Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(12):1630-1635. doi:10.7150/ijbs.26932 This issue Cite
Review
The Biological Roles of Translation Initiation Factor 3b
1. Center for Translational Medicine, the First Affiliated Hospital of Xi'an Jiaotong University
2. Key Laboratory for Tumor Precision Medicine of Shaanxi Province, the First Affiliated Hospital of Xi'an Jiaotong University
Received 2018-4-27; Accepted 2018-8-5; Published 2018-9-7
Abstract
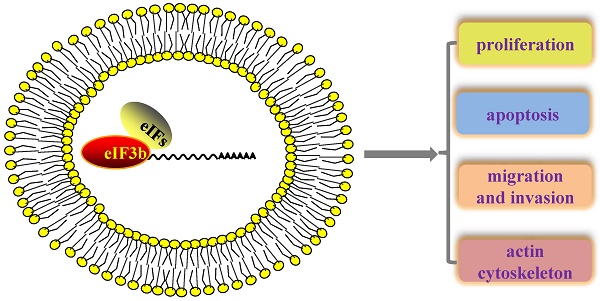
Translation has important roles in almost all physiological and pathological processes, and translation initiation factors are particularly relevant to the translation initiation step, which is the most important step in translation regulation. Translation initiation factor 3b (eIF3b), a key subunit of the largest translation initiation factor 3 (eIF3), is widely considered a scaffold protein that acts to ensure the accuracy of translation initiation. A series of recent finds has revealed that eIF3 is closely related to oncogenesis. However, the concrete mechanism by which eIF3b is involve in carcinogenesis remains elusive. Here, we summarize a series of research findings regarding the relationship between eIF3b, translation and cancer.
Keywords: eIF3b, translation initiation, cancer.
1. Introduction
The process of gene expression is complex but precisely controlled by multi-level regulation. Transcription and translation are the most basic and main regulatory mechanisms of gene expression. The rate of protein synthesis is determined primarily by translation, and the phase of mRNA translation is divided into three steps: initiation, elongation and termination. Translational control occurs predominately at the initiation step, which involves translation initiation factors.
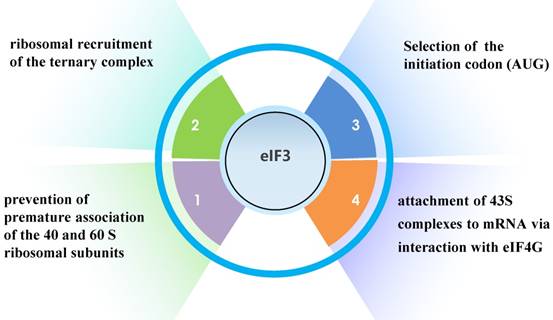
Translation initiation factor 3 (eIF3), the largest, most complex translation initiation factor, is involved in almost the entire process of translation initiation regulation, including ribosomal recruitment of the ternary complex, attachment of 43S complexes to mRNA via interaction with eIF4G, selection of the initiation codon (AUG), and prevention of premature association of the 40 and 60 S ribosomal subunits (Fig. 1) [1].
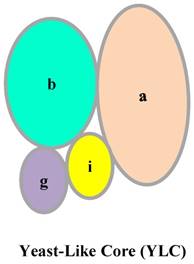
In mammals, eIF3 is an approximately 800- kDa complex, composed of 13 subunits: eIF3a, eIF3b, eIF3c, eIF3d, eIF3e, eIF3f, eIF3g, eIF3h, eIF3i, eIF3j, eIF3k, eIF3l, and eIF3m [2]. eIF3a, eIF3c, eIF3e, eIF3k, eIF3l, and eIF3m, which contain a PCI (proteasome, COP9/signalosome, eIF3) domain, and eIF3f, and eIF3h, which have an MPN (Mpr1-Pad1-N-terminal) domain, make up the octameric structural core of eIF3 [2]. Some studies have indicated that the remaining subunits, eIF3b, eIF3d, eIF3g, and eIF3i, have a stable but flexible connection with the octameric structural core [3,4,5]. However, another study proposed that eIF3a, eIF3b, eIF3g, and eIF3i in humans form the Yeast-Like Core (YLC) (Fig. 2) [6]. The YLC is essential for the regulation of a series of cellular processes by the eIF3 complex, and notably, the eIF3b subunit of the YLC is essential for the formation of the integral eIF3 complex [6]. In HeLa cells, the expression of eIF3b is hampered, resulting in downregulation of all eIF3 subunits. However, eIF3i and eIF3j levels are not markedly changed [6]. These findings illustrate the importance of the eIF3b subunit in the eIF3 complex.
The composition of eIF3 is conserved in most eukaryotes, certain plants, and protists [7]. However, the eIF3 complex of the budding yeast Saccharomyces cerevisiae consists only of TIF32, PRT1, NIP1, TIF35 and TIF34, which corresponds to eIF3a, eIF3b, eIF3c, eIF3g, and eIF3i, respectively [8].
In summary, eIF3b is widely regarded as a major scaffold protein that plays essential roles in translation regulation, cell growth, and oncogenesis.
The functions of eIF3.
Structure of YLC.
2. Structure of eIF3b
eIF3b is also known as Prt1 homolog, P110, P116, eIF-3-Eta, eIF3S9 and HPrt1. The highly conserved gene has been cloned and mapped to the chromosome region 7p 22.3. The gene has 19 exons and 18 introns and the encoded 92- kDa protein consists of 814 amino acids.
(1) Domains of eIF3b
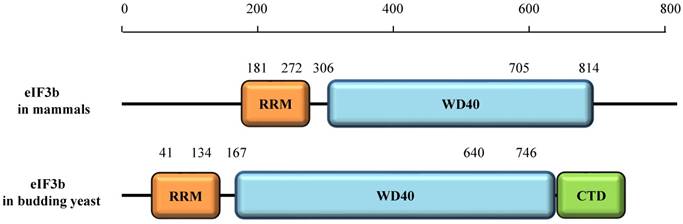
The eIF3b protein in mammals contains two domains. The N-terminal domain (NTD) consists of a structurally uncanonical RNA recognition motif (RRM) and the WD40β domain (Fig. 3) [2]. Interestingly, the sheet area of eIF3b-RRM in humans (residues 185-268), is mostly negatively charged [9]. The typical RRM, a common structural motif, contains conserved ribonucleoprotein1 (RNP1) and ribonucleoprotein2 (RNP2) sequences, which include conserved aromatic residues necessary for interaction with RNA [10,11]. Surprisingly, the RNP2 of eIF3b has a shortage of aromatic residues [9]. Also, helix 1 of the uncanonical RRM is composed of 14 amino acids, which in contrast to the helix of typical RRM, which does not exceed 10 amino acids [9]. Previous studies showed that eIF3b cannot interact with RNA in vitro [12,13], and these results indicate that the atypical RRM of eIF3b is not directly related to RNA binding. Notably, eIF3b can only bind RNA in the presence of the whole eIF3 complex. Nonetheless, the structure of the yeast eIF3b-RRM (PRT1-RRM) corresponds to the canonical RNA recognition motif, which is vital for eIF3 complex integrity and key for interaction with ribosomal subunits [9]. Similarly, the eIF3b-RRM in yeast also cannot bind with RNA in vitro [14].
The WD40β domain of eIF3b (residues 306 -705), a nine-bladed β-propeller fold, is highly conserved from yeast to humans [2,16]. The WD40 domain, which consists of about 40 amino acid residues in a single repeat, is one of the top interacting domains in eukaryotic genomes [17], and its name is derived from the conserved tryptophan-aspartic acid (W-D) dipeptide at its C terminus [18]. Each WD40 repeat consists of an antiparallel β-sheet with four -strands [18]. The WD40 domain may be related to a series of cellular processes, including cell cycle progression, apoptosis, transcription, signal transduction, chromatin dynamics regulation, cytoskeletal assembly, and others [18]. However, it generally regulates protein-protein interactions to mediate a series of signaling events [18].
The eIF3b protein in budding yeast contains three domains, the RRM, WD40β, and C-terminal domain (CTD). The CTD was shown to be essential for the interaction of eIF3i and eIF3g [19, 20].
(2) Modification of eIF3b
Although eIF3 has many functions mediated via multiple domains, the foundational mechanisms by which eIF3 exerts these functions remain vague. It is well known that protein phosphorylation is the most basic and, common key mechanism for regulating protein activity and function. Therefore, the activity of eIF3b may be directly influenced by the phosphorylation of eIF3b. Research has shown that targeting kinase or phosphatase activity upstream of eIF3b is a way to control eIF3b-mediated translation for tumor treatment, particularly via the phosphorylation of the eIF3 complex at seven phosphorylation sites (Ser-83, Ser-85, Ser-119, Ser-125, Ser-152, Ser-154, and Ser-164) and one acetylation site (Met-1) [36]. These observations provide evidence that post-translational modification of eIF3b contributes to the protein's function, but the underlying mechanisms require further investigation.
Domains of eIF3b protein in mammals and budding yeast.
Consistently, phosphorylation of other members of the eIF family has been linked to tumorigenesis. For example, inhibition of eIF2α phosphorylation can promote tumorigenesis through the overexpression of PKR (protein kinase RNA-activated) [37,38]. By contrast, another study reported that eIF2α phosphorylation may prevent cancer development and progression. Thus, the role of eIF2α phosphorylation in cancer development is complex, and may depend on the specific physiological context [39]. Similarly, studies have reported opposing roles for the phosphorylation of eIF4G in tumor regulation [40,41,42,43]. Although the mechanisms appear complex, these findings do provide evidence that phosphorylation of the eIFs is associated with tumorigenesis. Future discovery of an upstream kinase of eIF3b as well as an inhibitor of that kinase could offer an important breakthrough in cancer.
3. Functions of eIF3b
(1) eIF3b and translation
Previous research indicated that eIF3b is the master scaffolding subunit. The combination of four subunits (eIF3a, eIF3g, eIF3i, and eIF3j) supports the accuracy of the scanning-arrested structure, which is essential for rigorous AUG recognition [9,21,22]. Similarly, PRT1, a homolog of eIF3b in yeast, binds TIF32 (eIF3a), TIF35 (eIF3g), TIF34 (eIF3i), HCR1 (eIF3j), and Pci8 (eIF3e), which highlight the conserved nature of eIF3b [19,23,24]. Another study demonstrated that knock-down of eIF3b brings about a large reduction in the ratio of polysomes to monosomes (P/M), which reflects translation rates [6]. Moreover, another study demonstrated that eIF3b has molecular function of scanning for the start codon. eIF3b, is relocated to the 40S intersubunit face, where it establishes an interaction network with the eIF2-Met-tRNAiMet ternary complex [55].
A groundbreaking study demonstrated the interaction among P311, the transforming growth factor (TGF) 5' untranslated regions (UTRs) and eIF3b in an RNA-independent manner can promote the translation of TGF-1, 2, and 3 in NIH-3T3 cells [15]. P311 is known to have important roles in cell differentiation, cell migration, and blood vessels formations. Another study showed that eIF3b is essential for the interaction of AUF1 (AU-rich element RNA-binding protein 1) with the cryptochrome1 (Cry1, mouse core clock gene) 3'UTR, which regulates the translation of Cry1 mRNA [25]. Interestingly, AUF1 also interacts with eIF3b in an RNA-independent manner [25]. Encouragingly, another RNA-binding protein CERKL (Ceramide Kinase Like), which is associated with an autosomal recessive form of retinitis pigmentosa, was also found to bind to eIF3b by immunoprecipitation experiments in HEK-293T cells [26]. While our understanding of the underlying mechanism remains incomplete, these findings hint that it may involve ribosomes. The interaction of CERKL and eIF3b also may not be an RNA-mediated indirect interaction, that is, the RNA-mediated indirect interaction may not be related to the RRM domain of eIF3b and instead may be related to the WD40β domain of eIF3b, because it often acts as a rigid scaffold to mediate protein-protein interaction. Chaperonin containing TCP-1 (tail-less complex polypeptide 1) (CCT) can interact with newly synthesized eIF3b and promotes the accurate folding of eIF3h and eIF3i [27]. Also, direct interaction of the herpes simplex virus (HSV) immediate-early ICP27 protein with the eIF3b subunit in vitro largely affects viral gene expression [28].
The association of eIF3b and cancer.
Strikingly, two studies demonstrated that two proteins (DDX3 and S6K1) interact with eIF3b under while in the eIF3 complex [29,30]. A conserved RNA helicase involved in numerous cancers, DDX3, was immunoprecipitated together with several eIF3 subunits (excluding subunits eIF3j and, eIF3l) in HeLa cells and linked to translation regulation [29]. The eIF3 pre-initiation complex (PIC) is regarded as a dynamic scaffold for the translation initiation complex assembly by the regulation of mammalian target of rapamycin (mTOR) and S6K1 [30].
In summary, eIF3b does have a major influence on translation regulation.
(2) eIF3b and cell growth
The regulation of cell proliferation by eIF3b was studied in HeLa cells [6], and the results demonstrated that downregulation of the eIF3b subunit strongly diminished the capacity for cell growth. Although the mechanism by which eIF3b controls cell growth is poorly understood, two hypotheses have been proposed: (1) many proteins involved in growth-promoting signaling pathways are invalid translated, and an unbalanced state occurs between the growth-promoting pathway and the growth-retarding pathway when the entire protein translation level increases [31]; and (2) the translation initiation factor complex regulates the translation of many specific mRNAs involved in many significant cellular processes, including cell proliferation, apoptosis, DNA repair, and angiogenesis, and a berrant changes in the expression levels of these proteins can cause cancers [31].
As mentioned above, Prt1, which later becomes Prt1p, is the homolog of eIF3b in S. cerevisiae, and when Prt1 is mutated, it can inhibit global protein synthesis [32,33] and bring about G1 phase arrest [34,35]. In addition, emerging evidence shows a tight correlation between eIF3b and cell cycle progression. In several tumor types, eIF3b can regulate the cell cycle and eventual tumor developments.
(3) eIF3b and cancer
Accumulating evidence indicates that eukaryotic translation initiation factors are closely linked to oncogenesis and their expression levels vary among different tissues and cells.
eIF3a, eIF3c, and eIF3h are overexpressed in several human cancers, and eIF3e and eIF3f, which exhibit tumor suppressor characteristics, are downregulated in many human cancers [44]. These findings demonstrate a duality in eIF3 function that may be involved in regulating cancer development.
However, there is little evidence to indicate that whether altered levels of eIF3 subunits are a cause or a consequence of malignant phenotypes. One study showed that ectopic expression of eIFs in stably transfected NIH3T3 cells leads to several carcinogenic characteristics, including an increased protein synthesis rate, the ability for anchorage-independent growth, inhibition of apoptosis, and other malignant phenotypes [45].
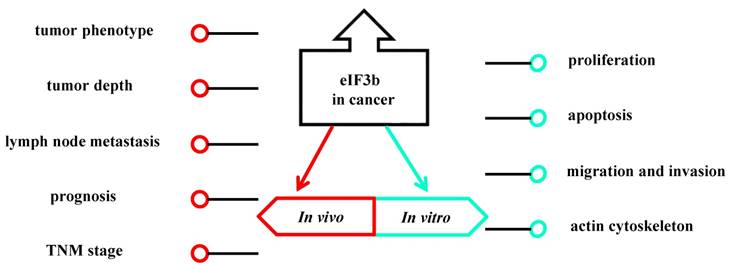
Also, high levels of eIF3b have been detected in several tumors, such as hepatocellular carcinoma (HCC) [46], osteosarcoma [47], clear cell renal cell carcinoma (ccRCC) [48], bladder cancer [49], prostate cancer [49], esophageal squamous cell carcinoma (ESCC) [50], glioblastoma [51], colon cancer [52], breast cancer [53] and gastric cancer [54]. In ccRCC, high expression of eIF3b is not only associated with an aggressive tumor phenotype, but also is an independent predictor of patient prognosis [48]. Overexpression of eIF3b in ESCC was found to be associated with lymph node metastasis, tumor depth, and advanced TNM stage, and ESCC patients with a high- level of eIF3b expression have a shorter overall survival and disease-free survival than those with lower eIF3b expression (Fig. 4) [50]. High eIF3b expression also associated with androgen independence and worse disease- specific -survival in prostate cancer patients [49]. Another study showed that eIF3b can inhibit cell proliferation by interfering with cell cycle progression and promoting apoptosis in vitro in ccRCC, ESCC, osteosarcoma, bladder cancer and glioblastoma. Moreover, in ccRCC and bladder cancer, downregulation of eIF3b can inhibit cancer cell epithelial-to-mesenchymal transition (EMT) by suppressing cancer cell migration and invasion.
Interestingly, eIF3b depletion in ccRCC cells damaged the actin cytoskeleton and focal adhesions via the down-regulation of integrin α5 protein levels [48]. Several studies have shown that integrins may promote tumor invasion. The impact of eIF3b deletion on ccRCC is mediated via the Akt pathway [48]. eIF3b depletion can lead to increased protein expression of tumor necrosis factor receptor superfamily member 21 (TNFRSF21) in osteosarcoma, and several studies have reported that TNFRSF21 is a key molecule in inflammation, immune regulation and malignant progression [48].
How alteration of the expression levels of different eIF3 subunits promotes tumorigenesis remains to be elucidated, but a possible explanation for these observations is that abnormal expression of eIFs might affect cell proliferation, apoptosis, migration, and invasion as well as, might change the translation efficiency of specific mRNAs, such as those encoding key proteins involved in the formation of various tumors.
Although the mechanism by which eIF3b specifically mediates tumor formation is unclear, we suspect that the relationship between the phosphorylation of eIF3b and tumorigenesis may be an important point in future research.
Future studies will need to focus on determining the signaling pathways involved in the regulation of eIF3b in order to develop eIF3b-targeting therapeutics for different human cancers.
4. Future perspectives
eIF3b is an important factor in translation initiation, with four proven functions in the course of translation initiation so far. However, the underlying mechanisms of eIF3b and other translation factors are still unclear. Moreover, the functions of eIF3b beyond its activity as a translation initiation factor should be investigated.
Several previous studies have focused on the elevated expression of eIF3b in almost all tumor types and how eIF3b contributes to tumorigenesis. However, only a limited number of studies have aimed to understand the underlying mechanism. eIF3b functions as an oncogene in many cancers. Generally speaking, translation initiation factors regulate the translation of all genes, including proto-oncogenes and tumor suppressor genes. We have yet to understand why eIF3b promotes the translation of proto-oncogenes and inhibits the translation of tumor suppressor genes to maintain its role as a proto-oncogene. Even more puzzling is how eIF3b regulates its activity as a translation initiation factor.
As mentioned above, the fact that downregulated eIF3b expression in HeLa cells can regulate almost all eIF3 subunits implies that eIF3b may determine the fate of the entire eIF3 family. Furthermore, eIF3 fulfills vital and multiple functions during translation initiation. Therefore, it is of great significance to using significant manpower and resources to the study of eIF3b. If we can identify drugs targeting eIF3b, this will greatly promote the development of cancer treatment. So far, more attention has been paid to eIF4, a translation initiation factor that is the target of many pathways, and advances have been made in therapeutic approaches to inhibit the eIF4 machinery [39]. The developed eIF4- targeting drugs have mostly been derived from the mTOR pathway [39]. One study has shown a shallow relationship between eIF3-PIC and mTOR, but the specific mechanism remains to be studied in depth [30].
In conclusion, research of eIF3b is only in its infancy, and no drugs have been developed for targeted therapy involving eIF3b. In the future, a more complete understanding of the functions of eIF3b in human cancers will provide answers needed to develop improved therapies.
Acknowledgements
This work was financially supported by grants from the National Natural Science Foundation of China (Nos. 81502620 and, 81702631).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Saletta F, Suryo Rahmanto Y, Richardson DR. The translational regulator eIF3a: the tricky eIF3 subunit!. Biochimica et biophysica acta. 2010;1806:275-86
2. des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491-5
3. Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA. et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108-19
4. Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA. et al. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539-43
5. Sun C, Todorovic A. et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). PNAS. 2011;108:20473-8
6. Wagner S, Herrmannova A, Sikrova D, Valasek LS. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic acids research. 2016;44:10772-88
7. Rezende AM. et al. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates - identification of conserved and divergent features based on orthologue analysis. BMC Genomics. 2014;11:1175
8. Phan L. et al. Identification of a Translation Initiation Factor 3 (eIF3) Core Complex, Conserved in Yeast and Mammals, That Interacts with eIF5. Molecular and cellular biology. 1998;18:4935-46
9. ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. The Journal of biological chemistry. 2007;282:8165-74
10. Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407-45
11. Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. The FEBS journal. 2005;272:2118-31
12. Katsura Asano TGK, William C. Merrick, and John W. B. Hershey. Conservation and Diversity of Eukaryotic Translation Initiation Factor eIF3. BIOLOGICAL CHEMISTRY. 1997;272:1101-9
13. Karen L. Block H-PV, and John W. B. Hershey. Characterization of cDNAs Encoding the p44 and p35 Subunits of Human Translation Initiation Factor eIF3. BIOLOGICAL CHEMISTRY. 1998;273:31901-8
14. Naranda T. et al. The 39-Kilodalton Subunit of Eukaryotic Translation Initiation Factor 3 Is Essential for the Complex's Integrity and for Cell Viability in Saccharomyces cerevisiae. Molecular and cellular biology. 1997:145-53
15. Yue MM. et al. Novel RNA-binding protein P311 binds eukaryotic translation initiation factor 3 subunit b (eIF3b) to promote translation of transforming growth factor beta1-3 (TGF-beta1-3). J Biol Chem. 2014;289:33971-33983
16. Liu Y, Neumann P, Kuhle B, Monecke T, Schell S, Chari A. et al. Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit. Structure. 2014;22:923-30
17. Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends in biochemical sciences. 2010;35:565-74
18. Xu C, Min J. Structure and function of WD40 domain proteins. Protein & cell. 2011;2:202-14
19. Asano K. et al. Complex Formation by All Five Homologues of Mammalian Translation Initiation Factor 3 Subunits from Yeast Saccharomyces cerevisiae. BIOLOGICAL CHEMISTRY. 1998. 1857 3-85
20. Herrmannova A, Daujotyte D, Yang JC, Cuchalova L, Gorrec F, Wagner S. et al. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic acids research. 2012;40:2294-311
21. Methot N. et al. The Human Homologue of the Yeast Prt1 Protein Is an Integral Part of the Eukaryotic Initiation Factor 3 Complex and Interacts with p170. BIOLOGICAL CHEMISTRY. 1997;272:1110-1116
22. Fraser CS. et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946-56
23. Valásek L. et al. Related eIF3 subunits TIF32 and HCR1 interacts with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891-904
24. Shalev A. et al. Saccharomyces cerevisiae protein Pci8p and human protein eIF3e/Int-6 interact with the eIF3 core complex by binding to cognate eIF3b subunits. J Biol Chem. 2001;276:34948-34957
25. Lee KH. et al. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 2014;42:3590-3606
26. Fathinajafabadi A. et al. CERKL, a retinal disease gene, encodes an mRNA-binding protein that localizes in compact and untranslated mRNPs associated with microtubules. PLoS One. 2014;9:e87898
27. Roobol A. et al. The chaperonin CCT interacts with and mediates the correct folding and activity of three subunits of translation initiation factor eIF3: b, i and h. Biochem J. 2014;458:213-224
28. Fontaine-Rodriguez EC. et al. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology. 2004;330:487-492
29. Lee CS. et al. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708-4718
30. Holz MK. et al. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569-580
31. Yin JY. et al. Translational control gone awry: a new mechanism of tumorigenesis and novel targets of cancer treatments. Biosci Rep. 2011;31:1-15
32. Feinberg B. et al. Analysis of Temperature-sensitive Mutant ts 187 of Saccharomyces cerevisiae Altered in a Component Required for the Initiation of Protein Synthesis. BIOLOGICAL CHEMISTRY. 1982;257:10846-10851
33. Evans DR. et al. Mutational Analysis of the Prt1 Protein Subunit of Yeast Translation Initiation Factor 3. MOLECULAR AND CELLULAR BIOLOGY. 1995;15:4525-4535
34. Dumas LB. et al. New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol Gen Genet. 1982;187:42-46
35. Evans DR. et al. Cell-cycle mutations among the collection of Saccharomyces cerevisiae dna mutants. FEMS Microbiol Lett. 1994;116:147-53
36. Damoc E. et al. Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry. Mol Cell Proteomics. 2007;6:1135-46
37. Donze O, et al.Hershey, Nahum Sonenberg. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. The EMBO Journal. 1995;14:3828-3834
38. Koromilas AE. et al. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685-1689
39. Silvera D. et al. Translational control in cancer. Nat Rev Cancer. 2010;10:254-266
40. Fukuchi-Shimogori T, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant Transformation by Overproduction of Translation Initiation Factor eIF4G. Cancer Res. 1997;57:5041-5044
41. Comtesse N. et al. Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. Int J Cancer. 2007;120:2538-2544
42. Bauer C. et al. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer. 2001;92:822-829
43. Rosenwald IB. et al. Expression of eukaryotic translation initiation factors 4E and 2alpha is increased frequently in bronchioloalveolar but not in squamous cell carcinomas of the lung. Cancer. 2001;92:8946-8956
44. Spilka R. et al. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340:9-21
45. Zhang L. et al. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790-5800
46. Golob-Schwarzl N. et al. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur J Cancer. 2017;83:56-70
47. Y J Choi. et al. Silencing of translation initiation factor eIF3b promotes apoptosis in osteosarcoma cells. Bone Joint Res. 2017;6:186-193
48. Zang Y. et al. Eukaryotic Translation Initiation Factor 3b is both a Promising Prognostic Biomarker and a Potential Therapeutic Target for Patients with Clear Cell Renal Cell Carcinoma. J Cancer. 2017;8:3049-3061
49. Wang H. et al. Translation initiation factor eIF3b expression in human cancer and its role in tumor growth and lung colonization. Clin Cancer Res. 2013;19:2850-2860
50. Xu F. et al. Eukaryotic translation initiation factor 3B accelerates the progression of esophageal squamous cell carcinoma by activating β-catenin signaling pathway. Oncotarget. 2016;7:43401-43411
51. Liang H. et al. Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol Sci. 2012;33:1057-1062
52. Wang Z. et al. RNA interference-mediated silencing of eukaryotic translation initiation factor 3, subunit B (EIF3B) gene expression inhibits proliferation of colon cancer cells. World J Surg Oncol. 2012;10:119
53. Lin L. et al. Molecular interaction between human tumor marker protein p150, the largest subunit of eIF3, and intermediate filament protein K7. J Cell Biochem. 2001;80:483-490
54. Chen G, Burger MM. p150 overexpression in gastric carcinoma: the association with p53, apoptosis and cell proliferation. Int J Cancer. 2004;112:393-398
55. Simonetti A. et al. eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition. Mol Cell. 2016;63:206-217
Author contact
![]() Corresponding author: Peijun Liu, Ph.D. & M.D., Professor, Email: liupeijunxjtu.edu.cn, Tel: 086-18991232306, Address: 277 YantaWestern Rd., Xi'an, Shaan Xi Province, China, 710061
Corresponding author: Peijun Liu, Ph.D. & M.D., Professor, Email: liupeijunxjtu.edu.cn, Tel: 086-18991232306, Address: 277 YantaWestern Rd., Xi'an, Shaan Xi Province, China, 710061

 Global reach, higher impact
Global reach, higher impact