Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(14):1935-1949. doi:10.7150/ijbs.27315 This issue Cite
Research Paper
A chemosensory protein MsepCSP5 involved in chemoreception of oriental armyworm Mythimna separata
1. College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China
2. College of Resources and Environment, Huazhong Agricultural University, Wuhan 430070, China
3. Department of Plant Protection, Faculty of Agriculture, Benha University, Banha, Qalyubia 13736, Egypt
4. College of Informatics, Huazhong Agricultural University, Wuhan 430070, China
Received 2018-5-17; Accepted 2018-9-29; Published 2018-11-1
Abstract

Chemosensory proteins (CSPs) have been suggested to perform several functions in insects, including chemoreception. To find out whether MsepCSP5 identified from Mythimna separata shows potential physiological functions in olfaction, gene expression profiles, ligand-binding experiments, molecular docking, RNA interference, and behavioral test were performed. Results showed that MsepCSP5 was highly expressed in female antennae. MsepCSP5 showed high binding affinities to a wide range of host-related semiochemicals, and displayed that 26 out of 35 candidate volatiles were highly bound (Ki < 10 µM) at pH 5.0 rather than pH 7.4. The binding sites of MsepCSP5 to candidate volatiles were well predicted by three-dimensional structure modeling and molecular docking experiments. Pursuing further, biological activities of M. separata to highly bound compounds elicited strong behavioral responses, such as alcoholic compounds displayed strong attractiveness whereas terpenes showed repellency to M. separata. The transcript expression level of MsepCSP5 gene significantly decreased after injecting target dsRNAs, and resulted in non-significant preference responses of M. separata to semiochemicals, such as 3-pentanol and 1-octene-3-ol. In conclusion, MsepCSP5 may involve in semiochemical reception of M. separata.
Keywords: chemosensory proteins, expression patterns, binding characteristics, behavioral response
Introduction
The perception of chemical cues from the external environment is a crucial olfactory mechanism for the insect's survival [1, 2]. It is stated that insects primarily rely on the olfactory system to distinguish unique chemical signals in their natural habitats, which is vital for host seeking, foraging, mating, oviposition and consequently existence [3-5]. At the initial stage of olfactory reception, the semiochemicals enter the pore tubules in the olfactory sensillum. These semiochemicals are transported by small and low molecular weight proteins via sensillar lymph to activate olfactory receptors (ORs) on the membrane of the olfactory sensory neurons [4, 6]. Next, the signals are transmitted to the brain for processing and coding by the central nervous system, and then translated into a behavioral response to the semiochemicals [6-8]. Insect olfactory system is highly sensitive and specific which mainly depends on the interaction of ligands between soluble proteins existing in insect sensillar lymph [odorant binding proteins (OBPs), chemosensory proteins (CSPs) and Niemann-Pick type C2 proteins (NPC2)], transmembrane proteins [olfactory receptors (ORs), ionotropic receptors (IRs) and sensory neuron membrane proteins (SNMPs)] [3, 6, 9-12]. It is now well established from a variety of studies that ligand-binding proteins are small soluble proteins that distinguish, bind, deliver odorants and trigger receptors [6, 13-16]. It is stated that CSPs possess six α-helices and four cysteine residues, making two disulfide bridges. These CSPs are distributed within chemosensory sensillar lymph as well as expressing their existence in non-sensory tissues [17-21]. They are also flexible to bind varied chemicals, such as short and long aliphatic chains [8]. Therefore, CSPs have been thought playing the central role in helping insects recognize the distinctive chemical signals and regulate their behaviors [2, 15]. For instance, CSPs in the female Glossina morsitans showed considerable expressions after sucking blood, suggesting that the CSPs are highly involved in host-seeking [22]. On the other hand, CSPs have been suggested as being responsible for growth and development [23]. Such as, CSP-P10 showed 30 times greater expression in regenerating legs compared to normal legs of Periplaneta americana [24]. However, the exact physiological functions and mechanism of CSPs remain elusive.
Mythimna separata (oriental armyworm, Lepidoptera: Noctuidae) is overwhelming and polyphagous pest of a variety of crops, such as wheat, maize and rice in Africa, Asia and Oceania. It is one of the most serious pests affecting the quality and quantity of crop yield [25, 26]. To control M. separata, molecular insights into olfactory mechanism are important to elucidate. Only a few studies on olfactory proteins have been conducted on M. separata [27-29]. A total of 126 olfactory genes including 13 GRs, 43 ORs, 3 SNMPs, 16 IRs, 37 OBPs and 14 CSPs were predicted from the genome of M. separata in previous studies, but their functions are still unknown [25, 30]. To date, attention to the functions of chemosensory protein genes has not been paid in M. separata. In order to study the functional roles of MsepCSP5 from M. separata in the present study, we evaluated expression profiles, purified protein to characterize the ligand's binding affinity, and finally molecular docking followed by RNAi technique combined with behavioral bioassay was employed to investigate gene silencing effects.
Materials and Methods
Insects and Tissues Collection
Mythimna separata used in the present study were obtained from Institute of Plant Protection, Hubei Academy of Agricultural Sciences, Wuhan, and kept under constant conditions (25±1°C, 70±5% relative humidity and 14:10h light:dark cycle). Adults were fed on 10% honey solution, and both sexes were kept in separate cages. Larvae (6th instar), pupae (5 days old), adults (1, 2, 3, 4 and 5 days mixed) and tissues (head without antennae, thorax, antennae, abdomen, legs, and wings) of both sexes of M. separata (1-5 days mixed) were collected for RT-qPCR analysis. Each tissue was collected with three replicates and stored at -80°C.
RNA Extraction and cDNA Synthesis
Total RNA from the whole larvae, pupae, adult and body tissues was extracted for RT-qPCR, and 24, 48, 72, 96, and 120 h old adults [non-injected control, ds-injected green fluorescent protein (dsGFP) and dsRNA injected (dsMsepCSP5)] for RNAi experiments using TRIzol reagent (Invitrogen, USA). The purity was examined by 1% agarose gel electrophoresis and the concentration was quantified using BioPhotometer Plus spectrophotometer (Eppendorf, Hamburg, Germany). We used 1µl of total RNA to synthesize single-stranded cDNA using the Prime-Script-II RT reagent kit with gDNA Eraser (TaKaRa Bio Inc., Shiga, Japan) according to the instruction manual. The product was either used directly for PCR amplification or stored at -20°C.
Sequence Analysis of MsepCSP5
From our previous cDNA library of M. separata [30], a complete sequence of MsepCSP5 was identified. The open reading frame was determined by ORF-finder (www.ncbi.nlm.nih.gov/gorf/gorf.html). Molecular weight of MsepCSP5 was determined using SWISS-PROT program (ExPASy server). Predication of signal peptides was performed using SignalP V-3.0 (www.cbs.dtu.dk/services/SignalP/). The sequence similarity of MsepCSP5 with homologos from other insect species was achieved on NCBI-BLAST (www.blast.ncbi.nlm.nih.gov/) and amino acid sequences were aligned on ClustalW and edited using Gene-Doc2.7 [31]. A phylogenetic tree based on neighbor-joining method was constructed using MEGA-6.0 program [32] with default setting and bootstrap support with 1000 replicates.
Real Time Quantitative PCR
Expression profile of MsepCSP5 was analyzed in larvae, pupae, adult and various tissues of M. separata by RT-qPCR using CFX96-Real-time thermal cycler (Applied Bio-systems, USA). The primer sequences were constructed using data base of NCBI (www.ncbi.nlm.nih.gov/tools/primer-blast/). RT-qPCR efficiencies were determined by constructing standard curve from 10-fold dilution series of template concentration with 3 parallel replicates. The resultant amplification efficiencies of primers were >90% in all experiments. We used β-actin gene (GQ856238) as an internal control for the normalization of MsepCSP5 expression. Each reaction contained 10μl of 2×Sybr Green qPCR Master Mix (Aidlab, China), 1μl of cDNA, 0.5μl of gene-specific primers and 8μl of sterilized ultra-pure water. Thermal cycling was performed at 95°C for 3min, then 40 cycles at 95°C for 10s and at 55°C for 30s.
Samples were analyzed three times as biological and three times as technical replicates to examine the reproducibility. The 2-ΔΔCT method was applied for the comparative quantification to differentiate mRNA expression levels [33].
Recombinant-Plasmid Construction
MsepCSP5 sequence was amplified by PCR with a forward primer (5′-CCGGAATTCATGAACAGCTTCACAGTTTTGTG-3′) containing an EcoR I-restriction site and a reverse primer (5′-CCGCTCGAGTTATTGCTTAAGTTTCCTAAGTTCC-3′) containing an Xho I-restriction site. The product obtained from PCR was ligated into a pTOPO-T vector, and subsequent product was transformed into E. coli (DH5α). Positive colonies were chosen based on PCR-confirmation, cultured in Luria-Bertani (LB) medium with 50μg ml-1 of kanamycin and then sequenced. The pTOPO-T plasmid containing target sequence was digested with Xho I and EcoR I restriction enzymes, ligated into pET30a, and transformed into E. coli. After DNA sequencing, transformation of E. coli BL21 (DE3) competent cells was carried out with the confirmed recombinant plasmid. The correct recombinant plasmid was used for obtaining mature protein.
Expression and Purification of Recombinant-Protein
The expression and purification of MsepCSP5 were conducted according to Zheng et al. [34] . For His-tag protein expression, we selected the positive clone after DNA sequencing, cultured in LB medium (5 mL) containing kanamycin (50 μg ml-1) and shaken (220 rpm) for 12 h at 37°C. After shaking, the culture was diluted in LB-medium (1000 mL) and grown until an optical density (OD600) of 0.4-0.6. For the enhancement of protein synthesis, the culture was further incubated with isopropyl-beta D-thiogalactopyranoside (IPTG; 0.1 mM L-1) at 37 °C for 4 h. The protein after expression was obtained as inclusion bodies. For purification, inclusion body protein was treated with 10ml of 8 M L-1 urea [CO(NH2)2] in 50 mM Tris-HCl buffer (pH 7.4) for solubilization and subsequently incubated for one hour in 10 mM DDT. Afterwards, 200μl cystine (100 mM L‑1) and 15 ml cysteine (5 mM L-1) were added. Prior to purification, a solution containing protein was dialyzed twelve times every two hour into Tris-HCl buffer (30 mM, pH 7.4). The purification of recombinant protein was accomplished passing through a Ni affinity chromatography column (Healthcare-GE, Uppsala, Sweden). The protein digestion was accomplished using the enterokinase enzyme, and subsequently incubated at 26 °C for 15 h to cleave off the Histidine-tag (His-tag). The digested protein was again passed through a column to get His-tag free protein. We verified the expression and purification of MsepCSP5 protein by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and quantified the concentration as explained by Cao et al. [35]. Purified protein was dialyzed in Tris-HCl buffer at two pH gradients (5.0 and 7.4) prior to fluorescence-binding assays.
Fluorescence Binding Assay
A total of 35 ligands were tested in fluorescence binding assay to assess the binding affinity of MsepCSP5 using a fluorescent probe i.e. N-phenyl-1-naphthylamine (1-NPN). A stock solution of each tested ligand (Sigma Aldrich, Louis, USA) was prepared in methanol (spectrophotometric-grade). The binding ability of 1-NPN was measure by adding 1-NPN (1 mM) into protein solution (2 μM L-1) diluted with Tris-HCl (30 mM), and final concentration was made 0 to 20 μM L-1 at room temperature. The mixture of MsepCSP5/1-NPN was excited at a specific wavelength (337 nm), and emission spectra ranging from 360 to 600 nm were recorded with a scanning speed of 300 nm min-1 using a RF-5301PC-fluorescence spectrophotometer (Shi-madzu, Kyoto, Japan) with slit width of 10 nm and light path quartz cuvette (1 cm) at temperature of 25°C. Ligand's binding with MsepCSP5 was performed with three independent replicates. Ligand's binding affinity (Ki) of MsepCSP5 was calculated as following:
Ki = IC50/(1+[1-NPN]/K1-NPN) (1)
where IC50 is the ligand's concentration, [1-NPN] is the free concentration of 1-NPN, and K1-NPN is the dissociation constant (Kd) of the complex MsepCSP5/1-NPN.
Three-Dimensional Structural Modeling and Ligand's Molecular Docking
The Delta-BLAST was executed on database website of NCBI (www.blast.ncbi.nlm.nih.gov/Blast.cgi) with MsepCSP5 amino acid sequence against protein data bank, PDB (www.rcsb.org), and SWISS-MODEL (www.swissmodel.expasy.org/) was used. The sequences from BLAST results were chosen based on highest resemblance and ClustalW2 was employed for multiple alignments. The top-hit sequence of protein was designated depending on query coverage, sequence homology, number of cysteines and phylogeny. The selected template of MbraCSP6 from M. brassicae (PDB:1KX8_A) for MsepCSP5 was used to construct a three-dimensional model [36]. We employed docking protocol executed in MOE V-2012.10 for molecular docking made by Chemical Computing Groups [37]. For further prediction of MsepCSP5 binding sites, ligands [2-heptanol, 3-pentanol, trans-2-hexen-1-ol, 1-octene-3-ol, methyl benzoate, ethyl acetate, (R)-(+)-α-pinene, (-)-limonene, (+)-3-carene and 2-undecanone] displaying high binding affinities to MsepCSP5 were selected for the docking into binding pocket of the three-dimensional structure. The parameters, taken into consideration for the calculation of interaction between ligand molecules and score of pertinent ligands, were as follows; rescoring-1 (Refinement:Force-field, London-d.G) and rescoring-2 (Placement:Triangle-Matcher, GBVI / WSA-d.G). The utmost appropriate docked structure of ligand /MsepCSP5 was elected considering minimum S-score and values of root mean square deviation (RMSD). S-score represents values that were calculated from built-in scoring functions of MOE considering the ligand's binding potential with receptor protein post docking. Root mean square deviation values are used for the comparison of docked conformation with reference conformation. Ligand possessing small RMSD and lowest S-scores can be developed as potential inhibitors [38].
Olfactometer Bioassay
In order to validate the olfactory orientation and behavioral response of M. separata to volatiles based on binding assays, olfactometer bioassay was performed [39]. A glass Y-tube olfactometer (arms: 3.0cm diameter by 26cm long, base: 4.0cm diameter by 25cm long) was used in this bioassay. The incoming air to the tube was allowed to pass via activated charcoal filter, then humidified with ultra-pure water. The humidified air coming from activated charcoal was divided into two tiny chambers; one holding tested volatile and the other holding liquid paraffin (control). Both tiny chambers were attached with arms of Y-tube. An inline flow-meter (Gilmont Instruments, USA) was used to keep the airflow constant (6.0 L min-1). Considering the nocturnal behavior of M. separata, the whole experiment was conducted after 1 to 2 h of sunset in the dark room; orange light was used, the temperature was 25±1°C and the relative humidity was 70%±5%. A 10µl of tested volatile was applied to 10×10mm filter paper (Whatman No.1) and was placed in one tiny chamber attached with one arm of olfactometer. A separate olfactometer was used for female and male moth. A three-day-old moth was released in the base of Y-tube and was given 10 min to respond to the volatile or control. To make sure that insects have no exposure to the tested volatiles, female and male moths were kept into isolated test tubes (50ml) and closed with sterilized cotton plug for approximately 30 min before the test. Thirty female and thirty male moths were used for each tested volatile in this behavioral bioassay.
Synthesis of Double Stranded RNA
The full-coding MsepCSP5 sequence was cloned into vector (pTOPO-T) and the amplification of target sequence was carried out using diluted plasmid as template. The amplification of MsepCSP5 sequence was performed by PCR using gene specific primers (Table S1) conjugated with T7 RNA-polymerase promoter (19-bases). The products generated by PCR of 384 bp for MsepCSP5 and 460 bp for green fluorescent protein (GFP) were purified and used as templates for dsRNA synthesis using T7-Ribo Max RNAi kit (Pro-mega, USA) following the instructions of the manufacturer. The precipitation of dsRNA was carried out using isopropanol, followed by resuspension in Nuclease-free water, and analyzed by Nanodrop-2000 spectrophotometer (ThermoScientific, Wilmington). The dsRNA quality was tested by agarose gel (1%), and saved at -80°C.
dsRNA Injection and Gene Expression Analysis
We injected dsRNA (1 µl of 2.5 µg µL-1) into 3rd and 4th abdominal segment of M. separata pupa (7 days old) using Eppendorf microinjection system TransferMan NK2. The injected pupae were kept under control conditions of 70% ± 5% relative humidity and 25 ± 1°C temperature until eclosion. RNAi had three treatments as follows: (i) non-injected (control), (ii) dsGFP-treated and (iii) dsMsepCSP5 treated. After eclosion, moths were kept into individual cages for each treatment. For RNAi analysis, three adults of M. separata from each sex were taken from each treatment at 24, 48, 72, 96, and 120 h of eclosion. The RT-qPCR for expression profiling was conducted under the concurrent conditions as described above.
The Y-tube bioassay (olfactometer) was also performed to examine gene knockdown effects after RNAi, and the procedure was same as described above. Three treatments (control, dsGFP and dsMsepCSP5) were tested in the bioassay for 72 h old moth of both sexes. For post RNAi bioassay, M. separata (30 female and 30 male) were used for each tested volatiles. 1-octen-3-ol and 3-pentanol were used for post RNAi behavioral bioassay and selection was based on the significant attraction of moths to the tested volatiles before RNAi.
Statistical Analysis
RT-qPCR data were analyzed performing one-way analysis of variance (ANOVA) using Tukey's HSD test to find out the significant differences of expression profiles among the treatments. A chi-square test (χ2) was applied in order to identify significant differences behavioral responses of M. separata. The significant differences were evaluated at p≤0.05. Data were analyzed utilizing SPSS version 16.0 for windows.
Results
Characterization and Sequence Analysis of MsepCSP5
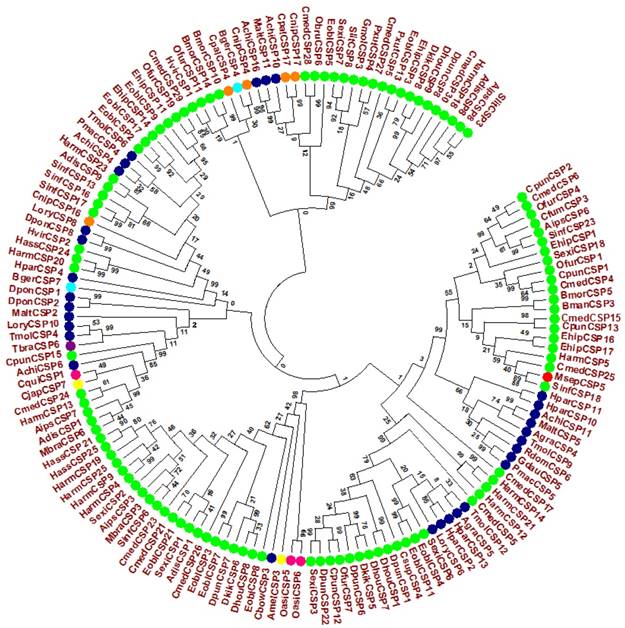
A full length cDNA encoding MsepCSP5 was cloned and verified by sequencing. Sequence analysis of MsepCSP5 revealed an Open Reading Frame (ORF) (Accession#JAV45876.1) contains 384 nucleotides encoding 127 amino acid residues, with 14.70 kDa molecular weight (Figure 1A). MsepCSP5 contained predicted signal peptides with 18 amino acid residues at N-terminus and an isoelectric point of 8.71. The sequence of MsepCSP5 was aligned with CSPs from other insect species of Lepidoptera and shared 60-80% similarities (Figure 1B). The alignment result revealed that 4 conserved cysteines were present in all CSPs. A neighbor-joining tree of 147 CSPs was built on the basis of 40% to 80% sequence homology with other CSPs including Lepidoptera (24 species), Coleoptera (11 species), Hemiptera (one species), Hymenoptera (two species), Dictyoptera (one species), Neuroptera (two species), Orthoptera (two species) (Figure 2). Phylogenetic-tree revealed that MsepCSP5 belongs to the same cluster of insect order locally suggesting that CSPs curtailed from broad gene duplication. Thus, these results proposed that MsepCSP5 originated from the same ancestors of insect order.
Tissue-Specific Expression of MsepCSP5
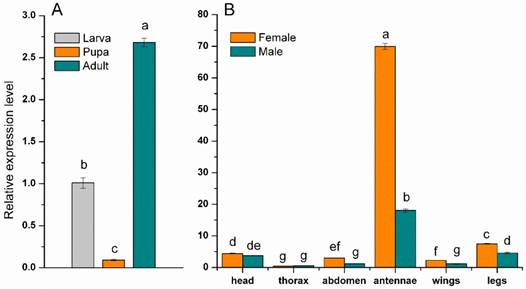
The expression of the MsepCSP5 was examined in specific tissues, sexes and developmental stages of M. separata to understand the physiological functions of protein. The resulting dataset showed that expression of MsepCSP5 was significantly (p≤0.05) larger in adult than the larva and pupa (Figure 3A). The MsepCSP5 displayed different expression patterns for both male and female moth, with some tissues displaying similar expression patterns, while others exhibited distinct trends (Figure 3B). MsepCSP5 was predominantly expressed 3.87-fold higher in female antennae than in male. MsepCSP5 was also ubiquitously expressed in legs, head, abdomen and wings tissues of female at relatively higher level than male. On the other hand, the tissues of thorax showed relatively similar expression of MsepCSP5 in both sexes, but the expression level was much lower as compared to the antennae. Based on the high expression pattern of MsepCSP5 in antennae, we selected this gene for further investigation to test its plausible role in chemoreception.
Characterization and sequence analysis of MsepCSP5. (A) Nucleotide and amino acid sequence of MsepCSP5. Stop codon is indicated by red star and 4 conserved cysteines are shown in red circles. Eighteen predicted signal peptides are represented by red underlined sequence. (B) Alignment of MsepCSP5 with amino acids from various insects. Red stars indicate conserved cysteines. The sky blue, purple and black colors denote 60%, 80% and 100% similarities. GenBank accession no. and names of insect species are as following: MsepCSP5, JAV45876.1, Mythimna separata; SinfCSP18, AGY49267.1, Sesamia inferens; AdisCSP1, AND82443.1, Athetis dissimilis; SexiCSP1, ABM67688.1, Spodoptera exigua; SlitCSP5, ALJ30216.1, Spodoptera litura; HarmCSP5, AEB54579.1, Helicoverpa armigera; OfurCSP7, BAV56811.1, Ostrinia furnacalis; SexiCSP2, ABM67689.1, Spodoptera exigua; HvirCSP1, AAM77041.1, Heliothis virescens; HarmCSP4, AEX07269.1, Helicoverpa armigera; AipsCSP3, AGR39573.1, Agrotis ipsilon; CmedCSP25, ALT31607.1, Cnaphalocrocis medinalis; EoblCSP21, ALS03846.1, Ectropis oblique; SlitCSP2, ALJ30213.1, Spodoptera litura; AdisCSP9, AND82451.1, Athetis dissimilis; CmedCSP23, ALT31605.1, Cnaphalocrocis medinalis; PxutCSP4, BAF91714.1, Papilio xuthus; DkikCSP5, AII01032.1, Dendrolimus kikuchii; DhouCSP7, AII01017.1, Dendrolimus houi; EhipCSP17, AOG12901.1, Eogystia hippophaecolus; CsupCSP4, AHC05675.1, Chilo suppressalis; CmedCSP21, AIX97840.1, Cnaphalocrocis medinalis.
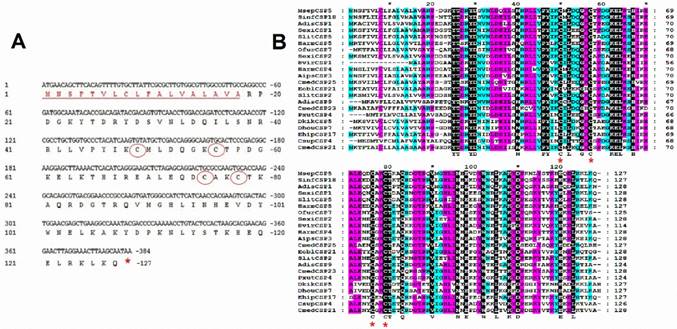
Evolutionary tree of MsepCSP5 and 146 sequences of CSPs from various insect's species. MsepCSP5 is denoted with a red circle and others insect species from different order are denoted as follow: Lepidoptera (green), Coleoptera (blue), Hemiptera (purple), Hymenoptera (yellow), Dictyoptera (Aqua), Neuroptera (orange), Orthoptera (pink), Species abbreviations are included for taxon identifications. Msep: Mythimna separata, Sinf: Sesamia inferens, Adis: Athetis dissimilis, Sexi: Spodoptera exigua, Hvir: Heliothis virescens, Harm: Helicoverpa armigera, Ehip: Eogystia hippophaecolus, Ofur: Ostrinia furnacalis, Cpun: Conogethes punctiferalis, Aips: Agrotis ipsilon, Eobl: Ectropis oblique, Cmed: Cnaphalocrocis medinalis, Bman: Bombyx mandarina, Bmor: Bombyx mori, Pxut: Papilio xuthus, Csup: Chilo suppressalis, Dhou: Dendrolimus houi, Hass: Helicoverpa assulta, Dkik: Dendrolimus kikuchii, Slit: Spodoptera litura, Cfum: Choristoneura fumiferana, Gmol: Grapholita molesta, Mbra: Mamestra brassicae, Dpun: Dendrolimus punctatus, Obru: Operophtera brumata, Achi: Anoplophora chinensis, Malt: Monochamus alternatus, Agra: Anthonomus grandis, Tmol: Tenebrio molitor, Cbow: Colaphellus bowringi, Gdau: Galeruca daurica, Hpar: Holotrichia parallela, Pmac: Pyrrhalta maculicollis, Lory: Lissorhoptrus oryzophilus, Rdom: Rhyzopertha dominica, Dpon: Dendroctonus ponderosae, Tbra: Triatoma brasiliensis, Amel: Apis mellifera, Cjap: Camponotus japonicas, Bger: Blattella germanica, Cnip: Chrysoperla nipponensis, Cpal: Chrysopa pallens, Oasi: Oedaleus asiaticus, Cqui: Culex quinquefasciatus.
Expression profiles of MsepCSP5. (A) Expression pattern in larva, pupa and adult, (B) Expression pattern in different body parts of female and male. Different letters on columns represent significant differences at p≤0.05.
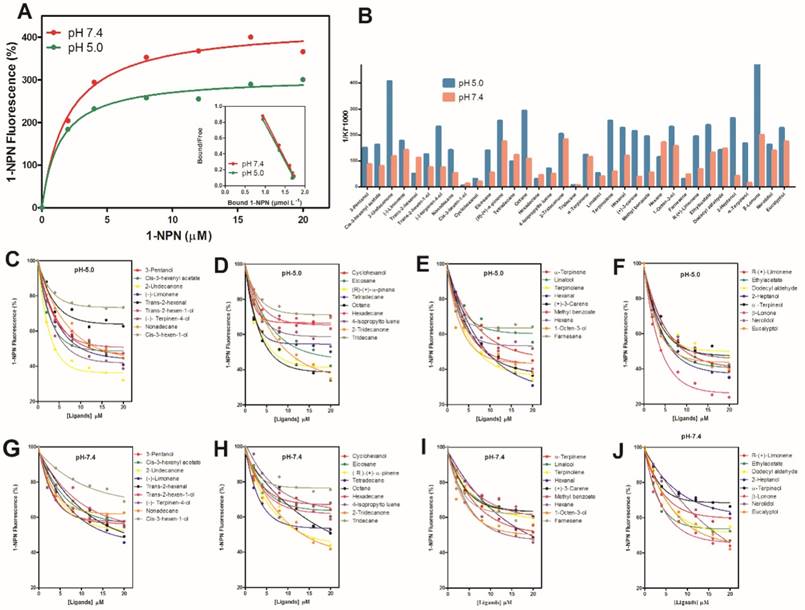
Fluorescence Binding Assay
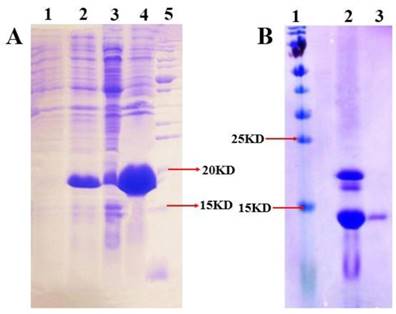
The expression and purification of MsepCSP5 (Figure 4) were examined by SDS-PAGE (15%). Binding assays were performed to examine MsepCSP5 binding properties to various ligands. The fluorescence binding assay displayed that MsepCSP5 strongly bound to fluorescent probe, N-phenyl-1-naphthylamine (1-NPN). The dissociation constants (Kd) of MsepCSP5/1-NPN complex calculated from Scatchard plots were 2.91 μM at pH 5.0 and 3.65 μM at pH 7.4. A saturation and linear Scatchard plot suggested single binding site in the protein with no allosteric effects (Figure 5A). The displacement curves of 1-NPN by ligands are shown in Figure 5. The values of IC50 and Ki of ligands were calculated (Table 1). Contemplating view point of binding and release mechanisms of ligands with soluble proteins in insect's body [40], we selected two pH levels (5.0 and pH 7.4) to simulate in vitro environment. The physiology of insect's antennal lymph is neutral where the odor molecules bind to protein. The nerve membrane is acidic which contains ion channel, where odor molecules are released from protein. Furthermore, the mainstream hypothesis of ligand binding and release is that proteins bind the odorants under neutral pH and release odorants under acidic condition [40]. Therefore, the binding assay at two different pH in this study contributed to understand the binding and release mechanism of MsepCSP5.
MsepCSP5 binding affinities with different group of volatiles
| Chemical Name | CAS No. | Purity (%) | pH 5.0 | pH 7.4 | ||
|---|---|---|---|---|---|---|
| IC50 (µM) | Ki (µM) | IC50 (µM) | Ki (µM) | |||
| Alcohols | ||||||
| 3-Pentanol | 584-02-1 | 98 | 14.05 | 6.71 | 29.79 | 11.79 |
| Trans-2-hexen-1-ol | 928-95-0 | 96 | 16.97 | 8.1 | 34.16 | 13.52 |
| Cis-3-hexen-1-ol | 928-96-1 | 97 | 726.82 | 347.15 | 226.79 | 89.75 |
| Cyclohexanol | 108-93-0 | 99 | 71.55 | 34.17 | 136.46 | 54 |
| Linalool | 78-70-6 | 97 | 41.3 | 19.72 | 65.32 | 25.85 |
| 2-Heptanol | 543-49-7 | 98 | 7.95 | 3.8 | 62.98 | 24.92 |
| α -Terpineol | 10482-56-1 | 90 | 12.6 | 6.02 | 197.86 | 78.3 |
| 1-Octen-3-ol | 3391-86-4 | 98 | 9.13 | 4.36 | 16.25 | 6.43 |
| Nerolidol | 7212-44-4 | 98 | 12.97 | 6.2 | 18.27 | 7.23 |
| Eucalyptol | 470-82-6 | 96 | 9.31 | 4.45 | 14.62 | 5.79 |
| Esters and Benzoates | ||||||
| Cis-3-hexenyl acetate | 3681-71-8 | 98 | 13.01 | 6.22 | 32.36 | 12.81 |
| Ethylacetate | 141-78-6 | 99.8 | 8.84 | 4.22 | 19.38 | 7.67 |
| Methyl benzoate | 93-58-3 | 99.5 | 10.85 | 5.18 | 47.47 | 18.79 |
| Aldehydes | ||||||
| Trans-2-hexenal | 6728-26-3 | 97 | 43.45 | 20.75 | 22.95 | 9.08 |
| Hexanal | 66-25-1 | 95 | 9.28 | 4.43 | 21.39 | 8.46 |
| Dodecyl aldehyde | 112-54-9 | 92 | 15 | 7.17 | 17.34 | 6.86 |
| Ketones | ||||||
| 2-Undecanone | 112-12-9 | 98 | 5.15 | 2.46 | 21.62 | 8.56 |
| β-Lonone | 79-77-6 | 96 | 4.38 | 2.09 | 12.77 | 5.06 |
| 2Tridecanone | 593-08-8 | 98 | 10.36 | 4.95 | 13.93 | 5.51 |
| Alkanes | ||||||
| Nonadecane | 629-92-5 | 99 | 14.97 | 7.15 | 49.25 | 19.49 |
| Eicosane | 112-95-8 | 99 | 15.18 | 7.25 | 47.84 | 18.93 |
| Tetradecane | 629-59-4 | 99 | 21.73 | 10.38 | 20.76 | 8.22 |
| Octane | 111-65-9 | 98 | 7.17 | 3.42 | 23.61 | 9.34 |
| Hexadecane | 544-76-3 | 99.8 | 74.29 | 35.48 | 58.27 | 23.06 |
| Tridecane | 629-50-5 | 99 | 477.83 | 228.22 | 522.48 | 206.76 |
| Hexane | 110-54-3 | 95 | 18.63 | 8.9 | 14.89 | 5.89 |
| Terpenes | ||||||
| (-)-Limonene | 5989-54-8 | 95 | 11.95 | 5.71 | 17.96 | 7.11 |
| ( R )-(+)- α -pinene | 7785-70-8 | 98 | 8.26 | 3.94 | 14.61 | 5.78 |
| 4-lsopropyltoluene | 99-87-6 | 98 | 30.24 | 14.44 | 52.31 | 20.7 |
| α -Terpinene | 99-86-5 | 85 | 17.15 | 8.19 | 22.17 | 8.77 |
| Terpinolene | 586-62-9 | 85 | 8.27 | 3.95 | 43.84 | 17.35 |
| (+)-3-Carene | 13466-78-9 | 90 | 9.79 | 4.67 | 67.6 | 26.75 |
| Farnesene | 502-61-4 | 98 | 74.65 | 35.65 | 54.42 | 21.54 |
| R-(+)-Limonene | 5989-27-5 | 99 | 10.85 | 5.18 | 37.92 | 15 |
| (-)-Terpinen-4-ol | 20126-76-5 | 95 | 9.07 | 4.33 | 34.37 | 13.6 |
SDS-PAGE analysis illustrating the expression and purification of MsepCSP5. (A) 1 and 2: induced and non-induced PET30/MsepCSP5; 3:PET30/MsepCSP5 supernatant; 4, insoluble inclusion bodies of PET30/MsepCSP5; 5: molecular marker, (B) 1, molecular marker; 2, MsepCSP5 digested protein with enterokinase enzyme; 3, purified MsepCSP5 protein without His-tag.
Thirty five ligands (plant volatiles) were selected to delineate the binding affinities (1/Ki×1000) of MsepCSP5 at pH 5.0 and 7.4 (Figure 5B). The pH strongly affected the ligand binding ability to MsepCSP5. When compared the binding ability of the volatiles at different pH values, 26 ligands had strong binding abilities at pH 5 (Figure 5C, D, E and F) except cyclohexanol, cis-3-hexen-1-ol, linalool, trans-2-hexenal, tetradecane, hexadecane, tridecane, 4-lsopropyltoluene and farnesene, however, numerous compounds displayed relatively low binding abilities at pH 7.4 (Figure 5G, H, I and J).
At pH 5, all of the ketones, esters and benzoates showed strong binding affinities (Ki < 10 µM) with the MsepCSP5. Additionally, most of alcoholic compounds, for instance, 3-pentanol, trans-2-hexen-1-ol, 2-heptanol, α-terpineol, 1-octen-3-ol, nerolidol and eucalyptol, presented high binding affinity to protein. 7, 4 and 1 compounds among terpene, alkane and aldehyde groups, respectively displayed strong binding affinity for MsepCSP5.
Fluorescence binding assay of MsepCSP5 with different ligands. (A) relative Scatchard plot analysis and binding curve of 1-NPN at pH 5.0 and 7.4, (B) Ligands binding ability of MsepCSP5 (presented by 1/Ki*1000) with 35 ligands at pH 5.0 and 7.4, (C, D, E and F) MsepCSP5 binding curves with various volatiles at pH 5.0, (G, H, I and J) MsepCSP5 binding curves with various volatiles at pH 7.4.
Three-Dimensional Modeling and Ligand's Docking
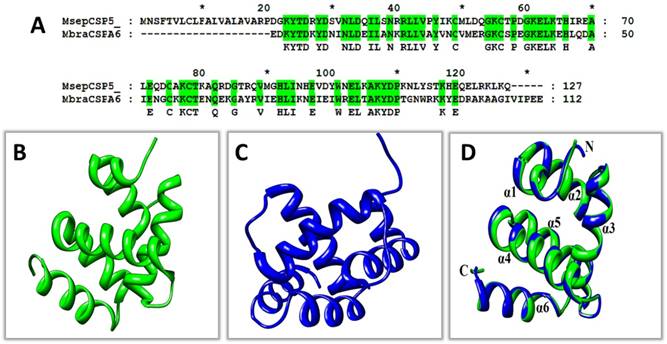
To advance the concept and understanding of interaction between MsepCSP5 and ligands, structural modeling and molecular docking were achieved using DELTA-BLAST. MsepCSP5 had 53% sequence similarity with MbraCSP6 (Figure 6A). Homology modeling of MsepCSP5 (Figure 6B) was created using MbraCSP6 (Figure 6C) as a template considering the following points: (1) highest sequence similarity between MbraCSP6 and MsepCSP5, (2) complete matching of MsepCSP5 and MbraCSP6 sequence and (3) both M. separata and M. brassicae belong to same order Lepidoptera. Homology modeling results displayed the best model with RMSD value of 0.226Å (Figure 6D) and Ramachandran Plot was used to confirm its quality by considering ψ and φ values. As illustrated in Figure 1S, 98% and 2% residues were present in the favored and near-marginal regions, respectively in a Ramachandran plot with the highest residues in α-helix region indicating that predicted model was reliable and acceptable.
3D modelling of MsepCSP5. (A) MsepCSP5 sequence alignment with MbraCSPA6 (B) 3D structure of MbraCSPA6 protein, (C) 3D structure of MsepCSP5 protein, (D) Structure of MsepCSP5 and MbraCSPA6 proteins after superimposition. Green color denotes the template MbraCSPA6 and blue color denotes MsepCSP5. N: N-terminal, C: C-terminal.
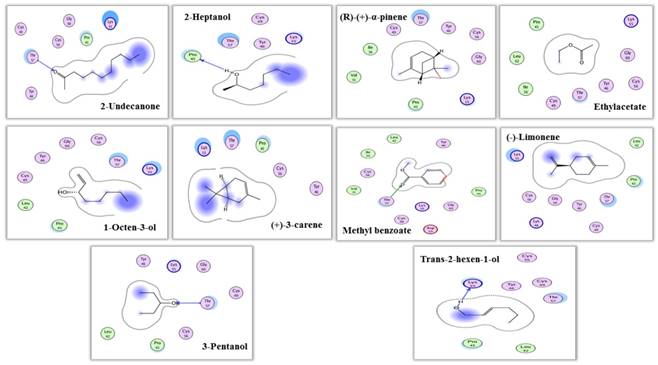
Interaction view (2 dimensional) of MsepCSP5. Hydrogen bonds are shown by arrows.
Molecular docking results of tested ligands for MsepCSP5
| PubChem IDs | Ligands | S-Score | RMSD | Residues interacting with H-bonding | Closer contact interacting residues |
|---|---|---|---|---|---|
| 8163 | 2-Undecanone | -17.05 | 1.30 | Thr-57 | Lys-55, Pro-45, Gly-60, Cys-56, Cys-49, Tyr-46 |
| 10976 | 2-Heptanol | -17.91 | 1.13 | Pro-45 | Cys-49, Thr-57, Tyr-46, Lys-55 |
| 6654 | ( R )-(+)-α-pinene | -17.42 | 0.80 | Thr-57, Tyr-46, Cys-56, Gly-60, Lys-55, Pro-45, Val-31, Ile-36, Cys-49 | |
| 8857 | Ethylacetate | -16.32 | 0.64 | Lys-55, Gly-60, Cys-60, Tyr-46, Thr-57, Cys-49, Ile-36, Leu-42, Pro-45 | |
| 18827 | 1-Octen-3-ol | -17.46 | 0.77 | Pro-45, Leu-42, Cys-49, Tyr-46, Gly-60, Cys-56, Thr-57, Lys-55 | |
| 26049 | (+)-3-carene | -16.13 | 1.82 | Lys-55, Thr-57, Pro-45, Cys-56, Tyr-46 | |
| 7150 | Methyl benzoate | -18.16 | 0.44 | Thr-57 | Tyr-46, Leu-42, Ile-36, Cys-56, Cys-56, Val-31, Lys-55, Gly-60, Asp-59 |
| 440917 | (-)-Limonene | -17.23 | 1.48 | Leu-42, Pro-45, Thr-57, Cys-49, Tyr-46, Gly-54, Lys-48, Cys-56, Lys-55 | |
| 11428 | 3-Pentanol | -16.62 | 1.20 | Thr-57 | Tyr-46, Lys-55, Gly-60, Cys-49, Cys-56, Pro-45, Leu-42 |
| 5318042 | Trans-2-hexen-1-ol | -16.31 | 0.91 | Lys-55 | Cys-56, Tyr-46, Cys-49, Thr-47, Leu-42, Pro-45 |
RMSD: root mean square deviation
Binding cavities of MsepCSP5. Green region represents hydrophobicity and red region represents hydrophilia. Oxygen atoms are shown by red atoms.
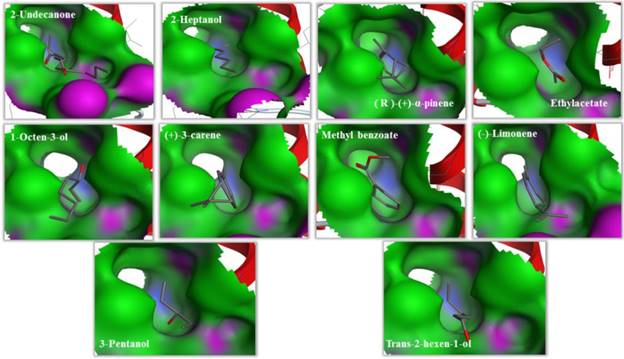
The predicted 3D model revealed that MsepCSP5 protein is globular in shape with 6 alpha-helices; α-1 (Leu33-Ser38), α-2 (Arg40-Leu51), α-3 (Pro58-Gln73), α-4 (Lys80-His96), α-5 (Val98-Tyr108) and α-6 (Ser115-Leu125) which formed binding pockets for ligand binding. The superimposition of both MsepCSP5 protein model and template structures showed six α-helices with 0.226Å RMSD value indicating similar folds of both model and template. To deepen the insights into the binding mechanism of MsepCSP5 and ligand, we selected 10 volatiles, based on the strong binding affinity in the binding assay, and docked them into the binding pockets of MsepCSP5. Docking results presented a cavity wall composed of several residues as follows: Lys-55, Pro-45, Gly-60, Cys-56, Cys-49, Tyr-46, Thr-57, Val-31, Ile-36, Cys-60, Leu-42, Asp-59, Gly-54, Lys-48 and Thr-47. Among these residues, Thr-57, Pro-45 and Lys-55 were hydrophilic with oxygen atoms located near the center of cavity. The interaction between six amino acid residues with 2-undecanone, nine amino acid residues with methyl benzoate, seven residues with 3-pentanol, and Thr-57 residue could make hydrogen bonding with these ligands (Figure 7). Six amino acid residues interacted with trans-2-hexen-1-ol and four residues with 2-heptanol, and Lys-55 and Pro-45 residues made hydrogen bonding with these two ligands, respectively. Remaining five ligands also interacted with various amino acid residues as shown in Table 2. Docking results showed strong binding of ligands in the core of MsepCSP5 (Figure 8).
Behavioral Response of M. separata to the Compounds Exhibiting Strong Binding Affinities with MsepCSP5
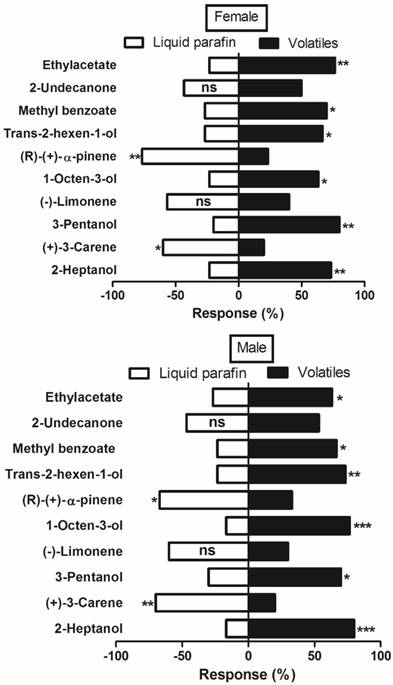
Olfactometer (Y-tube) bioassay was used to further elucidate binding mechanism of MsepCSP5 with ligands. A total of 10 volatiles belonging to different chemical groups; alcohol, ester and benzoate, ketones, and terpenes were tested for investigating M. separata male and female behavior response. The tested plant volatiles were chosen based on the strong binding affinity (Ki <10µM) with protein in the binding assays. The trends and responses of both sexes were similar to the tested volatiles (Figure 9). Male and female moth significantly elicited attraction to all tested alcoholic compounds. When males of M. separata were presented with a choice of trans-2-hexen-1-ol, 2-heptanol, 3-pentanol, and 1-octene-3-ol, the 2-heptanol elicited the highest attraction (80%, P=0.0004). However, females showed significantly (80%, P=0.001) greater attraction to 3-pentanol when compared to the three other alcoholic volatiles. Among the ester and benzoate volatiles, M. separata showed significant preference for the methyl benzoate (67% male, 70% female) and ethyl acetate (63% male, 77% female). In contrast, M. separata did not show attraction to the tested terpenes [(R)-(+)-α-pinene, (-)-limonene and (+)-3-carene]. 70% of males and 60% of females elicited significant preference for the control over (+)-3-carene. In case of (R)-(+)-α-pinene application, 67% of males and 77% of females exhibited significant preference for the control. Nonetheless, both male and female showed no obvious preference for the ketone volatile (2-undecanone).
RNAi-Based Silencing and Post-RNAi Behavior of M. separata
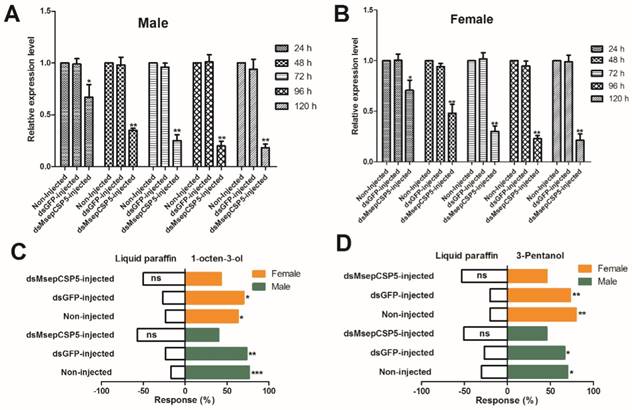
In order to test the silencing effects on target gene expression, we injected dsMsepCSP5 and dsGFP into M. separata pupae. Samples of male and female adults were collected at 24, 48, 72, 96 and 120 h of eclosion, and total RNAs were extracted for RT-qPCR analysis. MsepCSP5 gene expression in an individual sample was normalized with the M. separata β-actin gene. A down-regulation of MsepCSP5 expression was observed in dsRNA-treated insects, and this down-regulation was time-dependent (Figure 10A and 10B). A similar trend of reduction in the relative expression level was observed in both sexes with slightly higher reduction in male adults. In general, the mRNA abundance of MsepCSP5 was the highest at 24 h of eclosion and it decreased over time and reached the lowest level at 120 h of eclosion. In case of male, the relative expression level of MsepCSP5 decreased by 33% at day 1 and 82% at day 5 compared with the control (non-injected) and dsGFP. However, the relative expression level in the adult female was reduced by 29% and 79% at days 1 and 5, respectively when compared to the control and dsGFP.
In the olfactometer bioassay, the silencing of MsepCSP5 also resulted in behavioral changes of M. separata in response to selected volatiles (Figure 10C and 10D). Two compounds (1-octen-3-ol and 3-pentanol) that strongly attract M. separata before RNAi were selected. Moths treated with dsMsepCSP5 showed non-significant preference to tested volatiles.
Behavioral response of M. separata (male and female) to various volatiles by using Olfactometer (Y-tube) bioassay. Chi-square (χ2) test was employed to identify significant differences for the insects being attracted by volatiles. Number of stars represent p values at significant level; *** p≤0.001, **p≤0.01 and *p≤0.05.
Discussion
In insects, the olfactory system is important for odor detection, reproduction and survival [41]. CSPs are considered as carriers of odorants in insect chemoreception [2]. They are typically found in various insect tissues and perform divergent functions [42]. Here, we cloned MsepCSP5 from M. separata which shares high sequence similarity with other CSPs and possesses four conserved cysteine connected by two disulfide bonds between neighbor residues showing the typical characteristics of CSPs [43]. The similarity of MsepCSP5 with other CSPs supports the generally accepted concept that CSPs are conserved in nature [44]. A phylogenetic analysis displayed that MsepCSP5 originated from lepidopteran insects. Similar phylogenetic results of CSPs from diverse insect orders revealed that CSPs from the same species have close origin, suggesting that the variation in CSPs occurred within an insect order and curtailed from duplications within the order [45].
Expression patterns of chemosensory proteins in insect can provide clues to gene function which can improve the understanding of olfactory systems. In the present study, MsepCSP5 was widely distributed not only in developmental stages, but also in several body parts of M. separata. The relative expression of MsepCSP5 was higher in adults than larvae and pupae. It is widely held view that CSP genes can be expressed in chemosensory tissues and non-chemosensory tissues, indicating that CSPs might have diverse physiological functions in insects [46-48], such as embryo development in A. mellifera [49], reproduction in S. exigua [50], and limb regeneration in P. americana [24]. Results of tissues analysis displayed that MsepCSP5 was mainly expressed in antennae compared to others tissues, suggesting a major role in chemodetection. MsepCSP5 in antennae also showed female-biased expression patterns, 3.87-folds higher than male, conjecturing that it might play an important role in host recognition. In addition, MsepCSP5 with expression in legs and wings suggested to some extent that it was perhaps functionally associated with contact chemoreception and gustatory function [21, 51, 52].
The functions of MsepCSP5 were explored further by fluorescence binding assays with 35 volatiles mostly released by rice plants [53-55]. The results demonstrated some common and different binding pattern, and about 74% of tested ligands displayed strong biding with MsepCSP5 at pH 5.0. Most terpene compounds, except farnesene and 4-lsopropyltoluene, showed high binding to MsepCSP5. Similar results of ligand binding were also documented by He et al. [52] and Ming et al. [56]. The strong binding of MsepCSP5 with terpene compounds suggests the olfactory role of MsepCSP5 in binding and transporting the plant volatiles. MsepCSP5 displayed high binding to dodecyl aldehyde and hexanal, while showed moderate binding to trans-2-hexenal. Prior studies have noted the strong binding of alcoholic compounds, such as the binding of HoblCSP2 and LstiGOBP2 to cis-3-hexen-1-ol [39, 57-60]. The binding of MsepCSP5 with aldehydes indicates its plausible role in chemoreception. Tested ketone volatiles were highly bound to MsepCSP5 and this outcome is similar to Waris et al. [61] who documented high binding of NlugCSP8 with 2-tridecanone. The strong binding of MsepCSP5 with various compounds suggests that it is likely involved in chemoreception of M. separata.
Effects of injected dsRNA on MsepCSP5. (A and B) Analysis of MsepCSP5 mRNA expression level in treated female and male M. separata after 24, 48, 72, 96 and 120 h post eclosion. “Non-injected” represents the insect without dsRNA injection; “dsGFP” represents the insect with green fluorescent protein injection; “dsMsepCSP5” represents the insect with injection of dsMsepCSP5. Asterisk on bars denotes significant differences at p≤0.01. Error bars indicate standard errors (n=3) (C and D) Post-RNAi behavior response of M. separata (female and male) towards 1-octen-3-ol and 3-pentanol. Number of stars represent p values at significant level; *** p≤0.001, **p≤0.01 and *p≤0.05.
To get further view of ligand binding mechanism, structural modeling and ligand's docking were achieved. 3D model displayed that MsepCSP5 has distinctive structure similar to other CSPs, consisting of six alpha-helices and four conserved cysteine forming two disulphide bridges which impose the helices organization [48]. This six α-helices formed binding pocket providing binding sites for hydrophobic ligands [62, 63]. Docking results demonstrated that several amino acid residues of MsepCSP5 could make strong hydrophobic interactions with ligands, most likely because the binding pocket of MsepCSP5 contains hydrophobic residues, which is consistent with the previous studies [64]. Zhang et al. [48] revealed that Ile-45 residue is essential for the binding of rhodojaponin-III with CSPSlit. Furthermore, Thr-57, Pro-45 and Lys-55 residues of MsepCSP5 showed hydrogen bonding with ligands indicating that some residues are vital for interaction of MsepCSP5. It has been established that hydrogen bonding attributes in the recognition of ligands [7]. Hydrogen bonds form network to hold the ligands in the central cavity. Overall results of molecular docking showed that numerous amino acid residues with specific recognition were involved in ligand binding to make a complex system.
It is stated that plant volatiles play key roles in regulating the behavior of phytophagous insects in terms of seeking for food, mate and oviposition [57]. Diverse response of insects to plant volatiles infers an ecological need for understanding olfactory system and evaluation of host plants. To further corroborate the results of ligand's binding, we performed behavioral bioassays of M. separata using the volatiles displaying high binding affinities to MsepCSP5 in fluorescence binding assay. In the behavioral assay, terpene volatiles [R)-(+)-α-pinene, (-)-limonene and (+)-3-carene] with high bindings to MsepCSP5 showed repellency to both sexes of M. separata, suggesting that terpenes are important parts of plant defense system. Terpene volatiles are demonstrated to be generally emitted by plants after herbivore attacking to directly repel herbivores by reducing their feeding and egg-laying activities [57, 65]. The responses of both sexes of M. separata were significantly attractive towards alcoholic compounds. This strong attraction of moths to alcoholic volatiles is similar to previous studies [39, 57-60]. Lihuang et al. [59] tested various alcoholic compounds and observed that female and male moths elicited a similar response to alcoholic volatiles, which are in line with our results. These results indicated that alcoholic compounds might play important roles in host seeking. The ester and benzoate compounds (ethyl acetate and methyl benzoate) also elicited strong positive responses to both sexes. Similar responses to these compounds were observed in the previous studies [66]. Understanding the insect olfactory response to plant volatiles can provide tactics for insect pest control by identification of semiochemicals attracting or repelling specific insect [57].
As discussed earlier, it is plausible that MsepCSP5 is involved in the chemoreception of M. separata. Therefore, RNAi experiments were carried out, and MsepCSP5 was down-regulated and its expression was hampered over time. Two volatiles, 1-octen-3-ol and 3-pentanol, were selected for post-RNAi behavioral bioassay based on their strong attraction for M. separata prior to RNAi. Y-tube bioassays for ds-MsepCSP5 injected adults displayed non-significant preference for 1-octen-3-ol and 3-pentanol volatiles indicating successful results for selected volatiles in post-RNAi behavioral bioassay. Recent studies also revealed that dsRNA-treated M. separata, D. helophoroides and A. bambawalei displayed non-significant responses to volatiles [16, 67, 68]. Based on our results, it could be concluded that MsepCSP5 is the imperative recognition protein for 1-octen-3-ol and 3-pentanol.
RNAi can also induce off-target effects in insects [69]. Therefore, various doses of dsRNA should be used in the experiments. Moreover, dose-response based analysis may assist to devise efficient dsRNA application recommendations with minimal risk of off-target effects. Using a control dsRNA (such as GFP) to evaluate the efficient results of target gene is also helpful to avoid off-target effects of RNAi [70].
Conclusion
In conclusion, MsepCSP5 might have a potential olfactory role in adults' perception, binding and transportation of chemical cues. Ligand binding specificity of MsepCSP5 as confirmed by 3D structural modeling and molecular docking demonstrated that several amino acid residues of MsepCSP5 could make strong hydrophobic interactions with ligands. Furthermore, the reduction in MsepCSP5 expression through RNAi displayed non-significant preference of M. separata to representative attractants. In short, the results propose that MsepCSP5 is possible to be involved in chemoreception of M. separata.
Supplementary Material
Supplementary figure S1 The Ramachandran plot of MsepCSP5 and table S1 Detail of the primers.
Acknowledgements
The present research work was financially aided by the National Key Research and Development Program (grant number: 2016YFD0200807), the Public Welfare Project from Ministry of Agriculture (grant number: 201403031) and Special Technical Innovation of Hubei Province (grant number: 2017ABA146).
Author Contributions
Aneela Younas and Man-Qun Wang conceived, designed and performed the experiments; Aneela Younas, Muhammad Irfan Waris and Muhammad Tahir ul Qamar analyzed the data; Xiang-Xian Chang contributed materials; Aneela Younas, Man-Qun Wang and Muhammad Shaaban wrote the paper; Hazem Abdelnabby revised the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Brito NF, Moreira MF, Melo ACA. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51-65
2. Pelosi P, Iovinella I, Zhu J, Wang G, Dani FR. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol Rev. 2018;93:184-200
3. Carraher C, Dalziel J, Jordan MD, Christie DL, Newcomb RD, Kralicek AV. Towards an understanding of the structural basis for insect olfaction by odorant receptors. Insect Biochem Molec. 2015;66:31-40
4. Wang Z, Yang P, Chen D, Jiang F, Li Y, Wang X. et al. Identification and functional analysis of olfactory receptor family reveal unusual characteristics of the olfactory system in the migratory locust. Cell Molec Life Sci. 2015;72:4429-43
5. Antony B, Soffan A, Jakše J, Abdelazim MM, Aldosari SA, Aldawood AS. et al. Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. Bmc Genomics. 2016;17:1-22
6. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
7. Sánchezgracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208-16
8. Zheng Y, Wang SN, Peng Y, Lu ZY, Shan S, Yang YQ. et al. Functional characterization of a Niemann-Pick type C2 protein in the parasitoid wasp microplitis mediator. Insect Sci. 2017;21:1-13
9. Gu SH, Sun L, Yang RN, Wu KM, Guo YY, Li XC. et al. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. Plos One. 2014;9:e103420
10. Gadenne C, Barrozo RB, Anton S. Plasticity in insect olfaction: to smell or not to smell? Annu Rev Entomol. 2016;61:317-33
11. Ishida Y, Tsuchiya W, Fujii T, Fujimoto Z, Miyazawa M, Ishibashi J. et al. Niemann-Pick type C2 protein mediating chemical communication in the worker ant. Proc Nat Acad Sci. 2014;111:3847-52
12. Zheng Y, Wang SN, Peng Y, Lu ZY, Shan S, Yang YQ. et al. Functional characterization of a Niemann–Pick type C2 protein in the parasitoid wasp Microplitis mediator. Insect Science. 2017 DOI: 10.1111/1744-7917.12473
13. Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol B. 1995;111:503-514
14. He P, Li ZQ, Zhang YF, Chen L, Wang J, Xu L. et al. Identification of odorant-binding and chemosensory protein genes and the ligand affinity of two of the encoded proteins suggest a complex olfactory perception system in Periplaneta americana. Insect Molec Biol. 2017;26:687-701
15. Liu G, Ma H, Xie H, Xuan N, Guo X, Fan Z. et al. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: role of CSP in insect defense. Plos One. 2016;11:e0154706
16. Yang RN, Li DZ, Yu G, Yi SC, Zhang Y, Kong DX. et al. Structural transformation detection contributes to screening of behaviorally active compounds: dynamic binding process analysis of DhelOBP21 from Dastarcus helophoroides. J Chem Ecol. 2017;43:1033-45
17. Jin X, Zhang SG, Zhang L. Expression of odorant-binding and chemosensory proteins and spatial map of chemosensilla on labial palps of Locusta migratoria (Orthoptera: Acrididae). Arthropod Struct Dev. 2006;35:47-56
18. Bin SY, Qu MQ, Li KM, Peng ZQ, Wu ZZ, Lin JT. Antennal and abdominal transcriptomes reveal chemosensory gene families in the coconut hispine beetle, Brontispa longissima. Sci Rep. 2017;7:2809-28122
19. Benton R, Vannice K, Gomez-Diaz C, Vosshall L. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149-55
20. Celorio-Mancera MDLP, Sundmalm SM, Vogel H, Rutishauser D, Ytterberg AJ, Zubarev RA. et al. Chemosensory proteins, major salivary factors in caterpillar mandibular glands. Insect Biochem Molec. 2012;42:796-805
21. Hua JF, Zhang S, Cui JJ, Wang DJ, Wang CY, Luo JY. et al. Functional characterizations of one odorant binding protein and three chemosensory proteins from Apolygus lucorum (Meyer-Dur) (Hemiptera: Miridae) legs. J Insect Physiol. 2013;59:690-6
22. Liu R, He X, Lehane S, Lehane M, Hertzfowler C, Berriman M. et al. Expression of chemosensory proteins in the tsetse fly Glossina morsitans is related to female host-seeking behaviour. Insect Molec Biol. 2012;21:41-8
23. Liu Z, Smagghe G, Lei Z, Wang JJ. Identification of male- and female-specific olfaction genes in antennae of the oriental fruit fly (Bactrocera dorsalis). Plos One. 2016;11:e0147783
24. Nomura A, Kawasaki K, Kubo T, Natori S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int J Dev Biol. 1992;36:391-8
25. He Y, Feng B, Guo Q, Du Y. Age influences the olfactory profiles of the migratory oriental armyworm Mythimna separata at the molecular level. Bmc Genomics. 2017;18:1-19
26. Liu Z, Wang X, Lei C, Zhu F. Sensory genes identification with head transcriptome of the migratory armyworm, Mythimna separata. Sci Rep. 2017;7:1-14
27. Chang XQ, Zhang S, Lv L, Wang MQ. Insight into the ultrastructure of antennal sensilla of Mythimna separata (Lepidoptera: Noctuidae). J Insect Sci. 2015;15:124-132 doi: 10.1093/jisesa/iev103
28. Wang X, Liu M, Wu C. Electroantennogram of oriental armyworm Mythimna separata walker exposed to sex pheromone. Acta Entomologica Sinica. 1998;41:1-7
29. Jung JK, Seo BY, Cho JR, Kim Y. Monitoring of Mythimna separata adults by using a remote-sensing sex pheromone trap. Korean J Appl Entomol. 2013;52:341-8
30. Chang XQ, Nie XP, Zhang Z, Zeng FF, Lv L, Zhang S. et al. De novo analysis of the oriental armyworm Mythimna separata antennal transcriptome and expression patterns of odorant-binding proteins. Comp Biochem Physiol D. 2017;22:120-9
31. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-82
32. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis software. Mol Biol Evo. 2013;30:2725-9
33. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402-8
34. Zheng ZC, Li DZ, Zhou A, Yi SC, Liu H, Wang MQ. Predicted structure of a Minus-C OBP from Batocera horsfieldi (Hope) suggests an intermediate structure in evolution of OBPs. Sci Rep. 2016;6:33981-92
35. Cao B, Bao W, Wuriyanghan H. Silencing of target chitinase genes via oral delivery of dsRNA caused lethal phenotypic effects in Mythimna separata (Lepidoptera: Noctuidae). Appl Biochem Biotech. 2017;181:860-9
36. Tomaselli S, Crescenzi O, Sanfelice D, Ab E, Wechselberger R, Sergio A. et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochem. 2006;45:10606-13
37. Vilar S, Cozza GS. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8:1555-72
38. Qamar MTU, Kiran S, Ashfaq UA, Javed MR, Anwar F, Ali MA. et al. Discovery of novel dengue NS2B/NS3 protease inhibitors using pharmacophore modeling and molecular docking based virtual screening of the zinc database. Int J Pharmacol. 2016;12:621-32
39. Cao Q-J, Yu J, Ran Y-L, Chi D-F. Effects of plant volatiles on electrophysiological and behavioral responses of Cryptorrhynchus lapathi. Entomol Exp Appl. 2015;156:105-16
40. Li ZQ, Zhang S, Luo JY, Cui JJ, Ma Y, Dong SL. Two minus-C odorant binding proteins from Helicoverpa armigera display higher ligand binding affinity at acidic pH than neutral pH. J Insect Physiol. 2013;59:263-72
41. Chang H, Ai D, Zhang J, Dong S, Liu Y, Wang G. Candidate odorant binding proteins and chemosensory proteins in the larval chemosensory tissues of two closely related noctuidae moths, Helicoverpa armigera and H. assulta. Plos One. 2017;12:e0179243
42. Xin Y, Qi J, Zhou X, Mei YH, Guo HZ. Differential expression of chemosensory-protein genes in midguts in response to diet of Spodoptera litura. Sci Rep. 2017;7:796-807 doi: 10.1038/s41598-017-00403-5
43. Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Molec Life Sci. 2006;63:1658-76
44. Wanner KW, Willis LG, Theilmann DA, Isman MB, Feng Q, Plettner E. Analysis of the insect os-d-like gene family. J Chem Ecol. 2004;30:889-911
45. Vieira FG, Julio R. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genom Biol Evo. 2011;3:476-90
46. Gu SH, Wang SY, Guo YY. Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition. Plos One. 2012;7:e42871
47. Guo W, Wang X, Ma Z, Xue L, Han J, Yu D. et al. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. Plos Genet. 2011;7:e1001291
48. Zhang Y, Dong X, Liu J, Hu M, Zhong G, Geng P. et al. Molecular cloning, expression and molecular modeling of chemosensory protein from Spodoptera litura and its binding properties with Rhodojaponin III. Plos One. 2012;7:e47611
49. Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evo. 2007;217:189-96
50. Gong L, Luo Q, Rizwanulhaq M, Hu MY. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. Bullet Entomol Res. 2012;102:600-8
51. Liu X, Luo Q, Zhong G, Rizwan-Ul-Haq M, Hu M. Molecular characterization and expression pattern of four chemosensory proteins from diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J Biochem. 2010;148:189-95
52. He P, Zhang J, Liu NY, Zhang YN, Yang K, Dong SL. Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stål. Plos One. 2011;6:e28921
53. Lou YG, Du MH, Turlings TC, Cheng JA, Shan WF. Exogenous application of jasmonic acid induces volatile emissions in rice and enhances parasitism of Nilaparvata lugens eggs by the parasitoid Anagrus nilaparvatae. J Chem Ecol. 2005;31:1985-2002
54. Yang K, He P, Dong SL. Different expression profiles suggest functional differentiation among chemosensory proteins in Nilaparvata lugens (Hemiptera: Delphacidae). J Insect Sci. 2014:14
55. Fujii T, Hori M, Matsuda K. Attractants for rice leaf bug, Trigonotylus caelestialium (Kirkaldy), are emitted from flowering rice panicles. J Chem Ecol. 2010;36:999-1005
56. Ming H, Peng H. Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the whitebacked planthopper, Sogatella furcifera. Comp Biochem Physiol B. 2014;174:1-8
57. Das A, Lee SH, Hyun TK, Kim SW, Kim JY. Plant volatiles as method of communication. Plant Biotech Rep. 2013;7:9-26
58. Light DM, Jang EB. Electroantennogram responses of the oriental fruit fly, Dacus dorsalis, to a spectrum of alcohol and aldehyde plant volatiles. Entomol Exp Appl. 1987;45:55-64
59. Lihuang K, Zhang Z, Kim K, Huang Q, Lei C. Antennal and behavioral responses of Mythimna separata (Walker) to three plant volatiles. Environ Sci Poll Res. 2017;24:24953-64
60. Ramachandran R, Khan ZR, Caballero P, Juliano BO. Olfactory sensitivity of two sympatric species of rice leaf folders (Lepidoptera: Pyralidae) to plant volatiles. J Chem Ecol. 1990;16:2647-52
61. Waris MI, Younas A, ul Qamar MT, Hao L, Ameen A, Ali S. et al. Silencing of chemosensory protein gene NlugCSP8 by RNAi induces declining behavioral responses of Nilaparvata lugens. Front Physiol. 2018;9:1-17
62. Ban L, Scaloni A, Brandazza A, Angeli S, Zhang L, Yan Y. et al. Chemosensory proteins of Locusta migratoria. Insect Molec Biol. 2003;12:125-34
63. Jansen S, Chmelík J, Zídek L, Padrta P, Novák P, Zdráhal Z. et al. Structure of Bombyx mori chemosensory protein 1 in solution. Arch Insect Biochem Physiol. 2007;66:135-45
64. Tian Z, Liu J, Zhang Y. Structural insights into Cydia pomonella pheromone binding protein 2 mediated prediction of potentially active semiochemicals. Sci Rep. 2016;6:22336-9
65. Schilmiller AL, Last RL, Pichersky E. Harnessing plant trichome biochemistry for the production of useful compounds. Plant J Cell Molec Biol. 2008;54:702-9
66. Obata T, Koh HS, Kim M, Fukami H. Constituents of plant hopper attractant in rice plant. Appl Entomol Zool. 1983;18:161-9
67. Li QL, Yi SC, Li DZ, Nie XP, Li SQ, Wang MQ. et al. Optimization of reverse chemical ecology method: false positive binding of Aenasius bambawalei odorant binding protein 1 caused by uncertain binding mechanism. Insect Molec Biol. 2018
68. Younas A, Waris MI, Tahir ul Qamar M, Shaaban M, Prager SM, Wang M-Q. Functional analysis of the chemosensory protein MsepCSP8 from the oriental armyworm Mythimna separata. Front Physiol. 2018;9:1-15
69. Mamta B, Rajam MV. RNAi technology: a new platform for crop pest control. Physiol Molec Biol Plants. 2017;23:487-501
70. Dowling D, Pauli T, Donath A, Meusemann K, Podsiadlowski L, Petersen M. et al. Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol Evol. 2016;8:3784-93
Author contact
![]() Corresponding author: mqwanghzau.edu.cn (Man-Qun Wang)
Corresponding author: mqwanghzau.edu.cn (Man-Qun Wang)

 Global reach, higher impact
Global reach, higher impact