Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(1):1-11. doi:10.7150/ijbs.27796 This issue Cite
Review
Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers
Faculty of Health Sciences, University of Macau, Macau SAR, China
Received 2018-9-11; Accepted 2018-10-15; Published 2019-1-6
Abstract
Cancer related exosomes are nano-size membrane vesicles that play important roles in tumor microenvironment. Emerging evidence indicates that exosomes can load unique cargoes, including proteins and nucleic acids that reflect the condition of tumor. Therefore, exosomes are being used as diagnostic and prognostic biomarkers for various cancers. In this review, we describe the current progresses of cancer related exosomes, including their biogenesis, molecular contents, biological functions, sources where they are derived from, and methods for their detection. We will also discuss the current exosomal biomarkers and the utilization of them for early diagnosis and prognostics in cancer.
Keywords: exosomes, cancer, biomarkers, therapy, diagnosis, prognosis
Introduction
Cancer has been the major cause of death in the world. The survival rate is poor because of lacking early diagnosis and personalized treatment. Indeed, detection of tumors at early stages has been recognizing as a vital component of cancer-control for many years. Any strategies to achieve it will depend on the correct cancer detection markers, which exist in patients' tumor tissue and serum at various forms such as DNA, miRNA and proteins [1].
Exosomes, which were first identified by Johnstone in 1987 [2], are composed of a lipid bilayer, like cystic structure, ranging from 40 to 150 nm in diameter. Exosomes could carry materials into cells and cart stuff away, including nucleic acids, proteins and enzymes, suggesting useful, even life preserving functions. Consequently, tumor molecular and genetic cargoes, at least in part, are abundant in the body fluids or other associated tissues which could be used as cancer diagnostic and prognostic biomarkers. In this review, we highlight and discuss current progresses on cancer related exosomes and the application of exosomal markers for cancer diagnosis and prognosis.
Biogenesis and molecular content of exosomes in cancer
The biogenesis of exosomes starts when the cell membrane internalizes to form an early endosome. During this process, cytoplasmic content is constantly banding with inward budding of endosomal membranes to form exosomes. The endosomal sorting complex required for transport (ESCRT) machinery is required during this process. Members of the ESCRT family, tumor susceptibility gene 101 (TSG101) and ALG-2-interacting Protein X (Alix) proteins involve in it and are regarded as the core components of exosome lately in the literature [3]. Besides, exosomes can also form in ESCRT independent way which contains flotillin-2, CD63, CD81, ceramide and cholesterol molecules. After then, the endosome which is named multi-vesicular body (MVB), fuses with cellular membrane and releases exosome. The Rab family of small GTPase proteins, especially Rab27a and Rab27b, controls this step [4, 5]. But mechanism that drives exosome formation and secretion is largely unknown because of different cell types and their status.
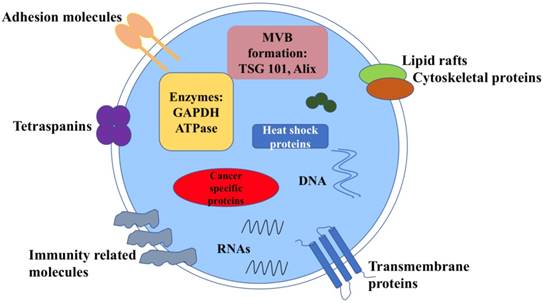
Exosomes carry various molecular contents which vary depend on the origin and their status [6] (Figure 1). For example, exosomes contains the characteristic proteins which belong to its biogenesis process, including TSG101, Alix, CD63, CD81 and Rab family proteins. All these molecules are often used as markers to confirm this vesicle. Exosomes also carry abundantly mRNA, DNA, microRNA(miRNA), long noncoding RNA (LncRNA) and other nucleic acid species inside. Besides, lipids are also an important part of the exosomes, such as cholesterol, phospholipids, glycerophospholipids and sphingolipids. They form the bilayer membrane structure and maintain its stable state [7]. Notably, in tumors, the molecular signature of tumor cells is enriched in exosomes that transfer between tumor cells and normal cells. Meanwhile, tumor cells may release more exosomes into microenvironment than normal cells, thus the circulating exosomes level is abundant.
Structure and contents of cancer derived exosomes. Exosomes contain lipid bilayer membrane structures, which carry typical transmembrane proteins and receptors, adhesion molecules, the lipid raft associated protein, immune regulator molecules and tetraspanins. Within the exosomal lumen, several proteins and nucleic acid can be found, including a vast array of different molecules depending on different types of cancer cells.
Functions of cancer related exosomes
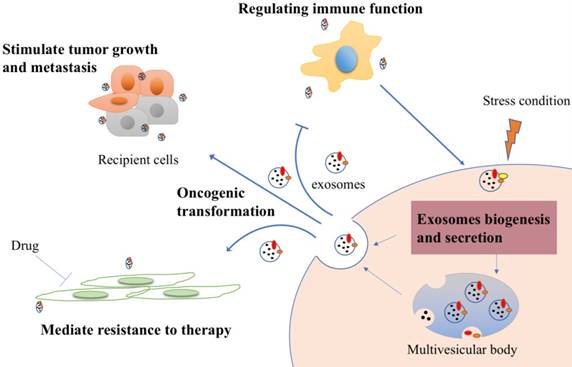
Exosome is a multifaceted regulator of cancer development. In multiple cancers, they usually harbor molecules derived from cancer, and are capable of changing the tumor microenvironment to effect neighbor cells or cells at specific distant sites. Therefore, they would be used as a tool to bridge normal cells to cancer cells and may facilitate the diagnosis of cancer [8]. To image the process of cell-to-cell communication, Suetsugu et al (2013) established cell and nude mouse models using green fluorescent protein (GFP)-tagged CD63. Labeled exosomes transferred from cancer cells to other cells in culture system. In vivo, GFP-exosomes are secreted into the tumor-surrounding tissue and circulate from primary tumors to metastatic niche [9].
First of all, exosomes play key roles in tumor growth and metastasis. Growth promoting gene could transported by exosomes and promote the proliferation of metastatic cancer cells. For example, epidermal growth factor receptor (EGFR)-containing exosomes favor this pattern to form liver-specific metastasis [10]. Scientists also found that exosomal miRNAs are capable of mediating silencing of downstream genes, which stimulate tumorigeneses in nontumorigenic epithelial cells [11]. Next, exosomes induce neoplastic proliferation or metastasis by evading cancer suppressors. Zhang L et al (2015) showed exosomes mediate tumor-suppressor phosphatase and tensin homolog (PTEN) transformation leads to PTEN loss in brain and enhance the outgrowth of brain metastasis [12]. Similarly, Putz et al (2012) reported PTEN protein could be directly brought out of cells and transferred to recipient cells as well as restore their function there [13]. It is also shown that exosomes are able to suppress glucose uptake by non-tumor cells to satisfy their own needs via secreting miRNA-122 high expressed-exosomes [14]. For another thing, exosomes are able to stimulate angiogenic activities [15]. In the study of Nazarenko et al (2010), when endothelial cells uptake tumor cells shuttled exosomal tetraspanin Tspan8, an important angiogenesis moderator, several angiogenesis-related genes of vascular endothelial growth factor (VEGF)-independent pathway would increase along with endothelial cell proliferation and migration [16]. Elucidation of the exosome uptake mechanisms is essential to understand the process of metastasis. A recent study demonstrated that exosomes derived from different tumors have their own “bias”. The theory is that exosomes from lung, liver and brain-tropic tumor cells are uptaken by resident cells at the pre-metastasis niche due to their specific integrin [17].
Second, exosomes generated by cancer cells can stimulate immune response. However, they have a dual function in this aspect. On the one hand, exosomes from nasopharyngeal carcinoma can facilitate Treg recruitment, increase their suppressive function, and induce CD4 T-cells conversion, leading to immune system evasion from host immune surveillance [18]. Using fluorescently labeled exosomes, Wen et al (2016) found that exosomes suppress immune response via directly inhibiting T-cell growth and decreasing Natural killer (NK) cell cytotoxicity [19]. Further, Mizyazaki et al (2018) reported that overexpressing estrogen receptor-binding fragment-associated antigen 9 (EBAG9) in exosomes and transferring them to T-cells can activate immune escape and cell migration more potently [20]. On the other hand, exposing tumor cells to stress may improve the antitumor immunity. After being heat stressed, exosomes from carcinoembryonic antigen (CEA)-positive tumor cells have higher immunogenicity, suggesting they could be used as a new effective vaccine for cancer immunotherapy [21].
In recent years, mounting evidence has suggested that exosomes play an important role of mediating resistance to therapy. Indeed, exosomes can transfer resistant phenotype from drug resistant cells to drug sensitive cells, through transferring of drug-efflux pumps and inclusive RNAs [22]. To detect how exosomal miRNAs contribute to the development of drug resistance, Fabbri et al (2015) designed a co-culture system using neuroblastoma cells and human monocytes. They found that miR-155 raised about four folds in this tumor cells since the exosomal transformation of monocytes, accompanied by increased cisplatin resistance via depressing its target shelterin component TERF1 [23]. Similarly, Qu et al (2016) showed that exosomes could also confer the sunitinib resistant phenotype to renal cancer cells. Activated exosomal LncARSR has been proved as the inducer of this process [24].
This cross-talk is also reflected between cancer stem cells and the normal cells, including fibroblast, endothelial cell and mesenchymal cell. Cancer stem cells (CSCs) are considered as cornerstone, which can contribute to tumor progression, therapy resistance and metastasis - and their associated exosomes - have been implicated in various cancer. It is now thought that exosomes may up-regulated mRNA, protein expression of related genes in recipient cells [25-27]. For example, pluripotent stem cell-derived mesenchymal stem cells derived exosomes enhance the proliferation, migration capacities of endothelial cells by activating phosphatidylinositol 3-kinases (PI3K)/ AKT pathway to exert a preventative effect [28]. As the research moves along, more puzzles will be unraveled. Some functions of exosomes in cancer are summarized in Figure 2.
Cancer derived exosomes-mediated cellular process signaling. Tumor exosomes carry proteins and nucleic acids and are released into tumor microenvironment. They could be uptaken by recipient cells to stimulate tumor growth and metastasis. After drug treatment, exosomes, if they derived from drug resistance cells, can also transmit the resistant feature to recipient cells to mediate resistance. Furthermore, exosomes can stimulate immune response to inhibit immune cells response in general, while increase immunogenicity to improve anti-tumor effect under some specific stress conditions.
Sources of cancer related exosomes
Body fluids derived exosomes
Oncologists first separated cancer cell derived exosomes from mice and patient's blood, which are most common sources for exosomes and reflect the function as pro- or anti-tumor effectors, including immune response, tumorigenesis, cell migration and invasion [29]. Platelet-derived extracellular vesicles (P-EVs) is the most abundant specie in human blood, accounting for two-thirds of peripheral blood extracellular vesicles [30]. Many studies highlight that P-EVs participate in several intracellular processes, including homeostasis, thrombotic and cancer progression [31, 32]. Based on the study of Liang et al (2015), miR-223 in the platelet-secreted extracellular vesicles from patients with hematogenous metastatic lung cancer is more abundant than healthy subjects [33]. Janowska et al (2005) also showed that P-EVs are capable to promote proliferation and invasion in lung carcinoma cell lines by activing mitogen activated protein kinase (MAPK) pathway or stimulating matrix metalloproteinases (MMPs) [34]. In addition, in response to the stress of inflammation, more P-EVs could be released [35]. Altogether, P-EVs might be useful for the development of novel cancer biomarkers and therapy strategies in the near future.
In the field of cancer research, however, two other sources of exosome have been discovered. For the urine exosomes, researchers demonstrated that exosomes could be detected from normal volunteers or patients with kidney disease [36]. For instance, after proteomic analyzing, 295 proteins were revealed, including multiple protein products of genes already known to be responsible for renal and systemic diseases, providing a potential tool to discovery of cancer in early stage [37]. In prostate cancer, McKiernan et al (2016) compared the efficiency of combining urine exosome gene expression assay with standard of care (SOC) or SOC alone. This novel assay plus SOC improve the accuracy and sensitivity of prostate cancer screening and diagnosis [38]. Beyond this, it is conceivable that urine exosomes could be used for low-cost screening for early detection of cancer and therapeutic monitoring. Nevertheless, because of glomerular filtration, only unique size (<100 nm) of exosomes could be detected [39, 40].
For the breast cancer patients, breast milk exosomes uniquely contribute to cancer detection. Qin et al (2016) detected the expression of six proteins linked to breast cancer in three milk samples (transitional, mature, and wean). The results showed that transforming growth factor β2 (TGFβ2) level increased in both wean and early involution, suggesting high TGFβ2-expressing breast milk exosomes have higher cancer risk [41]. Yu et al (2017) discovered the expression of miR-29b and miR-21 are high in milk-derived exosomes, which may activate the MAPK signaling and promote cell growth [42]. Besides, it was shown that human breast milk exosomes are connected with immune system. For example, exosomes, which are isolated from human colostrum and mature breast milk, may inhibit inflammatory factor production from allogeneic and autologous peripheral blood mononuclear cells as well as increase the number of microRNAs to participate in the process of immune regulation [43, 44]. Consistent with a prior research, about 67.82% well-characterized immune-related pre-miRNAs are detected in breast milk exosomes which could be transfer from mother's milk to the infant to develop immune system [44]. Hence, scientists begin to design the drug-loaded exosomes to improve chemotherapeutic drug efficacy and reduce toxicity. However, the discipline of cancer related exosomes in human breast milk is still not clear.
Tissue derived exosomes
Currently, the studying of this source of exosomes is still in the early stage. Gallart et al (2016) have successfully enriched the extracellular vesicle from tissues of central nervous system. They presented that more than 86% exosomal markers could be identified in brain exosomes [45]. Vella et al (2017) also reported this kind of exosomes, which are extracted from human brain tissue, maintain the same traits of brain homogenate by synthesizing proteomic and genomic profiles [46]. It suggests that exosomes released from tissue might act as a new effective resource. In Teng et al (2017) study, high level of tumor suppressor miRNAs, such as miR-193a, miR-18a and miR-155, were selectively packaged by exosomes and expelled from advanced colon cancer cells, thereby resulting in high accumulation in circulation [47]. Similarly, for human renal tissue, comparing with normal renal, carcinoma tissues highly express azurocidin to promote angiogenesis, but it was not related to tumor stage or grade [48]. However, regardless this deficiency, the analysis of such exosome derived cargoes still reveals the novel mechanism of tumorigenesis and is starting to be useful to monitor tumour procession as well as drug response. Furthermore, there are two further advantages. First, since they are directly isolated from tissue which effected by the complex tumor microenvironment, exosomes more accurately reflect the status of tumor or human body. For instance, although exosomes maintain cellular homeostasis by activating cytoplasmic DNA sensing response in vitro, liver tissue derived exosomes contribute more markedly in vivo [49]. Second, body fluid samples contain more contaminants, including serum abundant proteins, metabolite and even other derived exosomes. In contrast, they come directly from the origin and do not need to be transferred, thus allowing cancer cell exosomes avoid degradation and contamination. Meanwhile, the difference of post-mortem and storage time as well as the number of freeze-thaw cycles may challenge the quality and efficiency [46]. Nowadays, Mincheva and colleagues have established the protocol to isolate exosomes from tissue explants in 2016. They cut fresh tissue into small pieces and culture them for a short time. By centrifuging the collected culture medium, exosomes will be enriched [50]. But due to the different property of tissues, different culture medium or dispose method should be considered to maintain the integrity of exosomes and their cargoes.
Exosome based biomarkers in cancer
It appears that blood profiling from cancer patients has the value to capture tumor status. Indeed, since Gold demonstrated the role of glycoprotein antigen in detecting human colon cancer in 1965, lots of serum cancer biomarkers have been commonly used in practical clinical realities [51]. For example, carbohydrate antigen-and125 (CA-125) presents its potential for monitoring response to ovarian cancer diagnosis about 20 years ago. But it has low sensitivity, which is less than 50% for early-stage ovarian cancer and more than 20% false negatives value, and it also reveals high false positive value when the patients with benign gynecological diseases are tested [52]. Besides, there is a degradation reaction whose speed was uncontrollable in all kinds of serum tumor markers because they completely exposed to the blood. Therefore, although it is a good point to use a combination of more biomarkers to upgrade the overall sensitivity and specificity, the benefit is still far more away. Currently, to address this problem, circulating tumor cells (CTCs) have attracted interest. It carries the molecular and phenotypic information of tumors and can be easily obtained from blood. Nevertheless, they are extremely rare within an overwhelming background, which is 1 mL of whole blood contains about 5 million mononuclear cells, and only 1.43% of progressive breast cancer patients had more than 500 CTCs in 7.5mL blood [53-56]. Moreover, many issues regarding the mechanism of immune system or lymphatic spread remain unknown. But they are the crucial route for CTCs transfer, degradation, and even elimination [57]. In contrast, exosome contains the molecules that strongly reflect the parental property, and has the advantages that they are very abundant in blood, highly stable and could be analyzed in small volumes (100 μL) of frozen serum or plasma samples, which could be used as a new source of new biomarkers for personalized diagnosis and prognosis [58].
Exosomes carry multiple protein cargos, which have been explored from studies of human pancreatic cancer, lung cancer, colorectal cancer and melanoma (Table 1). For pancreatic cancer, Melo et al (2015) found glypican-1 GPC1+ exosomes show a higher proportion in pancreatic cancer patients (100%) than healthy people (average of 2.3%). Notably, they also found this marker revealed perfect sensitivity and specificity in early stage of pancreatic cancer [59]. Another signature in exosomes, macrophage migration inhibitory factor (MIF), was highly expressed in stage I patients with high liver metastatic potential [60]. In the case of treatment response of locally advanced pancreatic cancer, An et al. demonstrated that exosomal vimentin level was decreased after gemcitabine treatment, which means it can be used as a candidate for cancer prognosis [61]. Similarly to pancreatic cancer, EGFR is higher enriched in exosome from gastric cancer patients, thereby, making it a potential marker [10]. However, the abundance of exosomal specific proteins are very low, large volume serum or culture medium is required to isolate enough exosome to do the further studies neither proteomics or identification via western blot. Thus, exosome protein profile is still in infancy.
Genomic profile of the exosome, including miRNA, LncRNA, messenger RNA and mitochondrial RNA, appears to be a potential biomarker source to classify tumor types (Table 2). They can be amplified by PCR which makes up for the shortcoming of quantity, and therefore makes them as an excellent tool for cancer diagnose. For instance, Fang et al (2018) showed hepatoma cells exhibited high level of miR-103 and could deliver it into endothelial cells though exosomes to induce metastasis, suggesting secreted miR-103 has the capacity to be a predictive marker [62]. Shi et al reported exosomes-delivered miR-638 was obviously down-regulated in serum of hepatocellular cancer patients, with larger tumor size (> 5 cm) or at later TNM stage (III/IV) [63]. Furthermore, long noncoding RNAs, like lnc-sox2ot, lnc-h19 and LncRNA-ARSR, have been investigate in circulation exosomes and highly correlated with tumor stage and overall survival rate of patients [24, 64-68]. These findings demonstrate that it is possible to find exosomal biomarkers from circulation or other sources to evaluate cancer invasion and progression, with higher sensitivity and specificity.
Methods available for biomarker detection in cancer exosomes
To enhance specificity and availability of exosomal biomarkers, a variety of methods are developed. Over the past decade, mass-based proteomic analysis is used to identify protein species in exsomes. For instance, higher 27-Hydroxycholesterol level in estrogen receptor positive (ER+) breast cancer cell line (MCF-7) exosomes has been detected by liquid chromatography-mass spectrometry [69]. Furthermore, Hyun et al (2013) combined microscopy and mass spectrometry to map endogenous proteins in living cells. They successfully identified 495 proteins in mitochondrial matrix after labeling the peroxidase enzyme nearby the target proteins. This study outlines a new approach that may be valuable in the field of exosomes. However, because of the contaminant proteins, there are still some challenges to truly define the exosome proteomic map [70]. Researchers also developed tandem-mass-tag (TMT) quantitative proteomics approach and support vector machine cluster analysis. This approach highlights the protein abundance rather than protein presence. They thought the approach should be useful for identifying and confirming new protein candidates in exosome [71].
Exosomal protein markers in multiple cancers
| Tumor types | Body fluids | Enrichment method | Signature or pool in exosomes | Application | Ref. |
|---|---|---|---|---|---|
| Pancreatic cancer | Patients' blood | Ultracentrifugation | Glypican-1 | Diagnosis | [59] |
| Medium | Ultracentrifugation | CD151, ADAM1 and ADAM15 | Prognosis | [91] | |
| Patients' plasma | Ultracentrifugation | MIF | Prognosis and treatment | [60] | |
| Patients' serum | Ultracentrifugation | vimentin | Diagnosis and treatment | [61] | |
| Melanoma | Patients' plasma | Ultracentrifugation | TYRP2, VLA-4, HSP70, HSP90 and MET | Prognosis and treatment | [92] |
| Patients' serum | Ultracentrifugation | MIA and S100B | Prognosis and diagnosis | [93] | |
| Lung cancer | Urine | Ultracentrifugation | LRG1 | Diagnosis | [94] |
| Saliva | Ultracentrifugation | BPIFA1, CRNN, MUC5B, and IQGAP | Detection | [95] | |
| Medium | ExoQuick-TC™ kit | Vasorin | Treatment | [96] | |
| Patients' blood | Ultracentrifugation | LG3BP and PIGR | Diagnosis | [97] | |
| Glioblastoma | Medium | Ultracentrifugation | angiogenin, IL-6 and IL-8 | Detection and diagnosis | [98] |
| Gastric cancer | Medium | Ultracentrifugation | EGFR | Detection and diagnosis | [10] |
| Colorectal cancer | Patients' blood | Ultracentrifugation | CD147 | Detection and diagnosis | [99] |
| Patients' plasma | ExoCapTM kit | GPC1 | Diagnosis and treatment | [100] | |
| Ovarian cancer | Ascytic fluid | Ultracentrifugation | CD24 | Diagnosis | [101] |
| Ascitic Fluid | Ultracentrifugation | TGM2, U2AF1, U2AF2, and HNRHPU | Diagnosis | [102] | |
| Cholangiocarcinoma | Patients' blood | Ultracentrifugation | VNN1, CRP, FIBG, IGHA1, and A1AG1 | Diagnosis | [97] |
| Prostate cancer | Urine | Ultracentrifugation | PCA3 | Diagnosis and monitoring | [103] |
| Glioma | Cerebrospinal fluid | Ultracentrifugation | IL13QD | Detection and recurrence | [104] |
Exosomal nucleic acids markers in multiple cancers
| Tumor types | Source | Enrichment method | Signature or pool in exosomes | Application | Ref. |
|---|---|---|---|---|---|
| Pancreatic cancer | Patients serum | Ultracentrifugation | miR-1246, miR-4644, miR-3976 and miR-4306 | Diagnosis | [105] |
| Postoperative blood | LncRNA-Sox2ot | Prognosis | [64] | ||
| Patient plasma | Ultracentrifugation | miR-10b | Detection, prognosis | [106] | |
| Patients' blood | Ultracentrifugation | miR-10b, miR-21, miR-30c, miR-181a, miR-let7a | Detection | [107] | |
| Patients urine | Ultracentrifugation | PCA-3 mRNA | Diagnosis and monitoring | [103] | |
| Patients serum | Ultracentrifugation | KRAS DNA | Detection, prognosis | [108] | |
| Hepatocellular carcinoma | Serum | ExoQuick Kit | miR-18a, miR-221, miR-222, miR-224 | Diagnosis | [109] |
| Patients serum | miR-638 | Detection, prognosis and recurrence | [63] | ||
| Patients serum | Ultracentrifugation | miR-718 | Detection, prognosis and recurrence | [110] | |
| Medium | ExoQuick Kit | miR-122 | Potential treatment | [111] | |
| Medium and mice blood | Ultracentrifugation | miR-103 | Potential treatment | [62] | |
| Mice serum | Ultracentrifugation | CDYL circRNA | Diagnosis | [112] | |
| Medium | Ultracentrifugation | LncRNA-H19 | Treatment | [65] | |
| Renal cell carcinoma | Medium | Ultracentrifugation | LncRNA-ARSR | Potential treatment | [24] |
| Glioblastoma | Patients serum | Ultracentrifugation | EGFRvIII (mRNA) | Diagnosis and treatment | [98] |
| Patients serum | LncRNA-HOTAIR | Prognosis and diagnosis | [66] | ||
| Melanoma | Medium and mice plasma | Ultracentrifugation | BRAF(DNA) | Detection and treatment | [113] |
| Lung cancer | Medium and mice plasma | Ultracentrifugation | EGFR(DNA) | Detection and treatment | [113] |
| Patients serum | Ultracentrifugation | miR-1247-3p | Diagnosis | [114] | |
| Colorectal cancer | Patients serum | Ultracentrifugation | circ-KLDHC10 | Diagnosis | [112] |
| Patients serum | Ultracentrifugation | BRAF and KRAS(mRNA) | Detection | [115] | |
| Mice serum | Ultracentrifugation | miR-193a | Detection | [47] | |
| Patients serum | LncUEGC1 and LncUEGC2 | Diagnosis | [67] | ||
| Patients serum | Ultracentrifugation | miR-19a | Prognosis and recurrence | [116] | |
| Prostate cancer | Patients plasma | ExoQuick Kit | miR-1290 and miR-375 | Prognosis | [117] |
| Serum and urine | ExoMiR extraction kit | miR-141 and miR-375; miR-107 and miR-574-3p | Diagnosis | [118] | |
| Urine | EXOPRO Urine Clinical Sample Concentrator Kit | ERG, PCA3, and SPDEF (RNA) | Diagnosis | [38] | |
| Urine | Ultracentrifugation | miR-196a-5p and miR-501-3p | Diagnosis | [119] | |
| Ovarian cancer | Patients serum | modified magnetic activated cell sorting (MACS) procedure | miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214 | Diagnosis | [120] |
| Blood | Exosome Isolation Reagent | miR-21 | Diagnosis | [121] | |
| Urine | Ultracentrifugation | miR-30a-5p | Diagnosis and treatment | [122] | |
| Bladder cancer | Urine | miR-720/3007a, miR-205, miR-200c-3p and miR-29b-3p | Detection | [123] | |
| Gastric cancer | Patients serum | LncRNA HOTTIP | Diagnosis and prognosis | [68] |
In addition, exosomal enriched gene information is another potential biomarker source. Present technology such as RNA-sequencing and DNA sequencing [72] already enables analysis of genomic information of exosomes. Work carried out by Michal et al (2013) identified third putative Dis3p homologue (hDIS3L2), which causes altered levels of multiple mRNAs, while it is not a part of exosomal RNA [73]. Matthew et al (2017) reported that mounts of small noncoding RNA species were mapped via sequencing technology, including transfer RNAs, long noncoding RNA and small nucleolar RNAs. Then, they also confirmed the tRNAs took part in exosome-specific posttranscriptional modification [74]. It is believed that next generation sequencing is able to define the global or specific RNA profile of serum exosomes [75]. Similarly, DNA sequencing offers the opportunity to accurately detect profiling of the genomic landscapes of cancer cell derived exosomes, make an excellent tool to diagnose and judge the prognosis. A proof-of concept study by Lucas et al (2016) demonstrated this method and presented 95-99% of target regions covered at a mean depth of 133-490x, including multiple actionable mutations, such as notch receptor homologs (NOTCH1) and BRCA2 in patient exosomal DNA [76]. To summarize, the time has come for a better understanding of the characterization and the function of the exosomal genomics, which is beneficial for cancer detection.
Clinically relevant exosomal biomarkers for breast cancer
This pattern has attracted enormous attention of researchers resulting in the discovery of exosomal signatures with diagnostic and prognostic relevance for many types of cancer. Because cancer is a heterogeneous disease, here we choose breast cancer as an example.
Exosomal proteins
Protein values of exosomes from cancer patients are higher than normal and they reflect the cancer stage, especially the proteins derived from cancer cells (Table 3). In the study of Moon et al (2016), fibronectin level is significantly high in plasma at early stage of breast cancer patients, with sensitivity of 65.1% and specificity of 83.2% [77]. Further, levels of nephronnectin, a key factor known to promote adhesion and anchorage-independent growth, was found to be overexpressed in tumor tissue exosomes of breast cancer patients, associating with poor outcome in patients with luminal A subtype [78]. Besides, Khan et al (2014) showed survivin and its variant were detected in serum exosomes, and significantly higher in early stage breast cancer patients [79]. Another marker, extracellular matrix metalloproteinase inducer (EMMPRIN) has been enriched in tumor-derived microvesicles, also present high correlation with metastatic patients [80]. Based on these data, screening the different proteins of all breast cancer cells types, or looking for the same one of different cancer cells would increase the sensibility and specificity to promote cancer early detection, diagnosis and intervention.
Nucleic acids in exosomes
Besides from its specific protein, exosomes also carry a select set of functional circulating nucleic acids such as mRNAs, miRNAs, LncRNAs and DNA [81]. Exosomes are the main source of circulating miRNAs. So far, a great number of studies have analyzed the major miRNAs that show the perspectives to be diagnostic biomarkers in breast cancer. The results of weiying Zhou et al. indicate that miR-105 highly expressed in circulation at the premetastatic stage or distant metastatic stage. They also showed that this molecule and miR-181a predominantly existed in serum exosomes of stage II and III breast cancer patients [82]. Furthermore, increased invasion and proliferation are positively correlated with overexpression of exosomal miR-21 and miR-1246 in breast cancer [83]. In addition, other researchers also demonstrated that exosomes contain much miRNAs, such as miR-101, miR-372 and miR-130a-3p, which could be transferred from tumor to recipient cells, resulting in tumor progression and metastasis and presenting opportunity incorporating them into clinical study [84, 85].
Another non-coding RNA, long noncoding RNAs (LncRNAs), has a more complex structure which could bind proteins, RNA and DNA, and play important roles in life process ether normal or disease [86]. The use of exosomal LncRNAs as a potential biomarker in breast cancer was first reported by Lu et al (2015). In this study, LncRNA RP11-445H22.4 is highly expressed in cancer tissues and serum from breast cancer patients compared to healthy controls, with a sensitivity of 92% and a specificity of 74%. Notably, they found that the level of circulating this molecule is more correlated with estrogen receptor and progesterone receptor than human epidermal growth factor receptor-2 state [87], indicating that LncRNA RP11-445H22.4 could be used as a novel complementary marker for breast cancer diagnosis and prognosis. The accumulating evidence for the unique nucleic acids signatures of exosomes is increasing (Table 4), but much work still awaits confirmation of their clinical responses in future studies.
Clinically relevant exosomal protein markers for breast cancer
| Proteins | Source | Detection method | Patient stages | Application | Diagnostic value/outcome | Ref. |
|---|---|---|---|---|---|---|
| Fibronectin | Plasma | ELISA | Early stage (0, I and II) | Diagnosis | Sensitivity of 67.7% and specificity of 72.0% | [77] |
| Nephronectin | Tissue | IHC microarray | Breast cancer (n=1393) | Prognosis | Sensitivity 70.8% | [78] |
| Survivin and Survivin-∆Ex3 | Serum | Western blot | Stage (II - IV) | Diagnosis and treatment | NA | [79] |
| FAK | Plasma | Western blot | In situ and stages I-III | Diagnosis and prognosis | NA | [124] |
| EGFR | Plasma | Western blot | In situ and stages I | Diagnosis and prognosis | NA | [124] |
| EMMPRIN | Blood | Flow cytometry | Metastasis (n=8) | Prognosis | Median 31.7 % [IQR 21.35-37.35] vs median 19.5 % [IQR 10.03-24.88] in control | [80] |
| Periostin | Plasma | Western blot | Primary breast cancer with lymph node metastasis at diagnosis; relapsed metastasis during the follow-up | Diagnosis | NA | [125] |
| HER2 | Plasma | Microfluidic chip | Breast cancer (n=19) | Diagnosis | NA | [126] |
| CD47 | blood | Flow cytometry | Breast cancer (n=60) | Diagnosis | P=0.004 | [127] |
| Del-1 | Plasma | ELISA | Patients underwent curative surgery (n=115) | Diagnosis and treatment | 94.8% patients showed a normalization of Del-1 lower than 0.5 after surgery and 10 patients showed Del-1> 0.4 | [128] |
| TRPC5 | Plasma | Flow cytometry | Unresectable metastasis (n=131) | Prognosis | P=0.0042 | [129] |
| UCH-L1 | Plasma | Flow cytometry | Resectable breast cancer (n=93) | Prognosis | P=0.009 | [130] |
Clinically relevant exosomal nucleic acid markers for breast cancer
| RNA/DNA | Source | Detection method | Patient stages and numbers | Application | Diagnostic value/outcome | Ref. |
|---|---|---|---|---|---|---|
| miR-105 | Serum | RT-qPCR | Pre-metastatic and metastatic stage (n=75) | Detection and diagnosis | P = 0.04 | [82] |
| miR-105 and miR-181a | Serum | RT-qPCR | Stage II, III (n=38) | Diagnosis | P < 0.01 | [82] |
| miR-101, miR-372 and miR-373 | serum | TaqMan MicroRNA assays | Invasive breast cancer (n=50) | Diagnosis | miR-373 -- higher in triple negative, estrogen-negative and progesterone-negative tumors | [84] |
| miR-1246 and miR-21 | Plasma | RT-qPCR | Breast cancer (n=16) | Detection | The AUC for miR-21 was 0.69 (95 % CI 0.50, 0.88; p = 0.048) and for miR-1246 was 0.69 (95 % CI 0.49, 0.89; p = 0.068) | [131] |
| ssODNs | Plasma | ADAPT | Patients with positive breast cancer biopsy (n=59) | Diagnosis | P<0.05 | [132] |
| miR-340-5p, miR-17-5p miR- 130a-3p, miR-93-5p | Plasma | PCR array | Recurrent (n=16) | Prognosis and recurrence | P<0.05 | [85] |
| NANOG NEUROD1 HTR7 KISS1R HOXC | Plasma | PCR array | With or without first relapse and death (n=173) | Prognosis | P<0.05 | [133] |
| LncRNA RP11-445H22.4 | Serum | qRT-PCR | Breast cancer (n=68) | Prognosis | p < 0.001, sensitivity was 92% and specificity was 74 % | [87] |
Conclusions and future perspectives
As mentioned above, targeting the exosomal cargos may express high diagnostic and prognostic potential. Indeed, numerous studies have attempted to explore the different profile and function of exosomes in health and disease, and to facilitate their clinical applications. But we are still on the early stage of exosomal research in cancer, especially breast cancer. Like other new biomarkers, cancer related exosomes also have disadvantages. The common method for exosome isolation is still ultracentrifugeation since it was first discovered. Although there are various alternative strategies that have emerged, including filtration, chromatography and bead isolation, the primary method remains purification by size [88]. This represents a disadvantage; some of protein fragments are enclosed in urine or serum during the isolated process, and therefore maybe directly disturb the results. Additionally, not only exosomes but also other extracellular vesicles such as microparticles and apoptotic bodies carry bioactive molecules [89, 90]. An outstanding question remains as to whether body fluids or tissue derived exosomes sufficient purenesses are needed as a result. Furthermore, for the exosomal content, the mechanism of specific markers enrichment is not fully understood. Most studies just analyzed one kind of exosomal contents at specific time point but neglected other factors. However, any ideal biomarkers must be unique and sensitive for tracing upon different stages of cancer. Thus, it is essential that scientists continue exploring the secret of this field and establish of standard operating procedures related to exosome analysis for successfully improve veracity and enrich repeatability into clinically meaningful tests. With more and more novel exosomal markers continuing to emerge, it will spark up hope for realistic tracking of exosome release and uptake, even combining cancer early detection and exosome-based drug delivery to improve patients' survival rate. Thus, beyond these considerations, further quantitative data which focus on the metastasis niche and its function in tumor development, as well as more subtle biological structure of exosomes will achieve the clinical utilization of exosome as a specific biomarker. The future application of exosomes as cancer early detection and prognostic biomarkers will certainly be a new weapon to win this fight with cancer.
Acknowledgements
We thank the members of the Deng laboratory for critical discussions. This work is supported by the Chair Professor Grant (CPG2017-00026-FHS), MYRG2016-00132-FHS and MYRG2016-00139 of University of Macau and FDCT grants (065/2015/A2 and 094/ 2015/A3) to Chu-Xia Deng. The authors declare no conflict of interest.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Emery JD, Shaw K, Williams B, Mazza D, Fallon-Ferguson J, Varlow M. et al. The role of primary care in early detection and follow-up of cancer. Nat Rev Clin Oncol. 2014;11:38-48
2. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-20
3. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
4. Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116-25
5. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19-30 sup pp 1-13
6. Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59:823-31
7. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. 2017;66:30-41
8. Anastasiadou E, Slack FJ. Cancer. Malicious exosomes. Science. 2014;346:1459-60
9. Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383-90
10. Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X. et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016
11. Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707-21
12. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC. et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100-4
13. Putz U, Howitt J, Doan A, Goh CP, Low LH, Silke J. et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal. 2012;5:ra70
14. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J. et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183-94
15. Zhang J, Chen C, Hu B, Niu X, Liu X, Zhang G. et al. Exosomes Derived from Human Endothelial Progenitor Cells Accelerate Cutaneous Wound Healing by Promoting Angiogenesis Through Erk1/2 Signaling. Int J Biol Sci. 2016;12:1472-87
16. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M. et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668-78
17. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M. et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-35
18. Mrizak D, Martin N, Barjon C, Jimenez-Pailhes AS, Mustapha R, Niki T. et al. Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J Natl Cancer Inst. 2015;107:363
19. Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S. et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer-Derived Exosomes. Cancer Res. 2016;76:6816-27
20. Miyazaki T, Ikeda K, Sato W, Horie-Inoue K, Inoue S. Extracellular vesicle-mediated EBAG9 transfer from cancer cells to tumor microenvironment promotes immune escape and tumor progression. Oncogenesis. 2018;7:7
21. Dai S, Wan T, Wang B, Zhou X, Xiu F, Chen T. et al. More efficient induction of HLA-A*0201-restricted and carcinoembryonic antigen (CEA)-specific CTL response by immunization with exosomes prepared from heat-stressed CEA-positive tumor cells. Clin Cancer Res. 2005;11:7554-63
22. Sousa D, Lima RT, Vasconcelos MH. Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol Med. 2015;21:595-608
23. Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T. et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107:1-13
24. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y. et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653-68
25. Qi X, Zhang J, Yuan H, Xu Z, Li Q, Niu X. et al. Exosomes Secreted by Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Repair Critical-Sized Bone Defects through Enhanced Angiogenesis and Osteogenesis in Osteoporotic Rats. Int J Biol Sci. 2016;12:836-49
26. Guo SC, Tao SC, Yin WJ, Qi X, Sheng JG, Zhang CQ. Exosomes from Human Synovial-Derived Mesenchymal Stem Cells Prevent Glucocorticoid-Induced Osteonecrosis of the Femoral Head in the Rat. Int J Biol Sci. 2016;12:1262-72
27. Liu X, Li Q, Niu X, Hu B, Chen S, Song W. et al. Exosomes Secreted from Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Prevent Osteonecrosis of the Femoral Head by Promoting Angiogenesis. Int J Biol Sci. 2017;13:232-44
28. Peuhu E, Kaukonen R, Lerche M, Saari M, Guzman C, Rantakari P. et al. SHARPIN regulates collagen architecture and ductal outgrowth in the developing mouse mammary gland. EMBO J. 2017;36:165-82
29. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 Chapter 3: Unit 3 22
30. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-79
31. Varon D, Shai E. Role of platelet-derived microparticles in angiogenesis and tumor progression. Discov Med. 2009;8:237-41
32. Tao SC, Guo SC, Zhang CQ. Platelet-derived Extracellular Vesicles: An Emerging Therapeutic Approach. Int J Biol Sci. 2017;13:828-34
33. Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L. et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer. 2015;14:58
34. Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J. et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752-60
35. Mause SF, Ritzel E, Liehn EA, Hristov M, Bidzhekov K, Muller-Newen G. et al. Platelet microparticles enhance the vasoregenerative potential of angiogenic early outgrowth cells after vascular injury. Circulation. 2010;122:495-506
36. Lv LL, Cao Y, Liu D, Xu M, Liu H, Tang RN. et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021-31
37. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368-73
38. McKiernan J, Donovan MJ, O'Neill V, Bentink S, Noerholm M, Belzer S. et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016;2:882-9
39. Scott RP, Quaggin SE. Review series: The cell biology of renal filtration. J Cell Biol. 2015;209:199-210
40. Haraldsson B, Nystrom J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev. 2008;88:451-87
41. Qin W, Tsukasaki Y, Dasgupta S, Mukhopadhyay N, Ikebe M, Sauter ER. Exosomes in Human Breast Milk Promote EMT. Clin Cancer Res. 2016;22:4517-24
42. Yu S, Zhao Z, Sun L, Li P. Fermentation Results in Quantitative Changes in Milk-Derived Exosomes and Different Effects on Cell Growth and Survival. J Agric Food Chem. 2017;65:1220-8
43. Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M. et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969-78
44. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q. et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118-23
45. Gallart-Palau X, Serra A, Sze SK. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol Neurodegener. 2016;11:41
46. Vella LJ, Scicluna BJ, Cheng L, Bawden EG, Masters CL, Ang CS. et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 2017;6:1348885
47. Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X. et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448
48. Jingushi K, Uemura M, Ohnishi N, Nakata W, Fujita K, Naito T. et al. Extracellular vesicles isolated from human renal cell carcinoma tissues disrupt vascular endothelial cell morphology via azurocidin. Int J Cancer. 2018;142:607-17
49. Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S. et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287
50. Mincheva-Nilsson L, Baranov V, Nagaeva O, Dehlin E. Isolation and Characterization of Exosomes from Cultures of Tissue Explants and Cell Lines. Curr Protoc Immunol. 2016;115:14 42 1-14 42 21
51. Gold P, Freedman SO. Demonstration of Tumor-Specific Antigens in Human Colonic Carcinomata by Immunological Tolerance and Absorption Techniques. J Exp Med. 1965;121:439-62
52. Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat Rev Cancer. 2010;10:371-8
53. Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L. et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13:688-95
54. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539-44
55. Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110-8
56. Dive C, Brady G. SnapShot: Circulating Tumor Cells. Cell. 2017;168:742-e1
57. Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341:1186-8
58. Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS. et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999
59. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-82
60. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-26
61. An MR, Lohse I, Tan ZJ, Zhu JH, Wu J, Kurapati H. et al. Quantitative Proteomic Analysis of Serum Exosomes from Patients with Locally Advanced Pancreatic Cancer Undergoing Chemoradiotherapy. Journal of Proteome Research. 2017;16:1763-72
62. Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin Y, Yuan Y. et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;00:1-17
63. Shi M, Jiang Y, Yang L, Yan SS, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. Journal of Cellular Biochemistry. 2018;119:4711-6
64. Li ZH, Jiang P, Li J, Peng MJ, Zhao X, Zhang X. et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822-38
65. Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F. et al. CD90+liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Molecular Cancer. 2015;14:155
66. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C. et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Molecular Cancer. 2018;17:14
67. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH. et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Molecular Cancer. 2018;17:84
68. Zhao R, Zhang YL, Zhang X, Yang YM, Zheng X, Li XH. et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Molecular Cancer. 2018;17:68
69. Roberg-Larsen H, Lund K, Seterdal KE, Solheim S, Vehus T, Solberg N. et al. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J Steroid Biochem Mol Biol. 2017;169:22-8
70. Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328-31
71. Clark DJ, Fondrie WE, Liao Z, Hanson PI, Fulton A, Mao L. et al. Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis. Anal Chem. 2015;87:10462-9
72. Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell. 2017;168:571-4
73. Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3'-5' degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855-68
74. Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R. et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A. 2017;114:E8987-E95
75. Selmaj I, Cichalewska M, Namiecinska M, Galazka G, Horzelski W, Selmaj KW. et al. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann Neurol. 2017;81:703-17
76. San Lucas FA, Allenson K, Bernard V, Castillo J, Kim DU, Ellis K. et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016;27:635-41
77. Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH. et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7:40189-99
78. Steigedal TS, Toraskar J, Redvers RP, Valla M, Magnussen SN, Bofin AM. et al. Nephronectin is Correlated with Poor Prognosis in Breast Cancer and Promotes Metastasis via its Integrin-Binding Motifs. Neoplasia. 2018;20:387-400
79. Khan S, Bennit HF, Turay D, Perez M, Mirshahidi S, Yuan Y. et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176
80. Menck K, Scharf C, Bleckmann A, Dyck L, Rost U, Wenzel D. et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J Mol Cell Biol. 2015;7:143-53
81. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426-37
82. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501-15
83. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL. et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Research. 2016:18
84. Eichelser C, Stuckrath I, Muller V, Milde-Langosch K, Wikman H, Pantel K. et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650-63
85. Rodriguez M, Silva J, Herrera A, Herrera M, Pena C, Martin P. et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget. 2015;6:40575-87
86. Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. N Engl J Med. 2016;374:2287-9
87. Xu N, Chen F, Wang F, Lu X, Wang X, Lv M. et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biol. 2015;36:7659-65
88. Ibrahim A, Marban E. Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu Rev Physiol. 2016;78:67-83
89. Rautou PE, Leroyer AS, Ramkhelawon B, Devue C, Duflaut D, Vion AC. et al. Microparticles from human atherosclerotic plaques promote endothelial ICAM-1-dependent monocyte adhesion and transendothelial migration. Circ Res. 2011;108:335-43
90. Dieude M, Bell C, Turgeon J, Beillevaire D, Pomerleau L, Yang B. et al. The 20S proteasome core, active within apoptotic exosome-like vesicles, induces autoantibody production and accelerates rejection. Sci Transl Med. 2015;7:318ra200
91. Jansen FH, Krijgsveld J, van Rijswijk A, van den Bemd GJ, van den Berg MS, van Weerden WM. et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics. 2009;8:1192-205
92. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-91
93. Alegre E, Zubiri L, Perez-Gracia JL, Gonzalez-Cao M, Soria L, Martin-Algarra S. et al. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin Chim Acta. 2016;454:28-32
94. Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32:1976-83
95. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S. et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J Proteome Res. 2018;17:1101-7
96. Huang A, Dong J, Li S, Wang C, Ding H, Li H. et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11:961-9
97. Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ. et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66:1125-43
98. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-6
99. Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W. et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano. 2018;12:671-80
100. Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z. et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. 2017;21:838-47
101. Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490-5
102. Shender VO, Pavlyukov MS, Ziganshin RH, Arapidi GP, Kovalchuk SI, Anikanov NA. et al. Proteome-metabolome profiling of ovarian cancer ascites reveals novel components involved in intercellular communication. Mol Cell Proteomics. 2014;13:3558-71
103. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603-7
104. Madhankumar AB, Mrowczynski OD, Patel SR, Weston CL, Zacharia BE, Glantz MJ. et al. Interleukin-13 conjugated quantum dots for identification of glioma initiating cells and their extracellular vesicles. Acta Biomater. 2017;58:205-13
105. Madhavan B, Yue SJ, Galli U, Rana S, Gross W, Muller M. et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. International Journal of Cancer. 2015;136:2616-27
106. Joshi GK, Deitz-McElyea S, Liyanage T, Lawrence K, Mali S, Sardar R. et al. Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing at Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS Nano. 2015;9:11075-89
107. Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86-93
108. Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V. et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741-7
109. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC. et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184
110. Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H. et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532-8
111. Lou G, Song X, Yang F, Wu S, Wang J, Chen Z. et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122
112. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
113. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-9
114. Fang T, Lv H, Lv G, Li T, Wang C, Han Q. et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191
115. Hao YX, Li YM, Ye M, Guo YY, Li QW, Peng XM. et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol Lett. 2017;13:3608-16
116. Summerer I, Unger K, Braselmann H, Schuettrumpf L, Maihoefer C, Baumeister P. et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113:76-82
117. Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R. et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33-41
118. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B. et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768-74
119. Rodriguez M, Bajo-Santos C, Hessvik NP, Lorenz S, Fromm B, Berge V. et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer. 2017;16:156
120. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21
121. Cappellesso R, Tinazzi A, Giurici T, Simonato F, Guzzardo V, Ventura L. et al. Programmed cell death 4 and microRNA 21 inverse expression is maintained in cells and exosomes from ovarian serous carcinoma effusions. Cancer Cytopathol. 2014;122:685-93
122. Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y. et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33:2915-23
123. Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015;14:194
124. Galindo-Hernandez O, Villegas-Comonfort S, Candanedo F, Gonzalez-Vazquez MC, Chavez-Ocana S, Jimenez-Villanueva X. et al. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch Med Res. 2013;44:208-14
125. Vardaki I, Ceder S, Rutishauser D, Baltatzis G, Foukakis T, Panaretakis T. Periostin is identified as a putative metastatic marker in breast cancer-derived exosomes. Oncotarget. 2016;7:74966-78
126. Fang S, Tian H, Li X, Jin D, Li X, Kong J. et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One. 2017;12:e0175050
127. Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R. et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502
128. Lee SJ, Lee J, Jung JH, Park HY, Lee CH, Moon PG. et al. y Exosomal Del-1 as a potent diagnostic marker for breast cancer: A prospective cohort study. Journal of Clinical Oncology. 2017;77:pp2781
129. Wang T, Ning K, Lu TX, Sun X, Jin L, Qi X. et al. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci. 2017;108:448-54
130. Ning K, Wang T, Sun X, Zhang P, Chen Y, Jin J. et al. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surg Oncol. 2017;115:932-40
131. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL. et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90
132. Domenyuk V, Zhong Z, Stark A, Xiao N, O'Neill HA, Wei X. et al. Plasma Exosome Profiling of Cancer Patients by a Next Generation Systems Biology Approach. Sci Rep. 2017;7:42741
133. Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8:69934-44
Author contact
![]() Corresponding author: cxdengmo
Corresponding author: cxdengmo

 Global reach, higher impact
Global reach, higher impact