Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(12):1715-1723. doi:10.7150/ijbs.27197 This issue Cite
Research Paper
TGF-β/SMAD4-Regulated LncRNA-LINP1 Inhibits Epithelial-Mesenchymal Transition in Lung Cancer
1. E-institutes of Shanghai Universities, Shanghai Jiaotong University School of Medicine, Shanghai 200025, China
2. State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China
3. Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
4. Beijing Key Laboratory of Noncoding RNA, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
5. University of Chinese Academy of Sciences, Beijing 100049, China.
Received 2018-5-10; Accepted 2018-7-28; Published 2018-9-7
Abstract
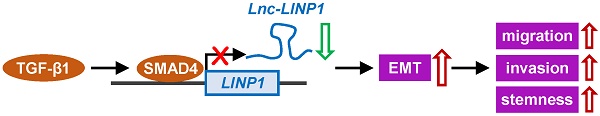
Lung cancer is the leading cause of cancer related deaths worldwide. TGF-β-induced epithelial-mesenchymal transition (EMT) is a key cell-intrinsic identity for tumor cell migration, invasion, and stemness acquisition in cancer metastasis. Long noncoding RNAs (lncRNAs) have not been fully investigated for their involvement in regulating TGF-β-induced EMT and metastasis in lung cancer. Here, we demonstrated that the transcription of lncRNA in nonhomologous end joining (NHEJ) pathway 1 (LINP1) was inhibited by TGF-β1 in a SMAD4-dependent manner. LINP1 suppressed EMT of lung cancer cells, thereby controlling cancer cell migration, invasion, and stemless. Moreover, LINP1 inhibited TGF-β-induced EMT and cell invasion in lung cancer cells. Our study reveals the role of LINP1 in the regulation of TGF-β-induced EMT in human lung cancer.
Keywords: lncRNA, LINP1, TGF-β, EMT, lung cancer
Introduction
Lung cancer is the most common cancer worldwide. Metastasis accounts for the poor prognosis and the high recurrence rate of lung cancer [1]. Epithelial-mesenchymal transition (EMT) has been considered as an important process for early metastatic dissemination from primary tumor, during which epithelium-derived cancer cells lose their epithelial properties and acquire mesenchymal characteristics that endow them with migratory, invasive, and stemness ability [2]. Among the signaling pathways involved in EMT process, transforming growth factor β (TGF-β) signaling is a major inducer of EMT in cancer progression [3]. In response to TGF-β, a heteromeric complex of transmembrane serine-threonine kinase receptors activates SMAD2/3, which associate with SMAD4 and then translocate into the nucleus. Activated SMADs complex interacts with a large group of transcription co-activators or co-repressors, forming many different complexes, which can either activate or repress target genes with high affinity and specificity [4]. For example, TGF-β/SMAD4 signaling can lead to the repression of its targets transcription by interacting with co-repressors including the homeodomain DNA-binding protein TGIF, the proto-oncogene products Ski, and SnoN, all of which can recruit HDACs to deacetylate histone [5, 6]. On the other hand, TGF-β can activate the expression of stress response factor ATF3 and further recruit it to the Id1 promoter, which is essential for Id1 transcription repression in epithelial cells [7].
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides. Recently, studies have shown that several lncRNAs mediate the function of TGF-β signaling in EMT. In hepatocellular carcinoma cells, TGF-β leads to the upregulation of lncRNA-ATB (activated by TGF-β), which can induce EMT and cell invasion [8]. In bladder cancer cells, TGF-β induces lncRNA-MALAT1 (metastasis associated lung adenocarcinoma transcript 1) expression, and as expected, lncRNA-MALAT1 promotes EMT, migration and invasion [9]. TGF-β also induces the expression of lncRNA-MEG3 (maternally expressed gene 3) and lncRNA-HIT (HOXA transcript induced by TGF-β) in lung cancer cells and mammary epithelial cells respectively, both of which can mediate TGF-β-induced EMT, invasion, and migration [10, 11]. On the other hand, TGF-β downregulates LINC01186 transcription level through SMAD3 in lung cancer cells, and LINC01186 inhibits cell migration, invasion, and EMT [12]. Nevertheless, lncRNAs have not been fully investigated for their involvement in regulating TGF-β-induced EMT and metastasis in lung cancer.
LncRNA in nonhomologous end joining (NHEJ) pathway 1 (LINP1) was first found to be overexpressed in human triple-negative breast cancer [13]. LINP1 enhances repair of DNA double-strand breaks by serving as a scaffold linking Ku80 and DNA-PKcs. Inhibition of LINP1 increases the sensitivity of response to radiotherapy in the breast and cervical cancer cells [13, 14]. A subsequent study reveals that LINP1 promotes cell proliferation, chemoresistance, and EMT in breast cancer cells [15]. However, the role of LINP1 in lung cancer remains unknown.
Here, we found that in human lung cancer cells, TGF-β1 inhibited LINP1 transcription in a SMAD4-dependent manner. The loss- and gain-of function experiments showed that LINP1 inhibited EMT process and consequently suppressed lung cancer cell migration, invasion, and stemness acquisition. Furthermore, LINP1 inhibited TGF-β-induced EMT and cell invasion in lung cancer cells.
Materials and Methods
Cell culture, primers, and reagents
A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, cat. no. SH30081.01, GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic/antimycotic solution (penicillin G, streptomycin) in a humidified incubator with 5% CO2. Cells were starved in media without FBS for 24 h, followed by the addition of recombinant human TGF-β1 (R&D Systems, 240-B-010) or vehicle.
Primers used for vector construction are listed below:
LINP1-full length-s: 5'- GTCTCCTTGACTCTGGGT -3'
LINP1-full length-a: 5'- TAAAATGTTGAGAAAGAAAG -3'
LINP1-promoter-s: 5'- TAAACACTCAAGGAAT -3'
LINP1-promoter-a: 5'- AGGGGAGGAACTG -3'
LINP1-promoter-mut (-1312 to -1300)-s: 5'-CTTTGGCAGGCGGATAGAAAACTCACACCA -3'
LINP1-promoter-mut (-1312 to -1300)-a: 5'-TGGTGTGAGTTTTCTATCCGCCTGCCAAAG -3'
LINP1-promoter-mut (-1118 to -1102)-s: 5'- TGGCTCCAGGGGGATCCCTAGGGCTTTC -3'
LINP1-promoter-mut (-1118 to -1102)-a: 5'- GAAAGCCCTAGGGATCCCCCTGGAGCCA -3'
LINP1-promoter-mut (-856 to -844)-s: 5'- GCTTACCTCTGTAAGTCCACCCTGGGTACC -3'
LINP1-promoter-mut (-856 to -844)-a: 5'- GGTACCCAGGGTGGACTTACAGAGGTAAGC -3'
Primers used for qRT-PCR are listed below:
LINP1-s: 5'- CGTTGGCATTTACTGAACC -3'
LINP1-a: 5'- CCCGCAGTTGGTCTGTCTT -3'
E-Cadherin-s: 5'- CGGTGGTCAAAGAGCCCTTA -3'
E-Cadherin-a: 5'- TGAGGGTTGGTGCAACGTCGTTA -3'
vimentin-s: 5'- AGATGGCCCTTGACATTGAG -3'
vimentin-a: 5'- TGGAAGAGGCAGAGAAATCC -3'
SNAIL-s: 5'- CACATCCGAAGCCACA -3'
SNAIL-a: 5'- AGAAGGTCCGAGCACA -3'
GAPDH-s: 5'- ACCCAGAAGACTGTGGATGG -3'
GAPDH-a: 5'- CAGTGAGCTTCCCGTTCAG -3'
Vector construction and transfection
pcDNA3.1(+) vector was constructed for LINP1 full length expression by BamH I and EcoR I (Takara) digestion. pGL4-luciferase reporter vector was constructed of wildtype LINP1 promoter (2 kb) or three SMAD binding sites mutants by SacI and XhoI (Takara) digestion. Transient transfections of vectors or siRNAs were carried out using jetPRIME (Polyplus Transfection) according to the manufacturer's instructions. SiRNAs against LINP1: XLOC-homo-1445: 5'-GGAUCGGGUUGUUGUUAAUTT-3' and siRNAs against SMAD4: 5'-AGGUGUGCAGUUGGAAUGU-3'.
Dual-luciferase reporter assay
pGL4-luciferase reporter vectors and siRNAs against SMAD4 were co-transfected using Lipo2000 regent (Invitrogen) on 5 × 104 293T cells. At 12 h after transfection, 293T cells were treated by 5 ng/ml TGF-β1 for 12 hours. At 12 h after TGF-β1 treatment, 293T cells were lysed in 200 µl lysis buffer, and the firefly luciferase and renilla activities were determined with a luminometer by the Dual Luciferase Assay System (Promega).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were carried out with chromatins prepared as reported previously (Takahashi et al., 2000) from A549 cells. DNA-protein complexes were immunoprecipitated using anti-SMAD4 antibody (#38454, Cell Signaling Technology) or Normal Rabbit IgG (#2729, Cell Signaling Technology). The specific primers for the SMAD4 binding of LINP1 promoter are as follows:
-1924F: 5'- CATTCCCACCAATAAGG -3'
-1727R: 5'- TAAGCCCAACAATCACT -3'
+654F: 5'- GCTGGTTCCGTAGTTT -3'
+917R: 5'- CAAGAAATGGAGTGCC -3'
ChIP-seq data analysis
The ChIP-seq data of SMAD4 in human lung cancer cell line H441 was downloaded from the GEO database (GSE51509) [16]. We downloaded the signal distributions in the wiggle (WIG) format in human genome (h18) obtained from the dataset. And we added the wig file to UCSC genome browser as a custom track to investigate whether there are peaks in the LINP1 promoter region.
Quantitative real-time PCR assay
Total RNAs were extracted from cells by Trizol reagent (Invitrogen) and phenol/chloroform methods. 2 μg RNA were reversely transcribed to cDNA using Super RT cDNA synthesis kit (Toyobo). Quantitative real-time PCR were performed using the 7500 Fast Real-time PCR System (Applied Biosystems) with the Real Master Mix containing SYBR Green (Toyobo) and unique primers above.
Western blot assay
Cells were lysed using the mammalian protein extraction reagent RIPA with phosphatase inhibitors and protein phosphatase inhibitors. Equivalent amounts (40 μg) of cell protein lysates were electrophoresed on an 10% SDS-polyacrylamide gel and transferred to a PVDF membrane. Then the membrane was incubated with primary antibodies of SMAD4 (1:1000, #38454, Cell Signaling Technology), E-Cadherin (1:1000, Ab76055, Abcam), N-Cadherin (1:1000, #13116, Cell Signaling Technology), vimentin (1:1000, #5741, Cell Signaling Technology), SNAIL (1:1000, #3879, Cell Signaling Technology), and GAPDH (1:1000, Ab37168, Abcam). After incubating with second antibodies, Western blots were imaged by Image Quant LAS 4000 mini (GE healthcare) and the results were quantified by Image Pro Plus 6.0.
Fluorescence immunohistochemistry
Cells on slides (Millicell EZ SLIDE) were fixed in 4% paraformaldehyde for 10 min at room temperature, followed by a standard permeabilization protocol. The slides were incubated with primary antibodies of E-Cadherin (1:50, 610181, BD), N-Cadherin (1:100, #13116, Cell Signaling Technology), and vimentin (1:100, #5741, Cell Signaling Technology). After incubating with second antibodies of Alexa Fluor®488-labeled anti-Mouse IgG (1:200, ZF-0512, Zhongshan Jinqiao) or Alexa Fluor®594-labeled anti-Rabbit IgG (1:200, ZF-0516, Zhongshan Jinqiao), slides were incubated with Hoechst (1:100) for nuclear staining and then imaged with a confocal microscopy (Zeiss, LSM880).
Wound-healing assay
For the wound-healing assay, 2 × 105 A549 cells were seeded in 6-well plate, cultured overnight, and transfected with pcDNA3.1-LINP1 or siRNAs. When cells reached approximately 85% confluence, the cells were treated with mitomycin (10 μg/ml) for 2 h. The monolayer cells were scratched with a sterile plastic tip. At 0 h and 12 h after scratch, images of the plates were acquired using a microscope.
Cell invasion assay
Cell invasion assay was performed using 24-well Matrigel invasion chambers (Corning). After 24 h of cell transfection, 1 × 105 cells with the cultured medium containing 1% FBS were seeded on the upper chamber. The bottom chamber was added 500 μl cultured medium with 10% FBS. After 24 h for overexpression or 12 h for knockdown assay, cells in the bottom chamber were fixed with methanol and stained with 0.1% crystal violet, and then counted using a microscopy for statistical analysis.
Sphere formation assay
The single A549 cell suspension was made by enzymatic disassociation, followed by filtration through a 40 μm cell strainer. Cells were washed in 1 × PBS twice, counted, and plated at clonal density in conditional medium, consisting of serum-free DMEM F12 supplemented with 0.4% BSA, 5 mM Hepes, penicillin/streptomycin, 20 ng/ml EGF (Life Technologies), and 10 ng/ml bFGF (Sigma). The number and size of spheres were documented for statistical analysis after 14 days.
Statistical analysis
The unpaired Student's t-test (two-tailed) was used to evaluate gene expression. Expression differences among three groups were analyzed by ANOVA. Statistical analysis was performed using GraphPad Prism software v5. Data are represented as mean ± SEM. All results are considered statistically significant at P < 0.05.
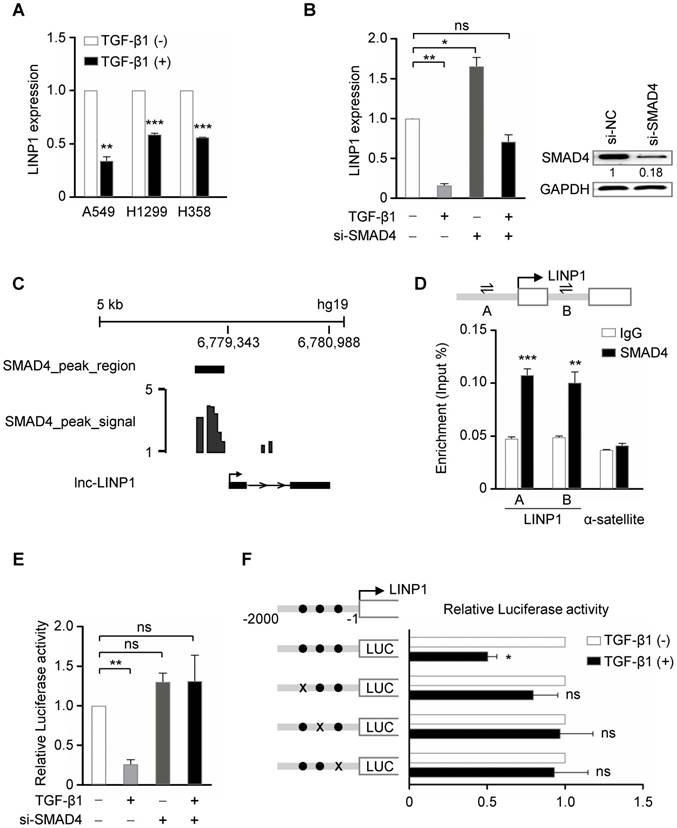
TGF-β represses lnc-LINP1 transcription by SMAD4 in human lung cancer cells. (A) qRT-PCR of LINP1 expression after the treatment of A549, H1299, and H358 cells with 5 ng/ml TGF-β1 or vehicle for 6 h. (B) qRT-PCR of LINP1 expression in A549 cells that were transfected with SMAD4 siRNAs, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. (C) Diagram of SMAD4 bindings at the LINP1 gene locus in the human lung cancer cell line H441. (D) ChIP assay detecting the binding of SMAD4 to the LINP1 gene locus in A549 cells. A schematic diagram (upper) shows the promoter region of the LINP1 gene locus, in which the transcriptional start site (TSS) is represented as an arrow, and A and B indicated the location of the specific primers respectively. The α-satellite locus was the negative control. (E) Dual-luciferase reporter assay of the luciferase activity of the LINP1 promoter containing the 2-kb region in 293T cells that were transfected with SMAD4 siRNAs, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. (F) Dual-luciferase reporter assay of the luciferase activity of the LINP1 wildtype promoter or three SMAD-binding-site-mutant promoters in 293T cells after the addition of TGF-β1 or vehicle for 12 h. The SMAD binding sites are represented as black dots and three mutants at -1312 to -1300, -1118 to -1102, and -856 to -844 loci are represented by black crosses respectively. The error bars indicate mean ± SEM and P value is calculated by unpaired Student's t-test, * P < 0.05, ** P < 0.01, *** P < 0.001, ns: not significant.
Results
TGF-β represses lnc-LINP1 transcription by SMAD4 in human lung cancer cells
A previous study has demonstrated that in breast cancer cells, EGF signaling, specifically through the activation of its downstream RAS-MEK-JNK pathway, induces LINP1 transcription by a consensus AP1-binding site at its promoter [13]. We first detected whether LINP1 is regulated by TGF-β pathway in lung cancer cells. In three lung cancer cell lines A549, H1299, and H358, TGF-β1 significantly inhibited LINP1 expression (Fig 1A). SMAD4 is a common mediator of the canonical TGF-β signal by forming a complex with phosphorylated receptor-activated SMADs (R-SMADs). In A549 cells, knockdown of SMAD4 increased LINP1 expression, and blocked the reduction of LINP1 after TGF-β1 treatment, suggesting that TGF-β regulated LINP1 expression in a SMAD4-dependent manner (Fig 1B). By analyzing the previously reported chromatin immunoprecipitation-sequencing (ChIP-seq) data from the human lung cancer cell line H441 [16], a strong enrichment of SMAD4 in the promoter region of LINP1 was observed (Fig 1C). In A549 cells, chromatin immunoprecipitation (ChIP) assay using anti-SMAD4 antibody confirmed that SMAD4 could specifically bind to the LINP1 promoter in vivo (Fig 1D). We further analyzed the LINP1 promoter using rVista2.0 and found three evolutionarily conserved putative SMAD binding sites within the 2 kb upstream of the transcription start site of LINP1. TGF-β1 treatment significantly reduced luciferase activity of the LINP1 promoter, while SMAD4 knockdown showed the ability to block the repression of luciferase activity by TGF-β1 (Fig 1E). Furthermore, mutations targeting any of these three SMAD binding sites eliminated the TGF-β1-dependent repression (Fig 1F). Together, the above data demonstrated that TGF-β signaling repressed LINP1 transcription by SMAD4 in lung cancer cells.
Lnc-LINP1 inhibits EMT of lung cancer cells
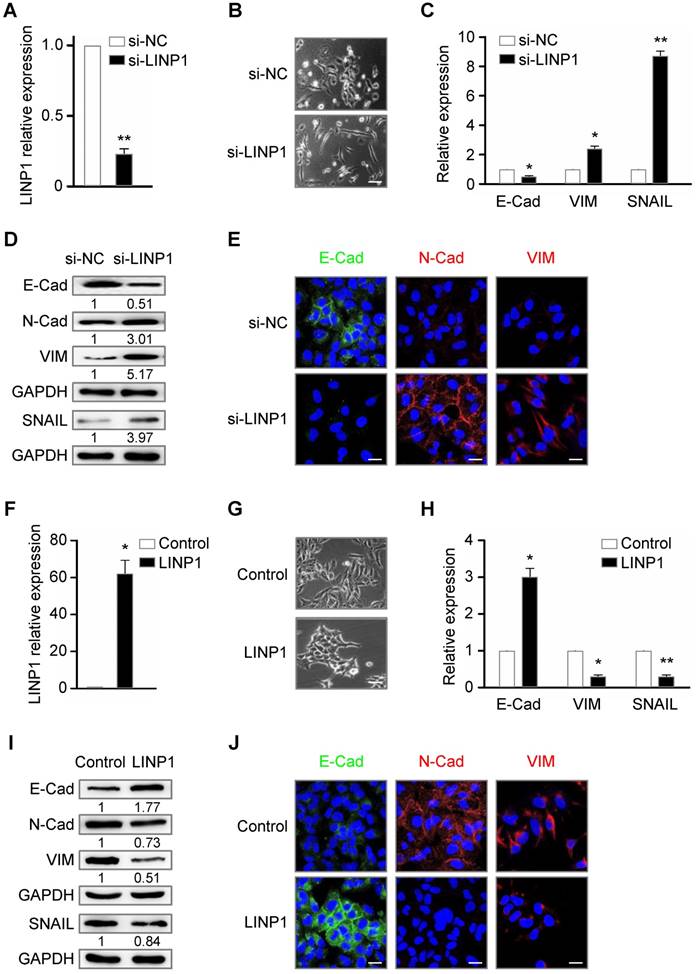
Given the role of TGF-β signaling in EMT process, we delineated the contribution of LINP1 to EMT in a lung cancer cell line A549. SiRNA specific for LINP1 effectively downregulated the endogenous level of LINP1 transcripts (Fig 2A). Knockdown of LINP1 rendered cancer cells acquiring a mesenchymal phenotype, with a weakened cell adhesion and a spindle-shaped morphology (Fig 2B). Epithelial cells express high level of E-Cadherin, while mesenchymal cells express N-Cadherin, vimentin, and SNAIL, a key transcriptional factor of EMT. Consistently, LINP1 knockdown significantly downregulated E-Cadherin expression as shown by the qRT-PCR and Western blot assays, and upregulated the expression of N-Cadherin, vimentin, and SNAIL (Fig 2C and 2D). Fluorescence immunohistochemistry staining also confirmed the downregulation of E-Cadherin expression and the upregulation of N-Cadherin and vimentin (Fig 2E). Conversely, LINP1 overexpressing in A549 cells led to a stronger cell adhesion, accompanied by the increase in E-Cadherin expression and the decrease in the expression of N-Cadherin, vimentin, and SNAIL (Fig 2F-2J). Taken together, these results suggested that LINP1 inhibited EMT of lung cancer cells.
Lnc-LINP1 represses the migration, invasion, and stemness of lung cancer cells
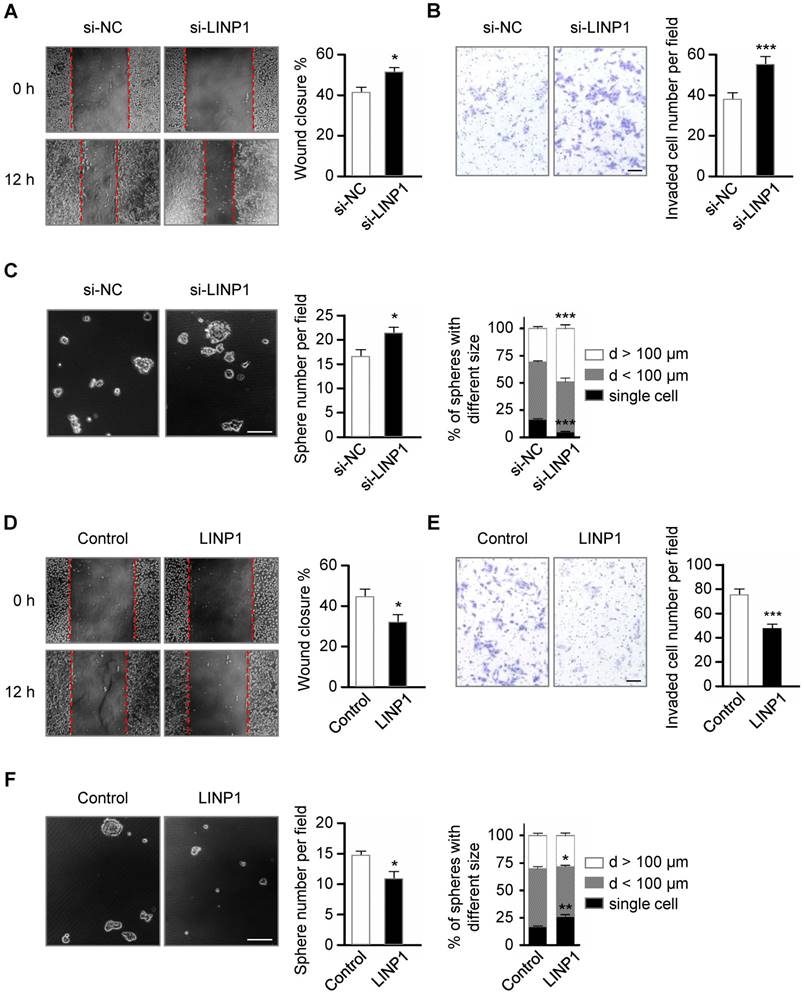
Tumor cells undergoing EMT become more migratory and invasive [2]. Next, we investigated the role of LINP1 in cancer cell migration and invasion in the lung cancer cell line A549. Scratch wound healing assay showed that LINP1 knockdown enhanced cell migration by 1.25-fold at 12 h (Fig 3A, P < 0.05). In the transwell assay, A549 cells transfected with LINP1 siRNA exhibited a 1.45-fold increase in the invasion through Matrigel (Fig 3B, P < 0.05). Previous studies have demonstrated that cancer cells following EMT induction show stem cell properties [17]. As expected, inhibition of LINP1 significantly promoted sphere formation both in the number and the size (Fig 3C). Conversely, when LINP1 was overexpressed in A549 cells, there was a significant decrease in cell migration, invasion, and sphere formation (Fig 3D-3F).
TGF-β regulates EMT partially through repressing lnc-LINP1
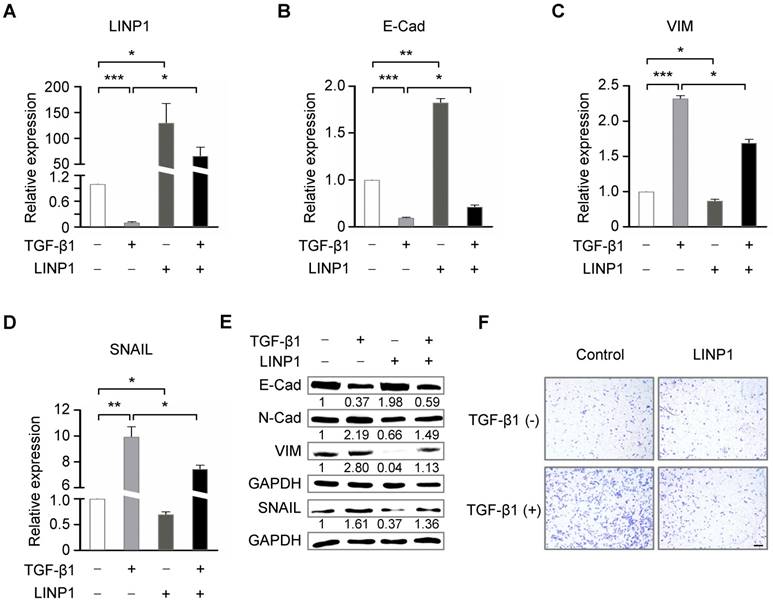
We next assessed the contribution of LINP1 inhibition to TGF-β-induced EMT. LINP1 overexpression blocked the downregulation of E-Cadherin and the upregulation of vimentin and SNAIL induced by TGF-β1 (Fig 4B-4E). Furthermore, LINP1 overexpression significantly abrogated TGF-β-induced cell invasion in A549 cells (Fig 4F). Taken together, these results suggested that LINP1 inhibited TGF-β-induced EMT and cell invasion in lung cancer cells.
Discussion
Over the past decade, accumulating evidence highlights the key roles of lncRNAs in various pathological processes including cancer metastasis. Recently, two studies have demonstrated that TGF-β-regulated lncRNAs play critical roles in EMT process of lung cancer cells. TGF-β upregulates lncRNA-MEG3 expression, which can promote EMT, migration, and invasion in lung cancer cells [10]. Another study reported that in lung cancer cell line A549, TGF-β suppresses LINC01186 expression by SMAD3, and LINC01186 regulates cancer cell migration, invasion, and EMT process [12]. In this study, we demonstrated the long noncoding transcript, LINP1, was significantly repressed by TGF-β/SMAD4 pathway. The loss- and gain-of-function assays of LINP1 in the lung cancer cell line A549 demonstrated that LINP1 inhibited EMT, thereby suppressing cancer cell migration, invasion, and stemness. Moreover, LINP1 repressed TGF-β-induced EMT and cell invasion in lung cancer cells.
Lnc-LINP1 inhibits EMT of lung cancer cells. (A) qRT-PCR of LINP1 expression in A549 cells that were transfected with negative control (NC) or LINP1 siRNAs for 24 h. (B) The morphological change of A549 cells that were transfected with control (top) or LINP1 (bottom) siRNAs for 24 h. (C and D) qRT-PCR and Western blot results of E-Cad (E-Cadherin), VIM (vimentin), and SNAIL in A549 cells with control or LINP1 siRNAs. (E) In situ fluorescence immunohistochemistry assay of E-Cadherin (E-Cad, green), N-Cadherin (N-Cad, red), and vimentin (VIM, red) in A549 cells with control or LINP1 siRNAs. Nucleus was dyed by Hoechst (blue). (F) qRT-PCR of LINP1 expression in A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1 for 24 h. (G) The morphological change of A549 cells that were transfected with pcDNA3.1-control (top) or pcDNA3.1-LINP1 (bottom) for 24 h. (H and I) qRT-PCR and Western blot results of E-Cadherin (E-Cad), vimentin (VIM), and SNAIL in A549 cells transfected with pcDNA3.1-control or pcDNA3.1-LINP1. (J) In situ fluorescence immunohistochemistry assay of E-Cadherin (E-Cad, green), N-Cadherin (N-Cad, red), and vimentin (VIM, red) in A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1. Nucleus was dyed by Hoechst (blue). The scale bars represent 20 μm. The error bars indicate mean ± SEM and P value is calculated by unpaired Student's t-test, * P < 0.05, ** P < 0.01.
Lnc-LINP1 represses the migration, invasion, and stemness of lung cancer cells. (A) Wound-healing assay of A549 cells transfected with negative control (NC) or LINP1 siRNAs. Wound closure was evaluated after 12 h of scratch. (B) Transwell invasion assay of A549 cells transfected with control or LINP1 siRNAs. The number of the invaded cells per field was counted after 12 h of transfection. The scale bars represent 50 μm. (C) Phase-contrast images of sphere formation in A549 cells transfected with control or LINP1 siRNAs. The scale bars represent 100 μm. Qualification of the number and the different sizes of spheres in A549 cells with control or LINP1 siRNAs. (D) Wound-healing assay of A549 cells transfected with pcDNA3.1-control or pcDNA3.1-LINP1. Wound closure was evaluated after 12 h of scratch. (E) Transwell invasion assay of A549 cells transfected with pcDNA3.1-control or pcDNA3.1-LINP1. The number of the invaded cells per field was counted after 24 h of transfection. The scale bars represent 50 μm. (F) Phase-contrast images of sphere formation in A549 cells transfected with pcDNA3.1-control or pcDNA3.1-LINP1. The scale bars represent 100 μm. Qualification of the number and the different sizes of spheres in A549 cells after transfection with pcDNA3.1-control or pcDNA3.1-LINP1. The error bars indicate mean ± SEM and P value is calculated by unpaired Student's t-test, * P < 0.05, ** P < 0.01, *** P < 0.001.
TGF-β regulates EMT partially through repressing lnc-LINP1. (A) qRT-PCR of LINP1 expression in A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. (B-D) qRT-PCR of E-Cadherin (E-Cad), vimentin (VIM), and SNAIL in A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. (E) Western blot assay of E-Cadherin (E-Cad), N-Cadherin (N-Cad), vimentin (VIM), and SNAIL in A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. (F) Transwell invasion assay of A549 cells that were transfected with pcDNA3.1-control or pcDNA3.1-LINP1, followed by 5 ng/ml TGF-β1 or vehicle for 12 h. The scale bars represent 100 μm. The error bars indicate mean ± SEM and P value is calculated by unpaired Student's t-test, * P < 0.05, ** P < 0.01, *** P < 0.001.
This study suggests a potential tumor suppressor function of LINP1, which is contradictory to the oncogenic function of LINP1 proposed by previous studies [13-15]. LINP1 has been shown to enhance repair of DNA double-strand breaks, and promote cell growth, EMT, and metastasis in breast and cervical cancers [13-15]. Our current work demonstrated that LINP1 functions as a tumor suppressor in lung cancer cells by inhibiting EMT-related migration, invasion, and stemness, indicating that the varied effects of LINP1 on tumor progression may be largely dependent on tumor cell context. Consistently, different functions of lncRNA-BANCR (BRAF-activated lncRNA) on tumor progression have been reported recently. LncRNA-BANCR enhances EMT and migration in colorectal cancer cells and melanoma cells, but inhibits EMT and migration in lung cancer cells [18-20]. Similarly, in gastric and lung cancer cells, lncRNA-SPRY4‑IT1 (SPRY4 intronic transcript 1) functions as a tumor suppressor by repressing cell proliferation, EMT, and metastasis, whereas in glioma cells, SPRY4‑IT1 acts as an oncogene [21-23]. How LINP1 plays context dependent functions in different tumor cell types needs to be further investigated.
Abbreviations
EMT: epithelial-mesenchymal transition; LINP1: lncRNA in nonhomologous end joining (NHEJ) pathway 1; TGF-β: transforming growth factor beta; SMAD4: SMAD family member 4; SNAIL: SNAIL family transcriptional repressor 1.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation (81772952, 81572717, 81272702, 31630093, 31430057) and National Key Research and Development Program of China (2016YFC1300600).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90
2. Thiery JP, Acloque H, Huang RY. et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871-90
3. Rosemary J. A, Akiko H. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790-811
4. Kamato D, Burch ML, Piva TJ. et al. Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation. Cell Signal. 2013;25(10):2017-24
5. Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114(Pt 24):4359-69
6. Massagué J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19(8):1745-54
7. Kang Y, Chen CR, Massagué J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11(4):915-26
8. Yuan JH, Yang F, Wang F. et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666-81
9. Yu F, Bing S, Mingyue T. et al. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531-41
10. Terashima M, Tange S, Ishimura A. et al. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J Biol Chem. 2017;292(1):82-99
11. Richards EJ, Zhang G, Li ZP. et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857-67
12. Hao Y, Yang X, Zhang D. et al. Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits migration and invasion through Epithelial-Mesenchymal-Transition in lung cancer. Gene. 2017;608:1-12
13. Zhang Y, He Q, Hu Z. et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016;23(6):522-30
14. Wang X, Liu H, Shi L. et al. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle. 2018;17(4):439-447
15. Liang Y, Li Y, Song X, Zhang N. et al. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol Ther. 2018;19(2):120-131
16. Isogaya K, Koinuma D, Tsutsumi S. et al. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res. 2014;24(8):994-1008
17. Mani SA, Guo W, Liao MJ. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704-15
18. Guo Q, Zhao Y, Chen J. et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8(2):869-875
19. Flockhart RJ, Webster DE, Qu K. et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006-14
20. Sun M, Liu XH, Wang KM. et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68
21. Xie M, Nie FQ, Sun M. et al. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250
22. Sun M, Liu XH, Lu KH. et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298
23. Liu H, Lv Z, Guo E. Knockdown of long noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation, metastasis and epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8(8):9140-6
Author contact
![]() Corresponding authors: tengyan0919com; yangxac.cn
Corresponding authors: tengyan0919com; yangxac.cn

 Global reach, higher impact
Global reach, higher impact