primers used to characterize induced pluripotency and mesenchymal stem cells
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') | Product size (bp) |
|---|---|---|---|
| Oct4 | CTT GCT GCA GAA GTG GGT GGA GGA | CTG CAG TGT GGG TTT CGG GCA | 169 |
| Sox2 | CCC CCG GCG GCA ATA GCA | TCG GCG CCG GGG AGA TAC AT | 448 |
| Nanog | AGT CCC AAA GGC AAA CAA CCC ACT TC | TGC TGG AGG CTG AGG TAT TTC TGT CTC | 161 |
| LIN28 | TGC ACC AGA GTA AGC TGC AC | CTC CTT TTG ATC TGC GCT TC | 189 |
| Klf4 | CCC ACA CAG GTG AGA AAC CT | ATG TGT AAG GCG AGG TGG TC | 169 |
| GAPDH | GGC AGC AGC AAG CAT TCC T | GCC CAA CAC CCC CAG TCA | 136 |
| GDF9 | TAG TCA GCT GAA GTG GGA CA | ACG ACA GGT GCA CTT TGT AG | 277 |
| GATA4 | TCC CTC TTC CCT CCT CAA AT | TCA GCG TGT AAA GGC ATC TG | 194 |
| HAND1 | TGC CTG AGA AAG AGA ACC AG | ATG GCA GGA TGA ACA AAC AC | 274 |
| βIII-tubulin | CAG AGC AAG AAC AGC AGC TAC TT | GTG AAC TCC ATC TCG TCC ATG CCC TC | 227 |
| MAP2 | GCA TGA GCT CTT GGC AGG | CCA ATT GAA CCC ATG TAA AGC C | 194 |
| GFAP | AGG GCT GAC ACG TCC AC | GCC TTA GAG GGG AGA GGA G | 132 |
| C/EPBα | AGG AAC ACG AAG CAC GAT CAG | CGC ACA TTC ACA TTG CAC AA | 141 |
| PPARr | AGCCTCATGAAGAGCCTTCCA | TCCGGAAGAAACCCTTGCA | 120 |
| Osteopontin | AGGAGGAGGCAGAGCACA | CTGGTATGGCACAGGTGATG | 150 |
| Aggrecan | ACA GCT GGG GAC ATT AGTGG | GTG GAA TGC AGA GGT GGT TT | 106 |
Lentivirus infection and iPSC generation
The iPSC was generated according to the previous report [5]. Ready-to-use highly efficient lentivirus kit containing OCT4, SOX2, KLF4 and c-MYC was used for iPSCs derivation. The kit was purchased from Cellexium Biomedica Incoporation, Taipei, Taiwan. The 1.5 x 104 cells of EPMSCs, CPMSCs, and EMSCs were seeded in one well of 12-well dish one day before transduction. The medium was replaced with virus-contained supernatant 200 μl with 800 μl culture medium supplemented with 1 μg/ml polybrene (Sigma, USA), and incubated for 24 hr. Twenty-four hours after transduction, a virus-containing medium was replaced with the 0.5 ml culture medium and replaced every other day. For iPSCs production, six days after transduction, the transfected MSCs were harvested by trypsinization and replated at 5 x 104 cells/100 mm dish on a MEF feeder layer. Next day, the medium was replaced with human ES cell medium [80% (v/v) knockout (KO) DMEM, 20% (v/v) KO serum replacement, 2 mM L-glutamine, 10 mM nonessential amino acid (all from Invitrogen), 50 μM β-mercaptoethanol (Sigma)] supplemented with 4 ng/ml bFGF. The medium was replaced every other day. Thirty days after transduction, colonies were picked up and transferred into 0.2 ml of human ES cell medium. The colonies were mechanically dissociated to small clumps by pipetting up and down. The cell suspension was transferred on MEF feeder in 24-well plates. We defined this stage as passage 1.
Characterization of embryo-like cells
Embryo-like cells were picked and fixed for imaging. The morphology of embryo-like cells was observed under inverted microscope (Olympus, Tokyo, Japan). Immunohistochemistry using GATA4 (1: 100, Chemicon) to identify the hypoblast, Cdx2 (1: 100, Chemicon) to identify the trophoblast, and Nanog (1:100) to identify the blastomere was performed. DAPI was used for counterstaining of cell nuclei. Cells were then observed by fluorescence microscopy (Olympus, Tokyo, Japan).
Human umbilical cord mesenchymal stem cells (HUCMSCs) culture
Detail of derivation of HUCMSCs as feeder cells for ES cells was reported in our previous report [28, 30]. Briefly, human umbilical cord was obtained from the pregnant women with informed consent approved by the institutional review board of Hualien Tzu Chi Hospital, Hualien, Taiwan. The Wharton's jelly was isolated from umbilical vessels and outlining amniotic membrane, and cut into pieces smaller than 0.5 cm3. The cut jelly was treated and incubated with trypsin/EDTA (Sigma, St. Louis, MO, USA) for 30 min at 37oC in a 95% air / 5% CO2 humidified atmosphere. The explants were then cultured in DMEM-LG containing 10% FBS and antibiotics, and left undisturbed for 5-7 days to allow the cells to migrate from the explants.
hES cells culture
The hES cells (TW1 cells line; P22) [30] were obtained from Biomedical Technology and Device Research Laboratories, Industrial Technology Research Institute and Initially cultured on MEFs, as directed by the supplier (FIRDI, Taiwan). Either MEFs or HUCMSCs after mitomycin-C deactivation were used as feeder cells for culturing the hES cells. They were plated at a density of 200,000 cells/9.4 cm2 well in six-well plates. The hES medium was replaced every other day.
Characterization of iPS cells (iPSCs)
For iPSC immunohistochemistry, Oct4, Sox2, Nanog, SSEA4, TRA-1-60 and TRA-1-81 (dilution 1: 50 to 1: 20) were used; all products were purchased from Chemicon, ES Cell Characterization Kit). Alkaline phosphatase staining was performed using the ES Cell Characterization kit (Chemicon).
Cells were fixed in 4% paraformaldehyde and permeabilized with PBS containing 5% skim milk (Becton, Dickinson, USA) and 0.1% Triton X-100 for 30 min. Cells were then incubated with mouse anti-human monoclonal antibodies overnight. After washing with PBS containing 0.5% Tween 20, cells were incubated with FITC-conjugated secondary antibodies for 30 min. Negative and positive control slides were prepared by incubating sections with isotype controls instead of the primary antibody. Cells were then washed 3 times with PBS and observed by fluorescence microscopy (Olympus, Tokyo, Japan).
Chromatin immunoprecipitation assay
The 1 × 107 cells were cross-linked with 1% formaldehyde for 5 min at room temperature and quenched by adding of 0.125 M glycine. The lysate of cells was sonicated to share chromatin-DNA complex. Chromatin, after centrifugation (13,000 rpm for 25 min at 4°C), was incubated over night with various antibodies. A chromatin sample, the same to that used in the ChIP, was regarded as the input. Immunoprecipitation was performed with Dynabeads Protein G (Invitrogen) - linked H3K4me3 (Upsate), H3K27me3 (Upstate) or normal rabbit IgG antibody as negative control. Elutes were used for quantitative PCR (Applied Biosystems) using gene specific primers design (Table 2). Data are expressed as percentage of input and all experiments were performed in triplicate. The data were normalized to the Pan H3 levels [5, 31, 32].
Primer sequence for ChIP.
| Primer | Forward sequence (5' to 3') | Reverse sequence (5' to 3') |
|---|---|---|
| Oct3/4 | TTG CCA GCC ATT ATC ATT CA | TAT AGA GCT GCT GCG GGA TT |
| SOX2 | GAG AAG GGC GTG AGA GAG TG | AAA CAG CCA GTG CAG GAG TT |
| NANOG | GAT TTG TGG GCC TGA AGA AA | GGA AAA AGG GGT TTC CAG AG |
| GATA6 | TGA GCG CAG TTC CGA CCC ACA GCC TG | GGG CGA GCG CGA GTC CGG GGT CTG |
| HAND1 | CCA TTG GCT CCC GGG AGA GGT TGA C | CCG GGC AAG GCT GAA AAT GAG ACG C |
In vitro Differentiation of iPSCs
Before differentiation, iPSCs were collected and resuspended in the iPSCs medium in the absence of bFGF. The iPSCs were then cultured in low attachment six-well plates as aggregates in suspension. After 5 days, 5 % FBS was added. Aggregated iPSCs usually formed an embryoid body (EB) after 7 - 10 days. The resultant EB was then plated on gelatin-coated chamber slides for fixation. After fixation, the differentiated iPSCs were studied by immunohistochemistry, using fluorescence-labeled antibodies specific for ectoderm (beta III tubulin; Tuj1 and glial fibrillary acidic protein), mesoderm (Brachyury), and endoderm (alpha fetal protein). All chemicals were purchased from Chemicon.
In vivo xenograft formation
The iPSCs were detached with mechanical slicing and pellets were resuspended in PBS with Matrigel (BD Bioscience, 1:1). These cells were injected into the subcutaneous tissue of the back of NOD-SCID mice (n=3). The iPSCs were kept on ice less than 45 mins for optimal viability prior to the injection. The tumor formation was followed up by palpation. Resulting tumors were dissected, fixed, and embedded in paraffin, for histological and immunohistochemical analyses (Hematoxylin & eosin, cytokeratin and vimentin, Tuj1, and AFP).
Results
The EPMSCs, EMSCs and CPMSCs had a MSC phenotype
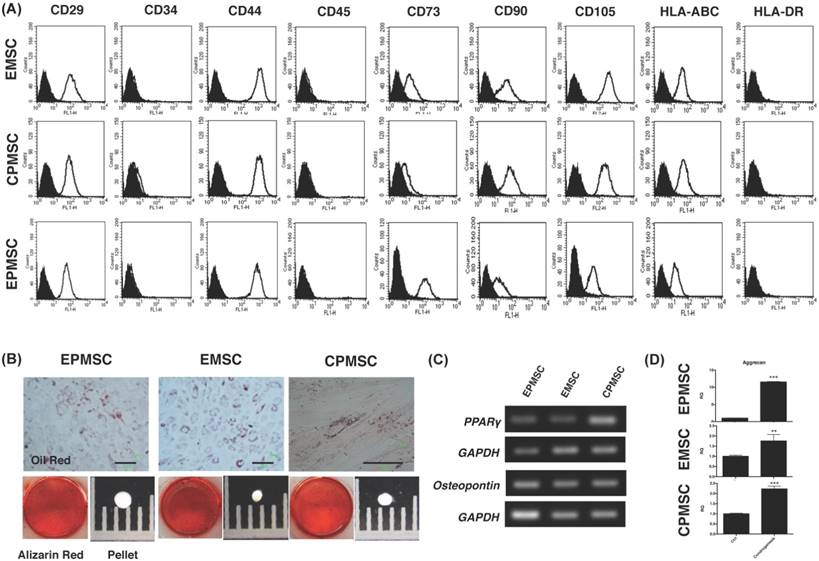
The derived EPMSCs, EMSCs and CPMSCs had a fibroblast-like appearance with a high degree of homogeneity and readily reached confluence in 14 days of culture. Like other MSCs, these MSCs expressed surface markers of CD29, CD44, CD73, CD90, CD105 and HLA-ABC (Fig. 1A), but did not express CD34, CD45 and HLA-DR (Fig. 1A).
These MSCs differentiated into adipocytes (positive of Oil red staining) and osteocytes (positive staining for Alizarin red) (Fig. 1B). They expressed PPARγ (adipocyte marker) and osteopontin (osteocyte marker) gene after the differentiation (Fig. 1C).
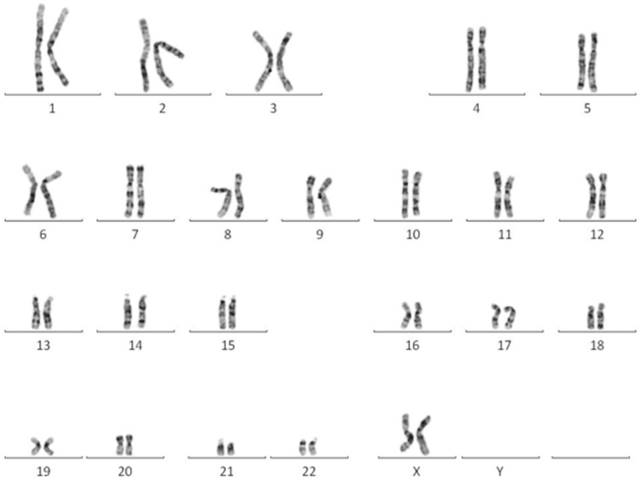
EPMSC revealed normal female karyotype
The karyotype of EPMSC revealed normal female 46, XX (Fig. 2).
iPSC induction from EPMSC and CPMSC resulted in embryo-like cells
While inducing iPSCs from EPMSCs and CPMSCs, to our surprise, various embryo-like cell clusters (we named “embryoids”) were found three days after introduction of iPSC cocktail. The embryoids showed different developmental stages from one cell to a hatching blastocyst (Fig. 3). They were found in approximately 50% of the cultured cells, and progressed in development by time until the hatching stage. Interestingly, the same iPSC induction did not produce any embryiod from normal endometrial tissues but could derive the same spectrum of embryoids from EPMSC and CPMSC.
The development process of single cell to iPSCs
To know the development of EPMSCs and CPMSC to iPSCs, we traced the development process from single cell after induction to form the iPSC sheet (Fig. 4). The same single cell developed into one cell (a), two cells (b), 8 cells (c), morula (d), blastocyst (e), hatching (f), and finally iPSCs sheets (g-i).
Embryo like cells expressed epiblast and trophectoderm markers
To know the embryo like cells expressed embryo specific markers (GATA4, Cdx2, and Nanog), we did the immunohistochemistry and confocal image. The resulted embryo like cells could express GATA4 (a hypoblast marker, Fig. 5A), Cdx2 (a trophoblast marker, Fig. 5B), and Nanog (a blastomere marker, Fig. 5). Cell nuclei was counterstained with DAPI (Fig. 5).
EPMSCs-derived iPSCs expressed pluripotency markers
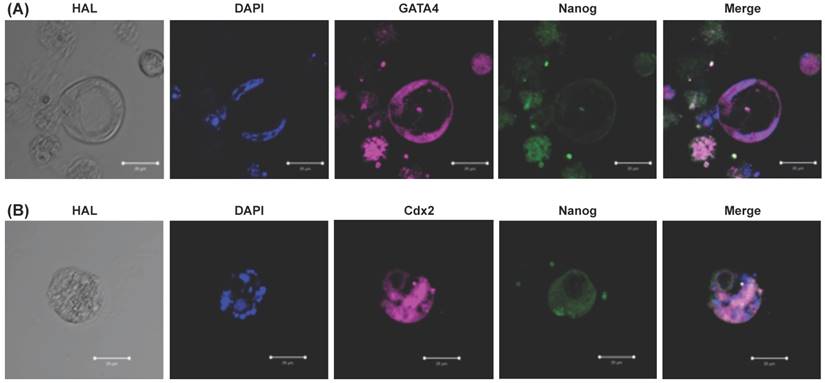
The EPMSC-derived iPSCs cultured on MEF or HUCMSC feeder formed colonies effectively, and continued to proliferate with a doubling time of 36 h. The morphology of colonies differed slightly from hESCs (Fig. 6A). The iPSC colonies were more flat than hESCs. However, the individual iPSCs morphology remained the same as hESCs. The cells remained round and small, with a high nucleus / cytoplasm ratio (Fig. 6A). The iPSCs expressed hESC-specific surface antigens, including alkaline phosphatase, SSEA-4, TRA-1-60, TRA-1-81, Nanog and Oct4 protein (Fig. 6B).
Immunophenotyping and in vitro differentiation of human endometrial polyp MSCs (EPMSCs), endometrial MSCs (EMSCs), and cervical polyp MSCs (CPMSCs). (A) Flowcytometry of rapidly dividing EMSCs, CPMSCs and EPMSCs showed negative for cluster of differentiation 34 (CD34), CD45 and human leukocyte antigen (HLA)-DR, and positive CD29, CD44, CD73, CD90, CD105 and HLA-ABC. (B) Upon adipogenic differentiation, the cells formed neutral lipid vacuoles and contained numerous Oil Red O-positive lipid droplets (B, upper panel). In osteogenic medium, the cells broadened to form a mineralized matrix, which was strongly stained with Alizarin Red S (B, lower panel) after 3-4 weeks of cultivation. In chondrogenic medium, the the cells formed pelles (B, lower panel). Scale bar: 100 μm (B, upper panel). (C) Expression of genes specific for adipogenic (peroxisome proliferator activated receptor; PPARγ) and osteogenic (osteopontin) differentiation was showed by RT-PCR analysis with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a positive control. (D) qRT-PCR showed aggrecan gene expression of the cells after chondrogenesis. **p<0.01, ***p<0.001.
RT-PCR showed the EPMSC-derived iPSCs expressed pluripotency genes including Oct4, sox2, Nanog, LIN28 and KLF4 at levels equivalent to or higher than those in the hESC line TW1 (Fig. 6C).
EMSC could form iPS after KSOM factor transfection
The EMSCs-derived iPSC cultured on MEF feeder could form colonies and expressed pluripotency markers such as Nanog, Sox2, and Oct4 (Fig. 7A). After EB formation, they could express three germ layer markers such as AFP (endoderm), Brachyury (mesoderm), Tuj1 and GFAP (endoderm) (Fig. 7B).
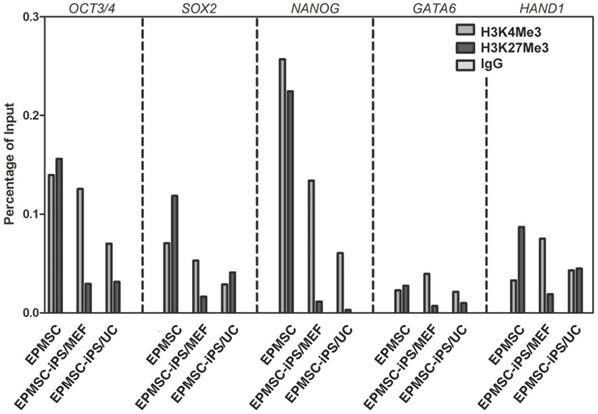
Histone modification on promoters of ES cell-specific genes in EPMSC-iPSCs
To investigate H3 methylation involved in transcriptional regulation in EPMSC-iPSCs after reprogramming, we performed chromatin immunoprecipitation on EPMSC and EPMSC-iPSC cultured on MEF or HUCMSC feeder (Fig. 8). We found an increase of the ratio of H3K4Me3 to H3K27Me3 in the promoter of pluripotent genes Oct3/4, Sox2, and Nanog and somatic genes GATA6 and HAND1 upon the iPSC induction from EPMSC (Fig. 8). Compared to the classical MEF-feeder, the HUCMSC feeder supported EPMSC-iPSC showed a same ratio in Oct3/4, Nanog and GTAT6 genes, but not in Sox2 and HAND1 gene.
EPMSC-derived iPSCs had the full capability of differentiation in vitro
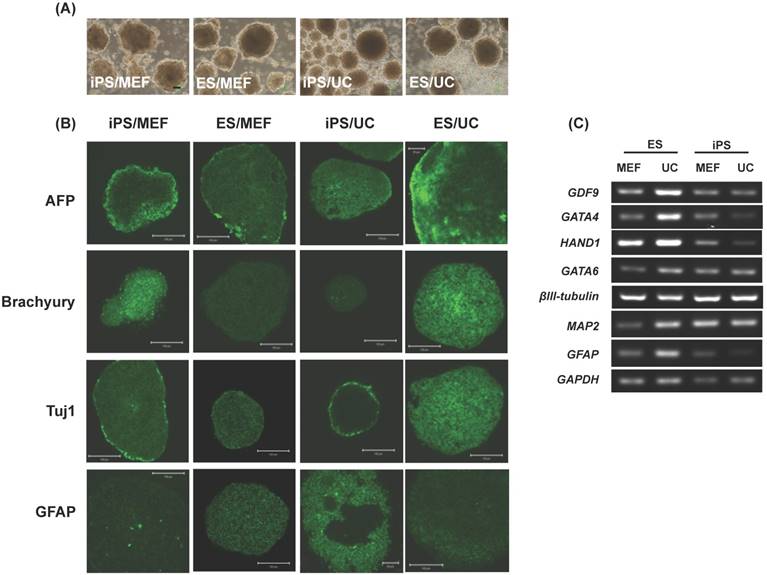
To determine the differentiation ability of EPMSC-iPSCs in vitro, we used floating cultivation to form embryoid bodies (EBs). After 5 days in suspension culture, both the MEF-fed and UCMSC-fed iPSCs formed a ball-shaped structure indifferent with the ESCs of the same feeders (Fig. 9A). Immunohistochemistry revealed these EBs were positive for markers of three germ layers, α-fetoprotein (AFP) for endoderm, Brachyury for mesoderm, and Tuj1 (βIII tubulin) and glial fibrillary acidic protein (GFAP) for ectoderm (Fig. 9B). RT-PCR confirmed the expression of the differentiation genes including GDF9 (germ cell marker), GATA4 (endoderm), heart- and neural crest derivatives-expressed protein 1 (HAND1, mesoderm), GATA-binding factor 6 (GATA6, endoderm), βIII tubulin (ectoderm), microtubule-associated protein 2 (MAP2, ectoderm) and glial fibrillary acidic protein (GFAP, ectoderm) (Fig. 9C).
A representative normal karyotype (46, XX) was observed in EPMSCs.
Early embryo-like cells developed from iPSC-induction of EPMSC and CPMSC. Representative pictures of EPMSC and CPMSC before and after induction to iPSC. (A) The original EPMSCs showed fibroblast-like cells with a spindle-shaped morphology (a). Three days after iPSC-induction, early embryo-like cells appeared, ranging from one-cell embryoid (b), two-cell embryoid (c), 6-cell embryoid (d), to blastocyst-like embryoid and hatching embryoid (f). Scale bar = 100 μm.
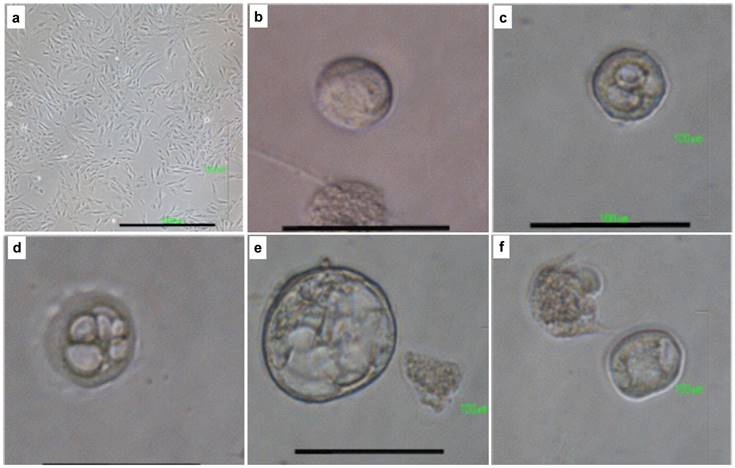
The development process from single cell of EPMSC to embryo-like cells and iPSCs. (a) one cell, (b) two cells, (c) 8 cells, (d) morula, (e) blastocyst, (f) hatching blastocyst, (g-i) iPSC sheets. Scale bar = 100 μm.
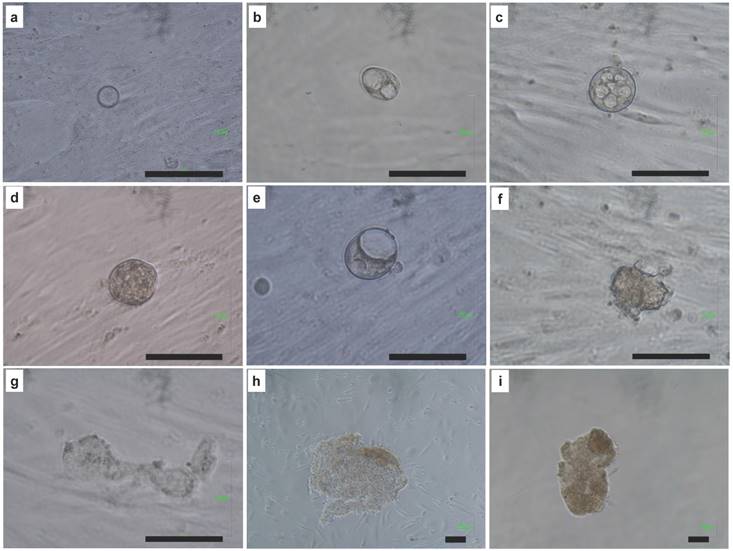
Confocal image and immunohistochemistry of resulted embryo like cells. (A) Embryo like cells were positive for GATA4 (pink color) which indicated a hypoblast marker. (B) Cells were positive for Cdx2 (pink color) which indicated a trophectoderm marker. Cells were positive for Nanog (green color) which indicated a blastomere marker. Cell nuclei was counterstained with DAPI (blue color). Scale bar = 50 μm.
Benign polyps of the uterus over-expressed C/EBPα
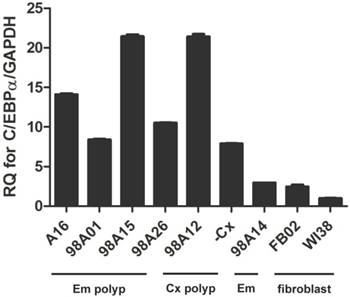
Recently, co-expression of C/EBPα was reported to increase efficiency to generate iPSC [16]. Therefore, we investigated how C/EBPa expressed in cells to make iPSC and embryo-like cells. The qRT-PCR was performed to investigate the C/EBPα expression in various tissue samples including endometrial and cervical polyps, endometrium, vaginal and lung fibroblasts. Fibroblast was used to generate iPSC frequently and was used as control. Uterine polyp and endometrium were used to generate iPSC in this experiment. High expression of C/EBPα was noted in four endometrial polyps and two cervical polyp cells, but not in endometrium and control fibroblasts (FB02 and WI38) (Fig. 10).
Morphology and expression of pluripotency genes in EPMSC-derived iPSCs compared to embryonic stem (ES) cells. (A) Morphology of the EPMSC-derived iPSCs of different magnifications at passage 4 on culture day 4 (a-c), as compared to ES cells of the passage 22 on culture day 4 (e-f). Scale bar = 1000 μm (a, d), = 100 μm (b, c, e, f). (B) Immunohistochemistry for Alkaline phosphatase (g), Stage-specific embryonic antigen (SSEA)-4 (h), TRA-1-60 (i), TRA-1-81 (j), Nanog (k) and octamer binding transcription factor 4 (Oct4) (l). Scale bar = 10 μm. (C) RT-PCR analyses of various pluripotency marker genes in EPMSC-derived iPS and ES cells cultured on different feeder layers. MEF: mouse embryonic fibroblast, UC: human umbilical cord mesenchymal stem cells.
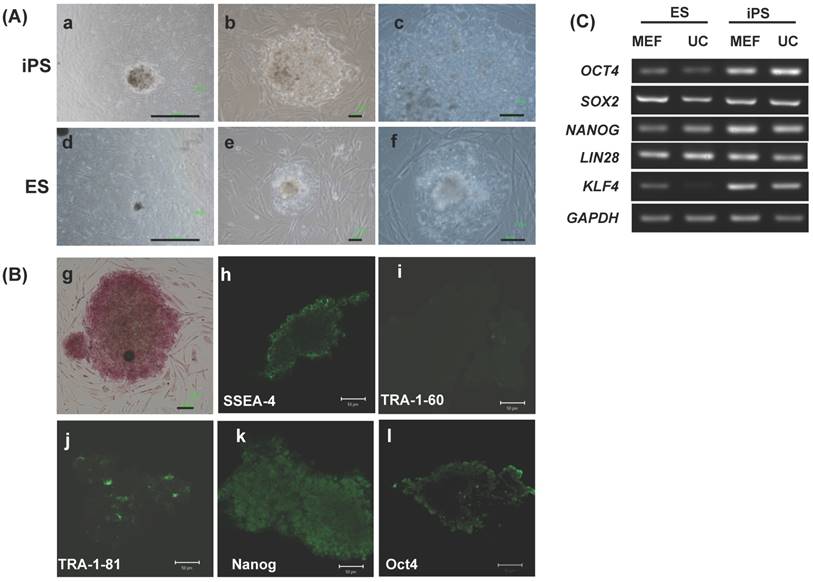
Morphology and expression of pluripotency genes in EMSC-derived iPSCs and embryoid body (EB). (A) Immunohistochemistry of EM-iPS for Nanog, Sox2, and octamer binding transcription factor 4 (Oct4). Scale bar = 100 μm. (B) Immunohistochemistry of EB showed positive for AFP (endoderm), Brachyury (mesoderm), Tuj1 and GFAP (ectoderm). Scale bar = 100 μm and 50 μm.
Analyses of histone markers at the promoter regions of pluripotency genes before and after iPSC induction of EPMSC (EPMSC-iPSC), and EPMSC-iPSC cultured on MEF or HUCMSC (UC) feeder on day 5. Chromatin immunoprecipitation of H3K4Me3 and H3K27Me3 markers was done with method described in M&M.
Embryoid body-mediated differentiation of human EPMSC-iPSCs. (A) Floating culture-derived embryonic body of ESCs (b, d) and EPMSC-iPSCs (a,c) grown at MEF or UCMSC (UC) feeders on Day 5. (B) Immunohistochemistry of various differentiation markers in the differentiated iPSCs and ESCs, including Alpha fetal protein (AFP), Brachyury, Beta III tubulin (Tuj1) and Glial fibrillary acidic protein (GFAP). Scale bar = 100 μm. (C) RT-PCR analyses of various differentiation markers of the three germ layers. MEF: mouse embryonic fibroblast, UC: human umbilical cord mesenchymal stem cells.
qRT-PCR analysis of CCAAT/enhancer binding protein-α (C/EBPα) expression in different cell lines. Primary mesenchymal cell cultures were derived from four endometrial polyp (Em polyp), two cervical polyp (Cx polyp), one normal endometrium (Em), as well as two fibroblast cell lines (FB02, WI38).
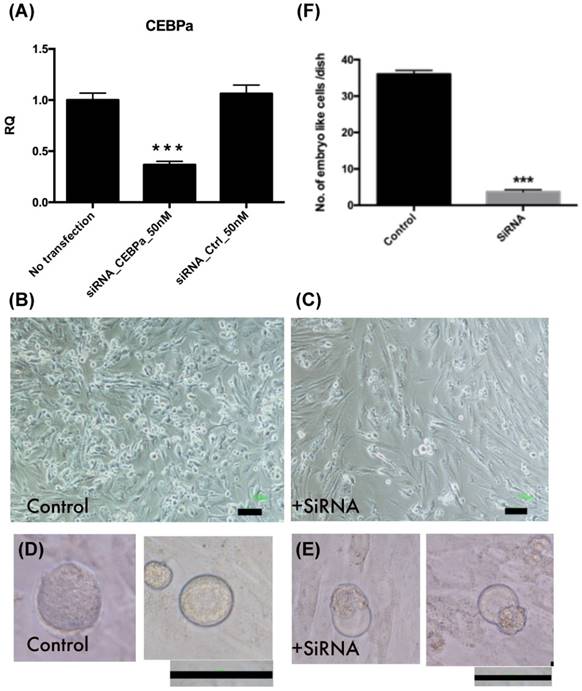
Inhibition of C/EBPα expression decreased embryo like cell formation
Endometrial polyp stem cells after C/EBPα siRNA transfection, the gene expression was significant decreased (p<0.001, Fig. 11A-C). After 4 factors transfection, embryo like cells (Fig. 11 D-E) are significantly decreased in siRNA transfected cells at day 4 (p<0.001, Fig. 11F).
Embryo like cells decreased after CCAAT/enhancer binding protein-α (C/EBPα) was downregulation. (A) The C/EBPα downregulation in endometrial polyp stem cells after siRNA transfection (p<0.001). (B-C) The cell morphology of control and siRNA transfected EPMSCs. (D-E) Four days after 4-factor transfection, the morphology of embryo like cells in both groups. (F) The amount of embryo like cells after 4-factor transfection in both groups (p<0.001). Scale bar =100 μm.
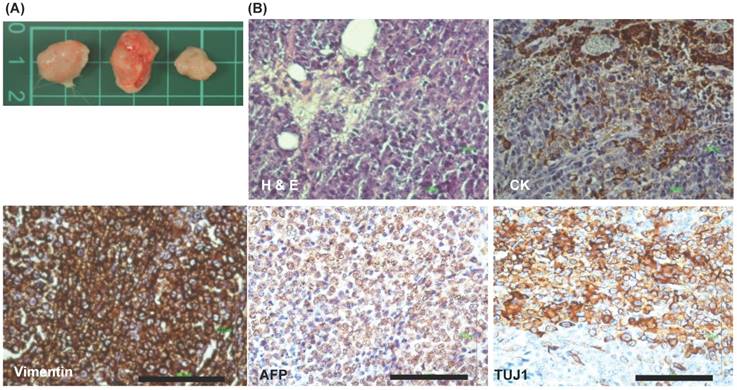
Growth of mesodermal and ectodermal tumors upon EPMSC-iPSCs injection in NOD-SCID mice
To clarify the in vivo three germ layers differentiation of EPMSC-iPSCs, these iPSCs were subcutaneously injected into NOD-SCID mice (immune-compromised mice) for generating teratoma. After iPSCs growth for 3 months in the mice, the tumors were developed. The incidence of tumorigenesis was 100% (3/3, Fig. 12A). Histology and IHC revealed partial positive for cytokeratin and strong positive for vimentin (mesoderm), AFP (endoderm) and Tuj1 (ectoderm) (Fig. 12B).
Xenograft experiments of iPSCs. (A) Xenograft tumor morphology. (B) Immunohistochemistry (IHC) of the xenograft tumors. H & E: hematoxyline & eosin, IHC showed positive for cytokeratin (CK, ectoderm), vimentin (mesoderm), AFP (endoderm), and Tuj1 (ectoderm). Scale bar = 100 μm.
Discussion
In this study, we demonstrated that iPSCs could be generated from EPMSCs, CPMSCs, and EMSCs by lentiviral transduction of the four transcription factors, Oct4, Sox2, Klf4 and c-myc. The established iPSCs are similar to hESCs in morphology, proliferation, feeder dependence, surface markers, gene expression, histone modification, and in vitro differentiation and in vivo tumorigenesis with three germ layer differentiation. Surprisingly, three days after induction, the embryo-like cells with various embryo developmental stages appeared. One cell, two cells, 4 cells, blastocyst and hatching blastocyst presented at the same time (Fig. 3). Approximately 50% cells could form various stages of embryoids. These embryoids could survive till 7 days after virus transduction. After that, a lot cells with broken cell membranes with hatched cell mass could be observed (Fig. 4). The embryo like cells expressed the markers for hypoblasts (GATA4), trophoblasts (Cdx2), and blastomere (Nanog, Fig. 5). However, this phenomenon was just observed in uterine polyp stem cells, but not in endometrial stem cells (EMSC).
There is no report of an embryo-like cell formation during iPSCs induction. The present study is the first to demonstrate that human uterine polyps could develop into the embryo-like cells in appropriate conditions. The generation of embryo-like cells from uterine polyps may provide a valuable model for identifying factors involving formation and differentiation of the embryo or embryo-like cell.
Models of embryo cell development in vitro have decades of history, but lack of a standard system to promote the formation of mature embryos. Endometrial polyps had excellent capability to differentiate into adipocyte, osteocyte and neuron [29]; they appeared to have better proliferation and clonogenicity than endometrial stem cells [29]. The polyps of Mullerian duct-derived organ may harbor some epigenetic markers making them vulnerable to reprogram to the earliest developmental stage. It may reflect during the induction pluripotency process, the cells can replay the embryogenesis process from early embryo to hatching blastocyst and became inner cell mass like pluripotency cells.
In Fig. 5, the NANOG overlaps with CDX2 and GATA4 partially in the immunostaining. We speculate the overlap might be indicated they were in transdifferentiation status.
C/EBPα could accelerate reprogramming iPSCs from B cells; iPSC like colony could be observed since day 4 after C/FBPα [16]. We found that C/EBPα expression was higher in uterine polyp stem cells than in endometrial stem cells and fibroblasts (Fig. 8). After downregulation the gene expression, the embryo like cells were significant decreased (Fig. 11). In this study, we found embryo like cells derived from uterine polyp stem cells but not from endometrial stem cells. C/EBPα may involve this embryogenesis phenomenon during this process.
We note the limitations to our study. We use small number of samples for the experiment. Further studies using large number of specimen should be performed. In addition, small embryo-like cell could not be easily picked-up and tracked for the further study. Single cell PCR may be able to use for detecting gene expression of the embryo-like cells in the future.
In conclusion, mesenchymal stem cells derived from endometrial polyps and cervical polyps could be dedifferentiated to early embryo-like cells by introducing genes of the iPSC cocktail. Polyps of the Mullerian duct derived organ may harbor some epigenetic markers making them vulnerable to reprogramming to the earliest developmental stage. This study provides a simple model to derive early human embryo-like cells by in vitro.
Acknowledgements
The authors thank Dr. Jon-Son Kuo for English editing. The funding source: Intramural project of Hualien Tzu Chi Hospital (TCRD 106-16).
Author contribution
DC: experiments design and preform, results interpretation, drafting the manuscript and final approval of manuscript. TY: results interpretation, drafting the manuscript and final approval of manuscript. HW: results interpretation, drafting the manuscript and final approval of manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704-12
2. Daneshvar N, Rasedee A, Shamsabadi FT, Moeini H, Mehrboud P, Rahman HS. et al. Induction of pluripotency in human umbilical cord mesenchymal stem cells in feeder layer-free condition. Tissue Cell. 2015;47:575-82
3. Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci U S A. 2010;107:5483-8
4. Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M. et al. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci U S A. 2010;107:17639-44
5. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-72
6. Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K. et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55-70
7. Papp B, Plath K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 2011;21:486-501
8. Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835-50
9. Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652-5
10. Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370-4
11. Nishino K, Toyoda M, Yamazaki-Inoue M, Makino H, Fukawatase Y, Chikazawa E. et al. Defining hypo-methylated regions of stem cell-specific promoters in human iPS cells derived from extra-embryonic amnions and lung fibroblasts. PLoS One. 2010;5:e13017
12. Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K. et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183-97
13. Brix J, Zhou Y, Luo Y. The Epigenetic Reprogramming Roadmap in Generation of iPSCs from Somatic Cells. J Genet Genomics. 2015;42:661-70
14. Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M. et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479-91
15. Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M. et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250-64
16. Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP. et al. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235-9
17. Qiu P, Bai Y, Pan S, Li W, Liu W, Hua J. Gender depended potentiality of differentiation of human umbilical cord mesenchymal stem cells into oocyte-Like cells in vitro. Cell Biochem Funct. 2013;31:365-73
18. Hua J, Pan S, Yang C, Dong W, Dou Z, Sidhu KS. Derivation of male germ cell-like lineage from human fetal bone marrow stem cells. Reprod Biomed Online. 2009;19:99-105
19. Nayernia K, Lee JH, Drusenheimer N, Nolte J, Wulf G, Dressel R. et al. Derivation of male germ cells from bone marrow stem cells. Lab Invest. 2006;86:654-63
20. Danner S, Kajahn J, Geismann C, Klink E, Kruse C. Derivation of oocyte-like cells from a clonal pancreatic stem cell line. Mol Hum Reprod. 2007;13:11-20
21. Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT. et al. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727-39
22. Nayernia K, Nolte J, Michelmann HW, Lee JH, Rathsack K, Drusenheimer N. et al. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell. 2006;11:125-32
23. Ramathal C, Durruthy-Durruthy J, Sukhwani M, Arakaki JE, Turek PJ, Orwig KE. et al. Fate of iPSCs derived from azoospermic and fertile men following xenotransplantation to murine seminiferous tubules. Cell Rep. 2014 in press
24. Easley CAt, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP. et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440-6
25. Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971-5
26. Ding DC, Shyu WC, Lin SZ. Mesenchymal stem cells. Cell Transplant. 2011;20:5-14
27. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD. et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143-7
28. Ding DC, Shyu WC, Chiang MF, Lin SZ, Chang YC, Wang HJ. et al. Enhancement of neuroplasticity through upregulation of beta1-integrin in human umbilical cord-derived stromal cell implanted stroke model. Neurobiol Dis. 2007;27:339-53
29. Ding DC, Chu TY, Chiou SH, Liu HW. Enhanced differentiation and clonogenicity of human endometrial polyp stem cells. Differentiation. 2011;81:172-80
30. Chang YL, Chen SJ, Kao CL, Hung SC, Ding DC, Yu CC. et al. Docosahexaenoic acid promotes dopaminergic differentiation in induced pluripotent stem cells and inhibits teratoma formation in rats with Parkinson-like pathology. Cell Transplant. 2012;21:313-32
31. Buro LJ, Chipumuro E, Henriksen MA. Menin and RNF20 recruitment is associated with dynamic histone modifications that regulate signal transducer and activator of transcription 1 (STAT1)-activated transcription of the interferon regulatory factor 1 gene (IRF1). Epigenetics Chromatin. 2010;3:16
32. Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E. et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332-8
Author contact
![]() Corresponding author: Dah-Ching Ding, M.D., Ph.D., Department of Obstetrics and Gynecology, Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, 707, Chung-Yang Rd., Sec. 3, Hualien, Taiwan. Tel: +886-3-8561825; Fax: +886-3-8577161; Email: dah1003com.tw
Corresponding author: Dah-Ching Ding, M.D., Ph.D., Department of Obstetrics and Gynecology, Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, 707, Chung-Yang Rd., Sec. 3, Hualien, Taiwan. Tel: +886-3-8561825; Fax: +886-3-8577161; Email: dah1003com.tw

 Global reach, higher impact
Global reach, higher impact