Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(12):1658-1668. doi:10.7150/ijbs.28482 This issue Cite
Research Paper
Epigenetic alterations of a novel antioxidant gene SLC22A3 predispose susceptible individuals to increased risk of esophageal cancer
1. Guangdong Key Laboratory for Genome Stability & Disease Prevention, Department of Pharmacology and Carson International Cancer Center, Shenzhen University School of Medicine, Shenzhen 518039, China;
2. Department of Stomatology, Shenzhen Hospital of Guangzhou University of Chinese Medicine, Shenzhen, 518033, China;
3. Department of Medicine and Therapeutics, Li Ka Shing Institute of Health Sciences, Chinese University of Hong Kong;
4. Shenzhen Huarui Translational Research Institute, Shenzhen, China;
5. Department of Medicine and Oncology, The Johns Hopkins University School of Medicine and Sidney Kimmel Comprehensive Cancer Center, Baltimore, Maryland, USA;
6. Department of Clinical Oncology, the First Affiliated Hospital, Zhengzhou University, Zhengzhou, China;
7. Department of Clinical Oncology, University of Hong Kong, Hong Kong
*These authors contributed equally to this work.
Received 2018-7-12; Accepted 2018-8-24; Published 2018-9-7
Abstract
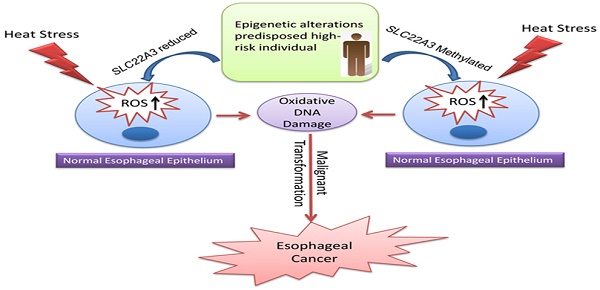
Esophageal squamous cell carcinoma (ESCC) occurs with the highest frequency in China, especially in the high-risk Northern Chinese. Recent studies have reported that SLC22A3 is significantly downregulated in non-tumor (NT) esophageal tissues from familial ESCC patients compared with those from sporadic ESCC. However, the mechanism of how SLC22A3 regulates familial ESCC remains unknown. In this study, post hoc genome-wide association studies (GWAS) in 496 cases with a family history of upper gastrointestinal tract cancers and 1056 controls were performed and the results revealed that SLC22A3 is a novel susceptibility gene for familial ESCC. Reduced expression of SLC22A3 in NT esophageal tissues from familial ESCC patients significantly correlates with its promoter hypermethylation. Moreover, case-control study of Chinese descendants from different risk areas of China revealed that the methylation of the SLC22A3 gene in peripheral blood leukocyte (PBL) DNA samples could be a risk factor for developing ESCC in this high-risk population. Functional studies showed that SLC22A3 is a novel antioxidant gene, and deregulation of SLC22A3 facilitates heat stress-induced oxidative DNA damage and formation of γ-H2AX foci in normal esophageal epithelial cells. Collectively, we show that epigenetic alterations of SLC22A3 predispose susceptible individuals to increased risk of esophageal cancer.
Keywords: Familial ESCC - SLC22A3 - DNA methylation - antioxidant gene -oxidative DNA damage
Background
Esophageal carcinoma (EC) is ranked as the sixth leading cause of cancer death [1]. Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of esophageal carcinoma [1, 2], affecting more than 450 000 people worldwide. Despite recent development in diagnosis and treatment, the overall 5-year survival rate for patients with ESCC remained 15% to 25% [3]. ESCC occurs with the highest frequency among the Chinese, especially the high-risk Northern Chinese, where the incidence rate is 121 per 100,000 people, which is over 20 times higher than that in low-risk regions [4]. Although environmental factors are critical to the varied distribution of ESCC, in a high-incidence area, only a minor proportion of the population develops ESCC, suggesting the importance of host susceptibility factors for those individuals who are at high risk to develop this disease [5-7]. Recently, the potential for individual susceptibility was further supported by genome-wide association studies (GWASs) on ESCC in Chinese populations [8-11], in which several genetic susceptibility loci for ESCC have been identified. Further, epidemiological surveys from China have shown that a family history of esophageal cancer significantly increased the risk of developing ESCC [6, 12], indicating there might be differences in genetic susceptibility between familial aggregated and sporadic cancer patients.
Recently, our group found that SLC22A3 is significantly downregulated in non-tumor (NT) esophageal tissues from familial ESCC patients compared with those from sporadic ESCCs [13], indicating that SLC22A3 may server as a potential tumor susceptibility gene in familial ESCC. The SLC22A3 gene encodes a cation transport protein belonging to the SLC22A family (SLC22A1-3 or OCT1-3), which is critical for drug transportation and cellular detoxification [14, 15]. Gene structure analysis found that the promoter region of SLC22A3 is located within a CpG island [16], suggesting that regulation of this gene may largely depend on methylation. More recent studies in prostate cancer revealed that the 6q risk variant is significantly associated with decreased SLC22A3 transcript levels; however, functional assays showed that SLC22A3 repression leads to reduced viability in both normal prostate and tumor prostate-derived cells [17]. Interestingly, unlike other prostate cancer risk variants, the association between the 6q risk locus and decreased SLC22A3 transcription is only observed in populations of European ancestry but not of Japanese and African ancestries, suggesting gene-gene or gene-environment interactions underlie this observation [17].
In the present work, we aimed to explore whether SLC22A3 promoter methylation is associated with its transcription and confers susceptibility to ESCC by performing BGS and analyzed the promoter region of the SLC22A3 gene in peripheral blood leukocyte (PBL) DNA samples from healthy volunteers and patients with ESCC. We also investigated the function of SLC22A3 in normal esophageal cells and its potential role in ESCC initiation.
Methods
Clinical Specimens
A case-control cohort from the Linzhou People's Hospital in Henan (ESCC High-risk Cohort) and the Sun Yat-Sen University Cancer Center in Guangzhou (ESCC Low-risk Cohort) were collected between 2005 and 2011. All participants provided informed consent. The High-risk Cohort consisted of peripheral blood leukocyte (PBL) DNA samples from healthy volunteers (n = 115) and patients with ESCC (n = 133, including 49 FH+ and 84 FH- cases). High-risk cases were chosen for this study based on three criteria: (a) the patient had resided in one of four geographic regions close to the Linzhou cancer hospital (Linzhou, Anyang, Xinxiang, and Hebi); (b) the patient had newly diagnosed cancer of the esophagus without previous treatment (i.e., no surgery, chemotherapy, or radiotherapy); and (c) histological diagnosis of ESCC was confirmed by pathologists. The Low-risk Cohort consisted of PBL DNA samples from healthy volunteers (n = 100) and patients with ESCC (n = 112). We also included low-risk cases in this study based on three criteria similar to those for high-risk cases, except Guangdong was the ancestral home for these patients. However, descendants from Chaoshan, a high-risk region in Guangdong province, were excluded from this study. The controls were matched to the ESCC cases by age, sex, and geographic region (Table S1). We also obtained snap-frozen specimens of tumors and surrounding non-tumorous esophageal tissues from patients with ESCC as described previously [13].
RNA extraction and qPCR
Total RNA was extracted by Trizol (Invitrogen) from ESCC cell lines. cDNA was produced using a PrimeScript RT Master Mix (TaKaRa), following the standard protocols provided by the manufacturer. Real-time quantitative PCR was performed using the SYBR-Green Prime Script RT-PCR kit (Takara). PCR was performed at 95°C for 15 s, 60°C for 1 min, and 72°C for 1 min for 40 cycles. Quantification was performed using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Housekeeping gene GAPDH was used as an internal control and mRNA expression was calculated using the comparative CT method (ΔΔ CT method). Primer sequences for qPCR are listed in Table S2.
Post hoc GWAS study
Based on the dataset of a previous esophageal cancer GWAS [8], we conducted genotyping data analyses by selecting 114 SNPs in a ~500-kb region covering the SLC22A3 gene on 6q25 from 496 cases with a family history of upper gastrointestinal tract cancers and 1056 controls. We then examined potential genetic relatedness based on pairwise identity by state for all the successfully genotyped samples using PLINK 1.07 software. The original script from EIGENSTRAT was modified to extract principal components for plotting, as described previously [18]. The association results for selected 6q25 SNPs are summarized in Table S3.
Bisulfite treatment and promoter methylation analysis
Bisulfite genomic sequencing (BGS) and real-time quantitative MSP (qMSP or MethyLight assay) were conducted to analyze the promoter region of the SLC22A3 gene. For BGS, at least 8-10 clones from each sample were selected for sequencing analysis using the ABI310 automatic sequencer (Applied Biosystems). The PCR for BGS was performed using an AmpliTaq Gold 360 PCR Master Mix Kit (Applied Biosystems). For the MethyLight assay, CpGenome Universal Methylated DNA (Chemicon) was used as a positive control. Quantitative MSP with a TaqMan probe was performed on an ABI7700 Sequence Detector System (Applied Biosystems) using FastStart Taq DNA Polymerase (Roche). Duplex PCR with a β-actin (ACTB) primer and probe sequences containing no CpGs was performed for normalization. A methylation index (MI), representing the ratio of densely methylated DNA in the sample at the target sequence relative to the fully methylated positive control, was calculated according to Mori et al. [19]: MI = (Ts ⁄ Tc) ⁄ (As ⁄ Ac), where Ts and Tc represent the levels of SLC22A3 methylation in the sample and the control DNA, respectively, and As and Ac correspond to the amplified ACTB level in the sample and the control DNA, respectively. Samples containing > 5% (MI > 0.05) fully methylated molecules were designated as methylated, and samples containing ≤ 5% were designated as unmethylated. The 0.05 cutoff gave the best discrimination between case and control. Primer sequences for BGS and MethyLight analyses are listed in Table S2.
Lentiviral transduction
Plasmids reconstructed with SLC22A3 shRNA (Sigma Aldrich NM_021977) and shRNA NTC were purchased from sigma Aldrich. Lenti-virus were produced using transfection reagent Lipofectamine® 3000 (Thermo) and were applied to infect ESCC cells according to the manufacturer's instructions. ESCC cells were seeded in antibiotic-free growth medium before infection with lenti-virus. When the cells reached 70-80% confluence, they were infected with the harvested lentiviral media. Polybrene (Sigma-Aldrich) was added in the lentiviral media to increase the infection efficiency. The stable cell lines were screened with puromycin in proper concentration. qPCR was used to confirm the transfection efficiency.
Immunofluorescence staining (IF)
Cells were cultured on coverslips to 80% confluence, and fresh medium was added before the experiments. Cells received heat-shock treatment at 43°C for 45 min, followed by recovery at 37°C for 30 min. Immunofluorescence staining was performed according to the standard protocol with primary antibodies (Anti-γ-H2AX abcam, 1:100) overnight at 4°C, and then incubated with Alexa fluor 594 goat anti-mouse IgG (Life, 1:500). Images were captured using a Leica DMRA fluorescence microscope (Rueil-Malmaison). Images were acquired by ZEN 2012 software. The γ-H2AX-positive cells refer to cells with ≥5 γ-H2AX foci. The data represent the means ± SD (n ≥ 50) from three independent experiments.
MitoSOX Red Staining
Oxidative stress in esophageal cells was assessed by staining with the mitochondrial superoxide indicator, MitoSOX Red (Molecular Probes). Briefly, cells were cultured on coverslips to 60-80% confluence, and fresh medium was added before the experiments. After heat-shock treatment (43°C for 45 min), MitoSOX Red was added to a final concentration of 2.5 μM, according to the manufacturer's recommendation. The cells were allowed to load MitoSOX for 15 min in a CO2 incubator at 37°C and were washed twice with Hank's Buffered Salt Solution (HBSS) containing calcium and magnesium before analysis. Slides were counterstained with DAPI in anti-fade solution. Images were captured using a Leica DMRA fluorescence microscope (Rueil-Malmaison).
Comet DNA damage assay
Cells received heat-shock treatment at 43°C for 45 min, followed by recovery at 37°C for 30 min. Cells were then collected and prepared using the OxiSelect Comet Assay Kit (Cell Biolabs). Comets were imaged by fluorescence microscopy. Percentage of Tail DNA, Tail moment (fraction of DNA in the tail × tail length) and Olive Tail Moment were analyzed with CASP software. The data were calculated from the measurement of 50 Comets for each sample.
Western blotting
Western blot analyses were performed according to the standard protocol with the following antibodies: anti-SLC22A3 (Abmart, 1:1000) anti-γ-H2AX (Abcam, 1:1000) and GAPDH (Abcam, 1:1000).
Statistical analysis
The statistical analyses were performed with SPSS standard version 17.0 (SPSS Inc), and P < 0.05 was considered as statistically significant. The SLC22A3 methylation levels in the cases and controls were compared using the Wilcoxon rank sum test with continuity correction. Correlations among SLC22A3 expression, DNA methylation was analyzed using Spearman correlation.
Results
SLC22A3 is a candidate susceptibility gene for familial ESCC
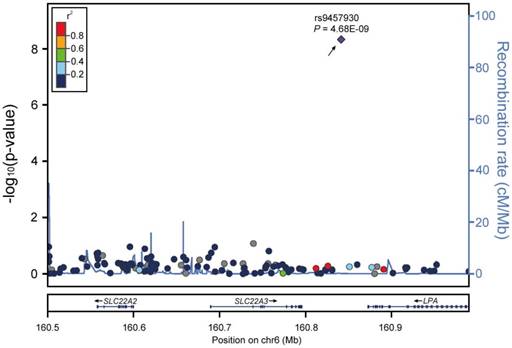
To test the association between SLC22A3 and ESCC in familial ESCC high-risk individuals, we performed a post hoc GWAS analysis to investigate the influence of common genetic variants around the SLC22A3 gene region on ESCC risk. We selected 114 genetic variants in a ~500-kb region surrounding the SLC22A3 gene on 6q25 from the dataset of a previous esophageal cancer GWAS in 496 cases with a family history of upper gastrointestinal tract cancers and 1056 healthy controls [8]. One SNP (rs9457930), located at ~45-kb downstream of the SLC22A3 gene, was found to be significantly associated with reduced ESCC risk in high-risk individuals with a family history of ESCC [P = 4.68e-09, odds ratio (OR) = 0.508, 95% confidence interval (CI) 0.504-0.512; Figure 1], indicating that SLC22A3 could be a protective factor and that deregulation of this gene may confer susceptibility to familial ESCC.
SLC22A3 is a novel tumor susceptibility gene in familial ESCC. Regional association plots for the SLC22A3 gene at 6q25.3. The -log10 P values (y axis) of 114 SNPs in 496 FH+ ESCC cases and 1056 controls are presented according to their chromosomal positions (x axis). Genetic recombination rates, based on CHB (Han Chinese in Beijing, China) and JPT (Japanese in Tokyo, Japan) samples from the HapMap Project, are represented by light-blue lines, and genes within the regions are depicted by arrows. The top genotyped SNP is labeled by rs ID, and the r2 values of the rest of the SNPs with the top genotyped SNP are indicated by different colors.
SLC22A3 downregulation was associated with its promoter methylation in ESCC
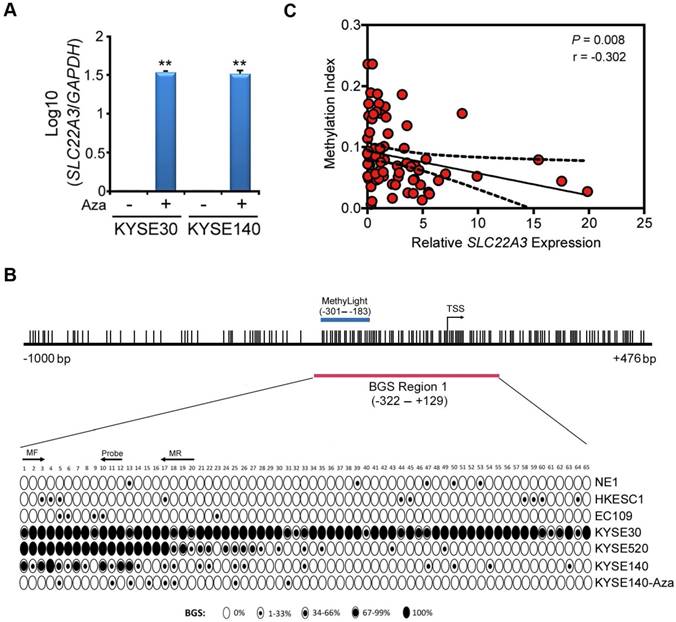
We have previously reported that SLC22A3 expression was downregulated in 7 ESCC cell lines compared to normal esophageal epithelial cell line NE1 [13]. Promoter hypermethylation, especially in tumor suppressor genes, is an important mechanism of transcript silencing in tumourigenesis [20, 21]. As SLC22A3 promoter region contains a CpG island, we speculated that SLC22A3 downregulation in ESCC is associated with its promoter methylation. Two ESCC cell lines (KYSE30 and KYSE140) without SLC22A3 expression were treated with 5-Aza-2'-deoxycytidine (5-Aza), a DNA methyltransferase inhibitor. After 5-Aza treatment, SLC22A3 expression was dramatically restored, suggesting that SLC22A3 methylation was associated with its downregulation in ESCC (Figure 2A). We next used a CpG-island searcher to study the 5'-region of the SLC22A3 gene (-1000 to +476) and found it was located right in a CpG island (Figure 2B). The methylation pattern of SLC22A3 was further analyzed in more detail, using high-resolution bisulfite genomic sequencing (BGS). A 451-base pair BGS region (BGS region 1: -322 to +129 relative to the transcription start site) composed of 65 CpG sites were analyzed (Figure 2B). In all cell lines studied, SLC22A3 BGS region 1 (proximal promoter) showed varying degree of methylation density and intra-sample heterogeneity of methylation (Figure 2B). Next, we designed a MethyLight assay overlaid on the 5' regions of the BGS region 1 (-301 to -183) and assessed the CpG sites from 1 to 20, as indicated in Figure 2B. A positive result would be obtained if there were methylation at the 3' ends of each primer (CpGs 3 and 17) and the probe binding site covering CpGs 10, 11, and 12. Intriguingly, SLC22A3 promoter methylation showed a significantly negative correlation with its mRNA level in 76 family history FH+ NT tissues (Spearman r = -0.302, P = 0.008; Figure 2C) [13], suggesting that promoter hypermethylation of SLC22A3 contributes to its gene silencing in familial ESCC.
SLC22A3 promoter methylation correlates with its reduced expression in ESCC cell lines and familial ESCC. (A) Restoration of SLC22A3 expression was observed in KYSE30 and KYSE140 cells after Aza-treatment. β-actin was used as an internal control. (B) A 1476-bp region containing the putative SLC22A3 promoter and its first exon was analyzed, and the region was found to be located within a CpG island (CGI). The locations of the two bisulfite genomic sequencing (BGS) regions and one quantitative methylation-specific PCR (MethyLight) region are indicated. The transcription start site (TSS) is denoted by an arrow. The CpG sites within the CGI are shown by vertical bars. For each sequenced cell line, the percentage methylation of each CpG was determined as the percentage of methylated CpGs from 10 randomly sequenced clones. Black or white circles, completely methylated or completely unmethylated CpGs, respectively; Concentric circles, partially methylated CpGs. (C) Correlations among SLC22A3 expression and DNA methylation in familial ESCC. Correlations between the relative expression levels of SLC22A3 and promoter methylation in NT specimens from 76 FH+ ESCCs. The data are expressed using Spearman correlation coefficients, linear regression lines (solid) and 95% CI lines (dotted).
SLC22A3 methylation confers susceptibility to ESCC in a high-risk population
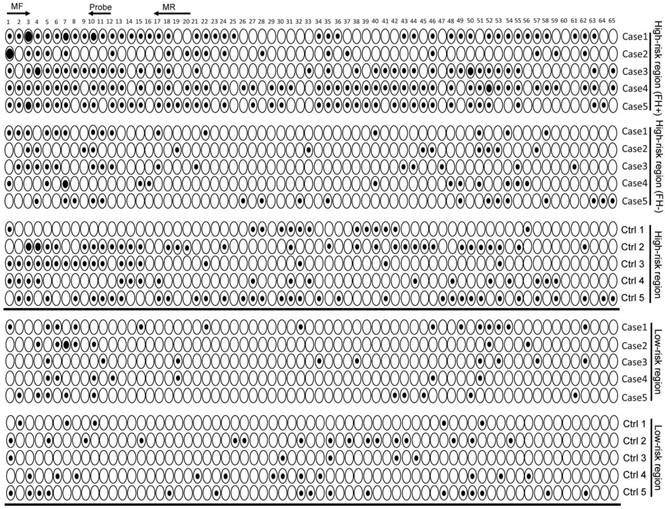
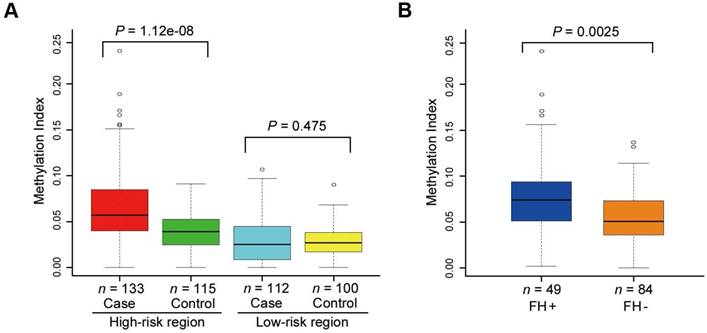
Our previous study showed that the SLC22A3 mRNA level was significantly lower in FH+ NT tissues compared with that in FH- NT tissues [13], indicating that methylation-mediated downregulation of SLC22A3 could be an earlier event in ESCC high-risk individuals. We first used BGS to obtain a detailed analysis of methylation in NT esophageal epithelia, which were isolated from randomly selected FH+ (n = 10) and FH- ESCC cases (n = 10). Interestingly, the SLC22A3 proximal promoter showed a distinctly different pattern of methylation in FH+ NT esophageal epithelia compared with that in FH- NT tissues (Figure S1). The entire BGS region 1 was largely lack of methylation in all ten FH- NT tissues, but heterogeneous methylation was found in 5 of 10 FH+ NT tissues. To determine whether this type of methylation is an early event, we next sequenced bisulfite-modified peripheral blood leukocyte (PBL) DNA from FH+ (n = 10) and FH- (n = 10) ESCC patients (Figure S2). Similar to the methylation pattern observed in NT esophageal epithelia, the blood DNA samples also showed inter-individual variations of SLC22A3 methylation, of which FH+ blood DNA samples showed a higher degree of methylation than FH- samples (Figure S2). To further prove that SLC22A3 hypermethylation is an early event that predisposes high-risk individuals to develop esophageal cancer, we extended the PBL DNA methylation study to case-control sample sets, including 133 cases and 115 controls recruited from an ESCC high-risk region of North China (high-risk cohort), as well as 112 cases and 100 controls from an ESCC low-risk region of South China (low-risk cohort). Initially, we conducted bisulfite sequencing to reveal the detailed methylation profile of SLC22A3 in high- and low-risk cohorts (n = 10 for each group). As shown in Figure 3, the highest degree of SLC22A3 methylation was found in the FH+ cases, while the lowest degree of SLC22A3 methylation was detected in the healthy controls from the low-risk region. From the results of the MethyLight assay, we found that the methylation level of SLC22A3 was significantly higher in ESCC cases than that in healthy controls from the high-risk region [Median Methylation Index (MI): 0.065 vs. 0.040; P = 1.12e-08; Figure 4A]. However, the methylation level of SLC22A3 showed no difference between cases and controls from the ESCC low-risk region (Median MI: 0.030 vs. 0.027; P = 0.475; Figure 4A). We further stratified the high-risk ESCC cases into FH+ (n = 49) and FH- (n = 84); the results showed that the methylation level of SLC22A3 was significantly higher in FH+ cases than that in FH- cases (Median MI: 0.080 vs. 0.056; P = 0.0025; Figure 4B). Moreover, a conditional logistic regression model revealed that SLC22A3 promoter hypermethylation was significantly associated with ESCC risk in the high-risk cohort (OR per 1% increment in SLC22A3 methylation = 1.352, 95% CI = 1.208-1.514) but not in the low-risk cohort (OR = 1.070, 95% CI = 0.9364-1.223; Table 1), indicating that early SLC22A3 hypermethylation contributes to ESCC susceptibility and may serve as an effective marker for earlier screening.
SLC22A3 promoter methylation status in ESCC high- and low-risk cohorts. Representative high-resolution methylation profiles obtained by bisulfite sequencing of BGS region 1 (-322 to +129), comprised of 65 CpG sites in the SLC22A3 promoter, in randomly selected cases and controls from ESCC high- and low-risk regions. For each sequenced blood leukocyte (PBL) DNA sample, the percentage methylation of each CpG was defined as the percentage of methylated CpGs from 8-10 randomly sequenced clones. Black circle, completely methylated CpG; White circle, completely unmethylated CpG; Concentric circle, partially methylated CpG.
Association between SLC22A3 methylation and risk of ESCC
| Group | SLC22A3 Methylation (1% increment) | ||||||
|---|---|---|---|---|---|---|---|
| OR(95% CI) | AdjOR*(95% CI) | ||||||
| High-risk region | |||||||
| Cases/Controls | 1.335(1.198-1.489) | 1.352(1.208-1.514) | |||||
| 133/115 | |||||||
| P-value | |||||||
| Likelihood ratio | 1.05E-09 | 2.06E-11 | |||||
| Wald test | 1.94E-07 | 4.88E-08 | |||||
| Score test | 2.89E-08 | 1.59E-09 | |||||
| Low-risk region | |||||||
| Cases/Controls | 1.058(0.9274-1.208) | 1.070(0.9364-1.223) | |||||
| 112/100 | |||||||
| P-value | |||||||
| Likelihood ratio | 0.397 | 0.089 | |||||
| Wald test | 0.399 | 0.098 | |||||
| Score test | 0.398 | 0.092 | |||||
SLC22A3 protects normal esophageal cells from heat stress-induced reactive oxygen species (ROS) and DNA damage
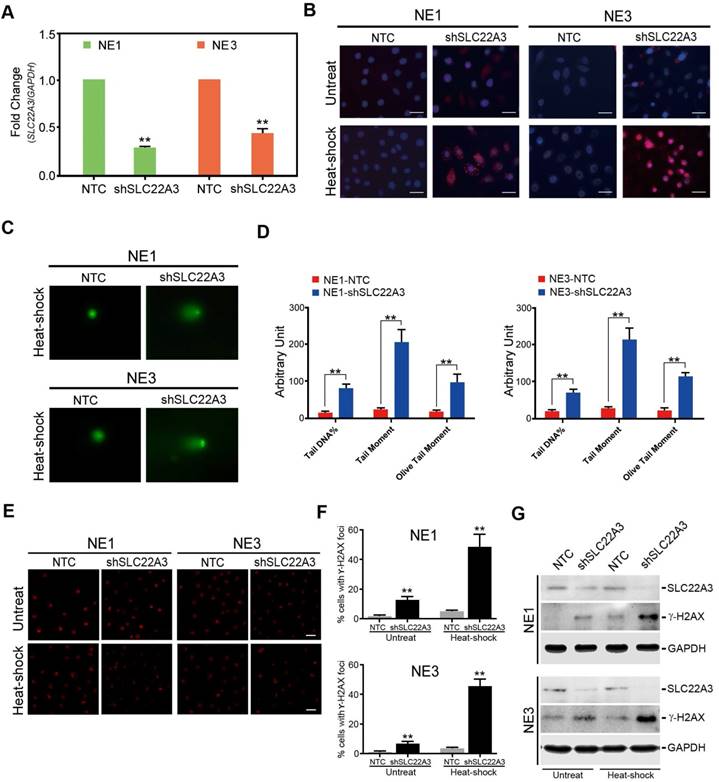
SLC22A3 is reported to be a heat stress responsive gene and plays a critical role in cellular detoxification [15, 22]. We explored whether SLC22A3 could protect normal esophageal cells from heat stress-induced ROS and DNA damage. To verify the antioxidant function of SLC22A3 in normal esophageal cells, we knocked down the endogenous SLC22A3 expression in NE1 and NE3 cells (NE1/NE3-shSLC22A3) by lentiviral transfection. Non-template short hairpin RNA-transfected cells (NE1/NE3-NTC) were used as control (Figure 5A). MitoSOX Red dye staining was performed to exam intracellular ROS levels in NE1/NE3-shSLC22A3 and NE1/NE3-NTC after heat shock. As shown in Figure 4B, after heat shock, a more significant increase in the mitochondrial ROS was detected in SLC22A3-repressed NE1/NE3 cells compared with control cells, evident by more mitochondrial fluorescence in the cell bodies. We next investigated whether the oxidative stress-induced DNA damage is enhanced in SLC22A3-repressed NE1/NE3 cells. Alkaline Comet Assay revealed that NE1/NE3-shSLC22A3 exhibited higher levels of DNA damage in response to heat-shock than the control cells (Figure 5C, D). In addition, the formation of γ-H2AX foci, a maker of DNA double stand breaks, was increased in heat-shock treated NE1/NE3-shSLC22A3 cells compared with control cells (Figure 5E, F). Western blot demonstrated that SLC22A3 expression was negatively correlated with the expression of DNA damage marker γ-H2AX in these cells (Figure 5G). All these data indicated that SLC22A3 may act as an antioxidant molecule and protect normal esophageal cells from heat stress induced ROS and DNA damage.
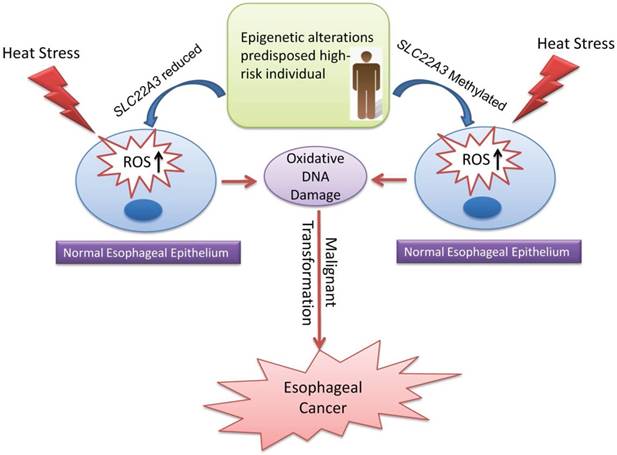
Collectively, we propose a disease model in which the epigenetic alterations of SLC22A3, predisposed in high-risk individuals with a family history, could accelerate the malignant transformation of normal esophageal epithelial cells under heat stress, leading to the development of ESCC (Figure 6).
Discussion
Multiple evidence has established SLC22A3 as a novel tumor susceptibility gene in high-risk individuals with a family history of ESCC in northern China. First of all, we identified SLC22A3 as one key gene that is associated with familial ESCC. A post hoc GWAS analysis revealed that one SNP (rs9457930), located at ~45-kb downstream of the SLC22A3 gene, is significantly associated with reduced ESCC risk in familial ESCC cases. This result is in line with our previous finding that SLC22A3 is downregulated in familial ESCC cases, indicating that SLC22A3 could be a protective factor and downregulation of this gene may confer susceptibility to ESCC. Secondly, epigenetic alterations of the SLC22A3 gene contribute to its significantly lower expression in FH+ ESCCs. Finally, population methylation analyses demonstrate that a gain of SLC22A3 promoter methylation is associated with an increased risk of developing ESCC in a northern Chinese population. Emerging evidence has shown that CpG island promoter hypermethylation of tumor suppressor genes is an early event in tumorigenesis, suggesting that epigenetic signatures are promising biomarkers for cancer risk and early neoplasia [23].
SLC22A3 promoter methylation levels in ESCC high- and low-risk cohorts. (A) Tukey boxplots represent the SLC22A3 promoter methylation indexes (MIs) for 133 ESCC cases and 115 age/sex matched controls from the high-risk region, as well as 112 ESCC cases and 100 matched controls from the low-risk region. Open circles, outliers; Rectangular box, range for the 25th to 75th percentile; Black line, median value. The measurement of the MIs was performed using real-time quantitative MSP (MethyLight), and the CpG-free ACTB genomic sequence was used for normalization. P values were calculated by the Wilcoxon rank sum test with continuity correction. (B) Tukey boxplots represent the SLC22A3 promoter MIs for the 133 high-risk ESCCs further stratified into cases with family history (FH+; n = 49) and cases without family history (FH-; n = 84). P values were calculated by the Wilcoxon rank sum test with continuity correction.
SLC22A3 protects normal esophageal cells from heat stress-induced ROS and DNA damage. (A) Expression of SLC22A3 in shSLC22A3 stably transfected NE1 cells and NE3 cells were confirmed by qPCR. Nontemplate shRNA-transfected cells (NE1-NTC and NE3-NTC) were used as controls. **, P < 0.001 by Student's t-test. (B) Representative images of MitoSOX Red dye staining in SLC22A3 knockdown NE1 cells and NE3 cells with and without heat-shock treatment. Scale bar = 20 μm. (C) Alkaline Comet assay showing DNA break formation after heat-shock treatment shSLC22A3 transfected NE1 cells and NE3 cells. NE1-NTC and NE3-NTC were used as controls. (D) Percentage of Tail DNA, Tail moment (fraction of DNA in the tail × tail length) and Olive Tail Moment were analyzed with CASP software. The data were calculated from 50 comets for each sample. **, P < 0.001 by Student's t-test. (E) Cells (NE1-shSLC22A3, NE1-shSLC22A3, NE1-NTC and NE3-NTC) that were treated with or without heat-shock were fixed and immunolabeled with an anti-γ-H2AX antibody. (Magnification: 40×). Images were performed by ZEN 2012 software. (F) Quantification of γ-H2AX-positive cells in (E). The γ-H2AX-positive cells refer to the cells with ≥5 γ-H2AX foci. The data represent the means ± SD (n ≥ 100) from three independent experiments. (G) Expression of SLC22A3 and γ-H2AX of these cells were examined by Western blotting.
With the rapidly developed methylation analysis technologies, cancer-specific hypermethylation events could be detected in more easily accessible biological material, such as sera, feces, urine, and sputum [24-27]. To date, several studies have reported that the epigenetic alterations detected in peripheral blood cell DNA could be a valuable predictive marker for people with a high risk for cancer [28-30]. Here, we also provide the first evidence that DNA methylation of the SLC22A3 gene in peripheral blood cells is associated with an increased esophageal cancer risk. However, to determine the diagnostic value of SLC22A3 methylation in an ESCC high-risk population, large-scale case-control cohorts need to be further investigated. Moreover, our methylation study also shows pronounced inter-allelic heterogeneity of methylation with varying degrees of methylation density at the SLC22A3 promoter in the NT tissues from ESCC patients. In line with previous observations, methylation heterogeneity or epigenetic mosaicism observed in cancer precursor tissues suggests that this epigenetic variability could reflect a type of genomic instability in the early development of cancer [31].
Proposed model illustrating the epigenetic alterations of SLC22A3 predispose in high-risk individuals, accelerate malignant transformation of normal esophageal epithelia under heat stress and lead to the development of ESCC.
Numerous evidence has proved that long-term consumption of hot food and drink was associated with an increased risk of esophageal cancer [32, 33]. In response to frequent heat stress, normal esophageal epithelia may accumulate DNA damage and become carcinogenic. In the SLC22A3 knockdown stable cell line (NE1/NE3-shSLC22A3), its intracellular ROS level increased leading to higher level of DNA break formation after heat shock, compared to that in NE1/NE3-NTC. ROS generation is often triggered by various forms of stress, including oxidative stress, heavy metal stress and heat stress [34, 35]. The unfavorable effects of ROS include oxidative damage to DNA, which is now generally believed to be a risk factor in carcinogenesis [36, 37]. DNA damage-related mutagenesis occurs in a wide range of human precancerous lesions, and is associated with early tumourigenesis [38, 39]. A recent study revealed that intraepithelial neoplasia (IEN) expressed high levels of γ-H2AX and shared similar genetic variant spectrum with ESCC, implying that defects in DNA damage repair and accumulation of mutation burden could contribute to the progression from IEN to ESCC [40]. Intriguingly, a previous epidemiological study found that frequent precursor lesions detected in an ESCC high-risk population in China are significantly associated not only with the consumption of burning hot beverages but also with a family history of esophageal cancer [41]. Taken together, these results demonstrated that SLC22A3 promoter hypermethylation may directly suppresses its gene transcription, resulting in increased susceptibility to ESCC in high-risk individuals. With the rapid development of methylation analysis technologies, detecting DNA methylation of SLC22A3 in sera, feces, urine, and sputum could be novel diagnostic and prognostic method for familial ESCC in the future.
Abbreviations
ESCC: esophageal squamous cell carcinoma
SLC22A3: solute carrier family 22 member 3
GWAS: post hoc genome-wide association studies
EC: esophageal carcinoma
NT: non-tumor
FH: family history
PBL: peripheral blood leukocyte
BGS: bisulfite genomic sequencing
IF: immunofluorescence staining
MI: methylation index
OR: odds ratio
CI: confidence interval
ROS: reactive oxygen species
γ-H2AX: phosphorylated (serine 139) H2A histone family, member X
CpG: 5'-C-phophate-G-3'
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Key R&D program of China (2017YFA0503900 and 2016YFC0904600), the National Natural Science Foundation of China (81772957), the Science and Technology Foundation of Shenzhen (KQTD20140630100658078), and the NCI grants (CA190040, UG3CA211457).
Ethics approval and consent to participate
This study was approved by the institutional review board of Shenzhen University. Informed consents were obtained for the original human work that produced the tissue samples.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65:87-108
2. Enzinger PC, Mayer RJ. Esophageal cancer. New England Journal of Medicine. 2003;349:2241-52
3. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010;127:2893-917
4. Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. International Journal of Cancer. 2002;102:271-4
5. Carter CL, Hu N, Wu M, Lin PZ, Murigande C, Bonney GE. Segregation analysis of esophageal cancer in 221 high-risk Chinese families. Journal of the National Cancer Institute. 1992;84:771-6
6. Hu N, Dawsey SM, Wu M, Bonney GE, He LJ, Han XY. et al. Familial aggregation of oesophageal cancer in Yangcheng County, Shanxi Province, China. International Journal of Epidemiology. 1992;21:877-82
7. Su H, Hu N, Shih J, Hu Y, Wang QH, Chuang EY. et al. Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Research. 2003;63:3872-6
8. Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nature Genetics. 2010;42:759-63
9. Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO. et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nature Genetics. 2010;42:764-7
10. Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nature Genetics. 2011;43:679-84
11. Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y. et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nature Genetics. 2012;44:1090-7
12. Chang-Claude J, Becher H, Blettner M, Qiu S, Yang G, Wahrendorf J. Familial aggregation of oesophageal cancer in a high incidence area in China. International Journal of Epidemiology. 1997;26:1159-65
13. Fu L, Qin YR, Ming XY, Zuo XB, Diao YW, Zhang LY. et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4631-40
14. Wu X, Kekuda R, Huang W, Fei YJ, Leibach FH, Chen J. et al. Identity of the organic cation transporter OCT3 as the extraneuronal monoamine transporter (uptake2) and evidence for the expression of the transporter in the brain. The Journal of Biological Chemistry. 1998;273:32776-86
15. Koepsell H. Polyspecific organic cation transporters: their functions and interactions with drugs. Trends in Pharmacological Science. 2004;25:375-81
16. Grundemann D, Schomig E. Gene structures of the human non-neuronal monoamine transporters EMT and OCT2. Human Genetics. 2000;106:627-35
17. Grisanzio C, Werner L, Takeda D, Awoyemi BC, Pomerantz MM, Yamada H. et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11252-7
18. Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P. et al. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nature Genetics. 2013;45:697-700
19. Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP. et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131:797-808
20. Weber A, Dietz A, Tischoff I, Tannapfel A. Role of epigenetics in the carcinogenesis of head and neck carcinomas - possible new targeted therapy? Laryngorhinootologie. 2007;86:9-13
21. Esteller M. Epigenetics in cancer. New England Journal of Medicine. 2008;358:1148-59
22. Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Molacular Biology of the Cell. 2004;15:2361-74
23. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nature Review Cancer. 2006;6:107-16
24. Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Research. 1999;59:67-70
25. Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L. et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. Journal of the National Cancer Institute. 2005;97:1124-32
26. Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W. et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. Journal of the National Cancer Institute. 2006;98:996-1004
27. Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K. et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Research. 2006;66:3338-44 Epub 2006/03/17. doi: 10.1158/0008-5472.CAN-05-3408
28. Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S. et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753-5
29. Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ. et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. New England Journal of Medicine. 2007;356:697-705
30. Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nature Genetics. 2004;36:497-501
31. Tycko B. Genetic and epigenetic mosaicism in cancer precursor tissues. Annals of the New York Academy of Sciences. 2003;983:43-54
32. Gao Y, Hu N, Han XY, Ding T, Giffen C, Goldstein AM. et al. Risk factors for esophageal and gastric cancers in Shanxi Province, China: a case-control study. Cancer Epidemiology. 2011;35:e91-9
33. Andrici J, Eslick GD. Hot Food and Beverage Consumption and the Risk of Esophageal Cancer: A Meta-Analysis. American Journal Preventive Medicine. 2015;49:952-60
34. Droge W. Free radicals in the physiological control of cell function. Physiological Reviews. 2002;82:47-95
35. Banerjee Mustafi S, Chakraborty PK, Dey RS, Raha S. Heat stress upregulates chaperone heat shock protein 70 and antioxidant manganese superoxide dismutase through reactive oxygen species (ROS), p38MAPK, and Akt. Cell Stress Chaperones. 2009;14:579-89
36. Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. Journal of Molecular Medicine (Berl). 1996;74:297-312
37. Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB Journal. 2003;17:1195-214
38. Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864-70
39. Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907-13
40. Liu X, Zhang M, Ying S, Zhang C, Lin R, Zheng J. et al. Genetic alterations in esophageal tissues from squamous dysplasia to carcinoma. Gastroenterology. 2017;153:166-77
41. Wahrendorf J, Chang-Claude J, Liang QS, Rei YG, Munoz N, Crespi M. et al. Precursor lesions of oesophageal cancer in young people in a high-risk population in China. Lancet. 1989;2:1239-41
Author contact
![]() Corresponding author: Li Fu, Guangdong Key Laboratory for Genome Stability & Disease Prevention, Department of Pharmacology and Carson International Cancer Center, Shenzhen University School of Medicine, Shenzhen, China; Email: gracelfuedu.cn
Corresponding author: Li Fu, Guangdong Key Laboratory for Genome Stability & Disease Prevention, Department of Pharmacology and Carson International Cancer Center, Shenzhen University School of Medicine, Shenzhen, China; Email: gracelfuedu.cn

 Global reach, higher impact
Global reach, higher impact