Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(1):69-80. doi:10.7150/ijbs.28129 This issue Cite
Research Paper
Identification and Functional Characterization of a New Splicing Variant of EZH2 in the Central Nervous System
School of Food Science and Engineering, Hefei University of Technology, No. 193 of Tunxi Road, Baohe District, Hefei, Anhui Province, China
*These authors contributed equally to the work.
Received 2018-6-26; Accepted 2018-9-26; Published 2019-1-6
Abstract
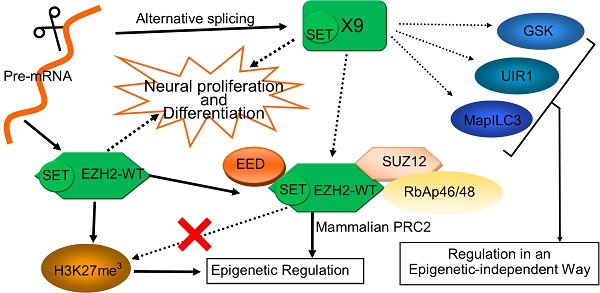
EZH2 plays vital roles in epigenetic regulation, neuronal development and cancer progression. Here a novel EZH2 variant, namely EZH2-X9 (X9 for short) resulting from alternative splicing, was isolated, identified and functionally characterized. X9 was highly expressed in the brains of SD rats, indicating a potentially distinguished role in the central nervous system (CNS). Owing to a transcript profiling, X9 was enriched in multiple brain regions at very early stage of life. Immunostaining validated the presence of the protein form of X9, which was localized similarly with the wild-type form, EZH2-WT. To investigate the functional consequence of X9, genetic intervention was performed in PC-12 cell line, a classic cellular model for neuronal development. It revealed that the depletion of either variant was sufficient to impair neuronal proliferation and differentiation significantly, an evidence that roles of X9 could not be complemented by EZH2-WT. Considering epigenetic regulation, X9 lost the capability to recruit the histone mark H3K27me3, but retained the cooperation with EED, as well as the repressive aspects in governing gene expression. Nonetheless, through profiling the genes affected, it's discovered that EZH2-WT and X9 markedly differed in their regulatory targets, as X9 intended to repress cell cycle- and autophagy-related genes, like GSK and MapILC3. Overall, a novel Ezh2 variant was characterized in the mammal CNS, providing insight with the structural and functional delineation of this key developmental switch, Ezh2.
Keywords: Ezh2, splicing variant, neuronal differentiation, H3K27me3
Introduction
The enhancer of zeste homolog 2 (Ezh2), a transcriptional repressor, is a catalytic subunit of the Polycomb repressive complex 2 (PRC2) [1] and induces the trimethylation of the histone H3 lysine 27 (H3K27me3) [2]. The methyltransferase activity of Ezh2 is required to recruit H3K27me3 in distinct genomic loci to control the expression of multiple functional genes [3]. EZH2 is essential in regulating the development and function of CNS (central nervous system). The timely expression of EZH2 maintained the balance of self-renewal and differentiation of neural stem cells (NSC) [4], with proliferation compromised by the removal of EZH2. In addition, EZH2 regulates the fate determination of NSC, wherein an enhanced presence of EZH2 pushes NSC towards the neuronal and oligodendrocytic lineage, at the expense of astrocytes [5, 6]. In terminally-differentiated neurons, EZH2 dictated and maintained the global consequences of cell differentiation [7].
Alternative splicing permits generation of diverse proteins from a single gene through excluding or including variable exons from the processed mRNA transcript [8]. Alternative splicing exponentially expands protein diversity and increases the plasticity of the system to meet the broad requirement of functional release and regulation [9, 10]. The most extensive alternative splicing occurs in brain tissues, supporting the complex functions of the nervous system [11]. It was found that transcripts from approximately 95% of multi-exon human genes are spliced in a variable way [12], and in most cases, the resulting transcripts are variably expressed and acted as distinct regulators in specific cell and tissue types [13]. Aberrant splicing incidents were regularly associated with the development of numerous diseases, like Alzheimers [14].
To date, several splicing variants of EZH2 have been identified and characterized. SRSF2 mutation led to a preferential inclusion of a “poison” cassette exon, which introduces a premature stop codon in the coding frame of EZH2 in the murine hematopoietic cells [15]; an EZH2β isoform was identified through skipping of exon4 in adult human tissues, with its physiological roles redundant with the original transcript [10]; very recently, in patients suffering from myelodysplastic syndromes, an EZH2 variant lacking of exon14 was discovered and considered distinct in regulating cancer cell proliferation and migration [16, 17]. Intriguingly, despite that a range of EZH2 variants were proposed through genomic sequencing and in silico splicing, no bona fide variant was empirically validated and characterized in the rodent neural cells, whereas EZH2 was considerably underscored.
Here we describe a new splicing variant of Ezh2, namely X9, in the brain tissues of SD rats. The properties of X9 were subsequently investigated with respect to tissue specificity, H3K27me3 recruitment and neural development. The findings contribute to the architectural and functional specification of Ezh2, a critical developmental switch, in the mammal CNS.
Results
Splice variant EZH2-X9 was identified in the brains of SD rats
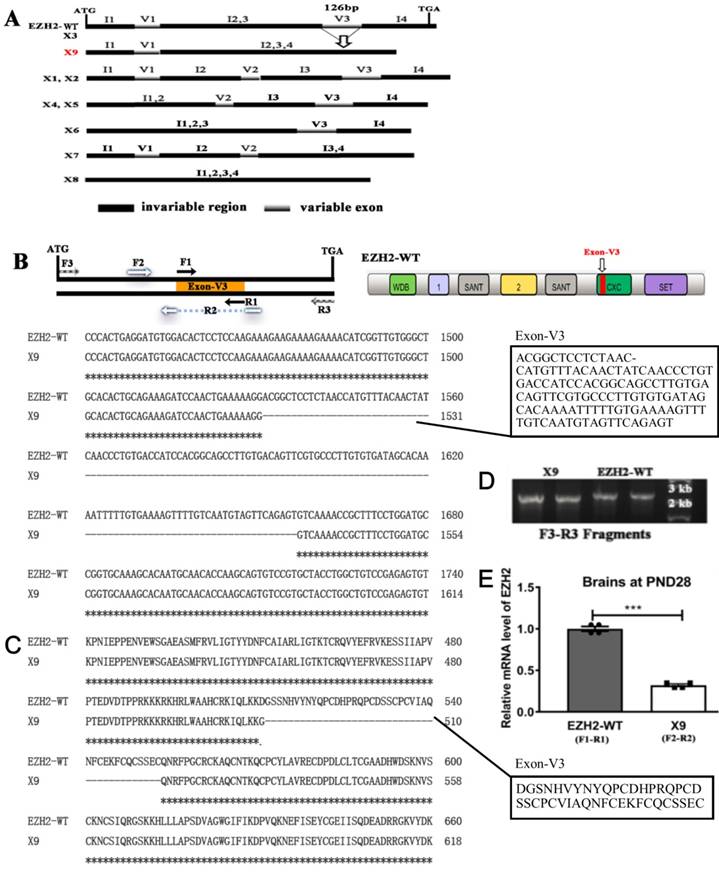
EZH2 plays fundamental roles in regulating the development of mammal CNS [18]. To date, nine EZH2 transcripts were identified in rats (Rattus norvegicus), namely EZH2, X1-X8, based on the genomic sequencing and in silico splicing (Fig. 1A). However, none of them were empirically substantiated except the wild-type isoform (EZH2-WT) in brains. In order to search for a genuine variant present in the CNS, the entire length of ORF fragments were amplified using the primer pair F3-R3 and subjected to nucleotide sequencing. Consequently, only two variants were obtained, that is, EZH2-WT and a new splicing variant called X9.
In light of homologous comparisons among the predicted EZH2 variants (WT, X1-X8), exon skipping of EZH2 occurred solely in three variable exons, V1, V2 and V3, while the coding regions staying unaffected among all the variants were indexed as the invariable regions I1-I4 (Fig. 1A). In comparison with EZH2-WT, V3 was skipped during the mRNA processing to generate X9, a variant structurally distinct from any other transcripts. According to the sequence alignment, V3 with the size of 126 bp differentiated the EZH2-WT and X9. Together with the predicted structure of V7 and V8, it might propose that specific exon skipping occurs at V3 (Fig. 1B). Of note, based on the domain organization of EZH2, V3 lies in the “CXC domain”, a cystein-rich region, suggesting that X9 might lose the corresponding activity assigned by CXC domain. The amino acid sequence of both variants surrounding V3 was shown in Fig. 1C. The predominant existence of EZH2-WT and X9 was verified through re-amplifying the respective transformant colonies (~50 TA subclones were checked) and showing the running band in the agarose gel (Fig. 1D). The blasting chromatograph of the new junction of X9 was shown in Fig. S1A. The position of DNA band is well suited with the anticipated size of EZH2-WT and X9, respectively, which demonstrates that EZH2-WT and X9 are the bona fide EZH2 transcripts expressed in the rat brains.
To compare the expression levels of EZH2-WT and X9, variant-specific primer pairs were designed and employed to perform the qPCR analysis. In the brains samples of rats in PND (postnatal days) 28, it was shown that EZH2-WT was transcribed in a significantly larger amount than X9 (Fig. 1E), with the calculation corrected by primer efficiencies (Fig. S1B,C).
EZH2-X9 was predominantly present in the nucleus
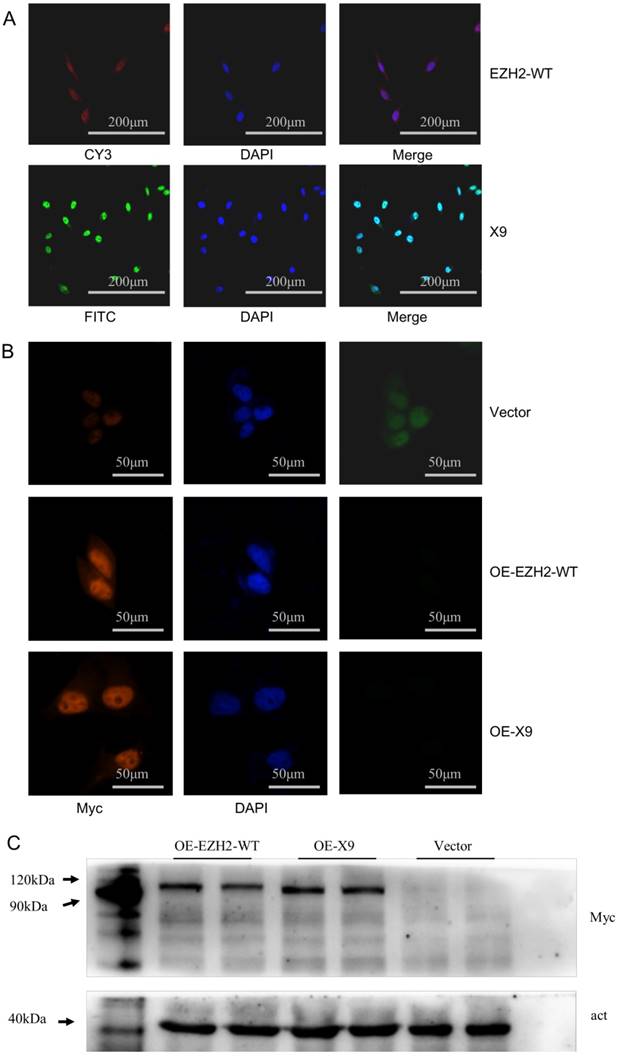
Intracellular location is an important indicator of potential function of a biological macromolecule. Given that Ezh2 could either exert its regulatory roles as a transcriptional repressor, or contribute to cellular fibrosis through its cytoplasmic partner [19], efforts were made to investigate the compartmentalization of X9. To this end, a FISH assay was carried out in PC-12 cells, paradigm neuronal cultures. The graphs were obtained through fluorescence detection with EZH2-WT and X9 labeled with Cy3 and FITC, respectively. It was then observed that the untransported and matured forms of X9 were predominantly distributed into the nucleus (Fig. 2A), reminiscent of the pre-mRNA, processed and matured forms of EZH2-WT.
In addition, overexpression trials were performed to validate the localization of protein isoforms. EZH2-WT or X9 was ligated into a GFP-Myc-containing vector respectively, resulting in the fusion proteins. Detected by immunostaining (Fig. 2B), the X9 protein still share, to a large extent, the cellular compartmentalization with EZH2-WT, that is, a predominant fraction was present in the nucleus while only a slight fraction retained in the cytosol. These data indicated that X9 might fulfill its cellular function through the transcriptional regulation.
To further validate the existence of the X9 protein, immunoblots of Myc in the EZH2-WT- and X9-overexpressing cells were performed. It revealed that the individual band corresponding to the respective cells was detected in the corresponding running positions (Fig. 2C). Taken together, X9 could be translated into protein and it primarily resides in the nucleus of neural cells.
Splice variant EZH2-X9 is identified in the brains of SD rats. (A) Schematic representation of EZH2 variants in rats. Invariable region refers to the exon combination that was not skipped by sequence comparisons, variable exon refers to the exon that was skipped by sequence comparisons; the arrow marks the skipping incident occurring in X9. (B) The position of primer pairs amplifying the individual variant, the location of exon-V3 in the protein architecture of EZH2-WT, and the sequence alignment of EZH2-WT and X9. The sequence of exon-V3 was boxed on the left. (C) Protein sequence alignment of EZH2-WT and X9 surrounding the exon-V3 region. (D) Agarose gel electrophoresis of PCR fragments using the primer pair F3-R3 against independent TA-cloning transformants. The size of DNA marker was marked at the adjacent lane. (E) Relative mRNA levels of EZH2 variants in the rat brain at PND28 (n = 6) The data are represented as Mean ± SEM; ***P< 0.001.
EZH2-X9 was predominantly present in the nucleus. (A) Representative FISH graphs of EZH2-WT and X9 on the coverslips of PC-12 cells. The probes used were EZH2-WT-CY3 and X9-FITC, respectively. Nuclei was stained with DAPI, and bars represent 200 μM. (B) IF staining of EZH2-WT and X9 on the coverslips of PC-12 cells. The antibody used was anti-Myc, against the Myc tag in the respective transfected cells. Nuclei was stained with DAPI, and bars represent 50 μM. (C) Immunoblots of Myc in the protein samples from OE-EZH2-WT, OE-X9-WT and Vector-transfected PC-12 cells. The protein marker was run alongside the samples and the immunoblot of β-actin was provided below.
EZH2-X9 was expressed in a spatiotemporal-specific way in rat brains. (A) Relative mRNA levels of EZH2-WT and X9 in various tissues of rats at PND 28 (n = 6). (B-D) Relative mRNA levels of EZH2-WT and X9 in various brain regions of rats at PND 0 (B), PND 14 (C) and PND 28 (D). The illustration of icons was listed as follows: OB-Olfactory Bulb; M-Primary Motor Cortex and Secondary Motor Cortex; V-primary visual cortex and secondary visual cx, lateral area and secondary visual cx, mediolat and V2MM secondary vis cx, mediomed; S-S1BF S1 cx, barrel field and S1FL S1 cx, forelimb region and S1HL S1 cx, hindlimb region and S1ShNc S1 cx, shoulder/neck and S1Tr S1 cx, trunk region and S1ULp S1 cx, upper lip region and S2 secondary somatosensory cortex; Hip-hippocampal fissure; Cpu-caudate putamen striatum; Cer -Cerebellum; R and L refer to right and left hemisphere, respectively (n = 6). (E) Immunoblots of EZH2-WT and X9 in the four brain regions at PND 0. Act is the internal reference protein, β-actin. The data are represented as Mean ± SEM; ***P< 0.001, **P<0.01, ***P< 0.001, ns, not significant.
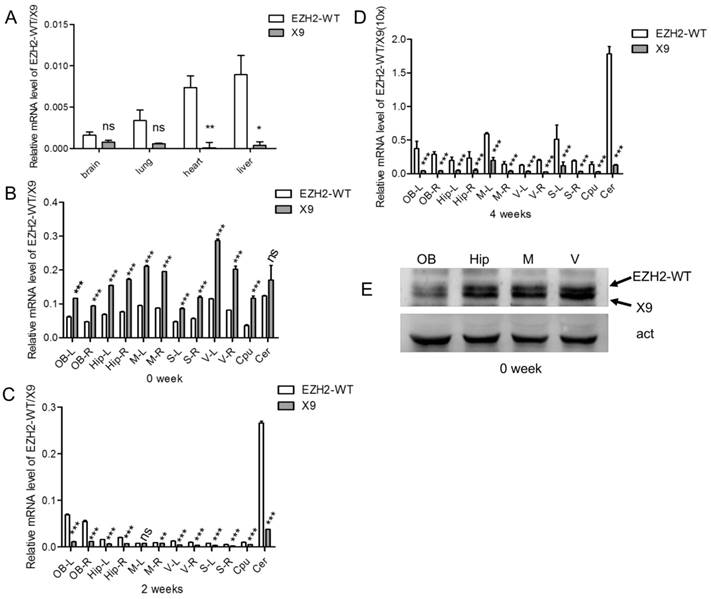
EZH2-X9 was expressed in a spatiotemporal way in rat brains
To investigate the tissue specificity of EZH2-X9, the samples of brain, lung, heart and liver were taken from rats at PND 28, and the transcript levels were subsequently examined. As shown in Fig. 3A, X9 was less abundant in all the tested tissues compared to EZH2-WT. Meanwhile, it's intriguing to discover that X9 was significantly enriched in the brain tissue, as its abundance relative to the total EZH2 mRNA was dramatically larger in the brains than other tissues. This finding suggested that X9 probably played essential roles in the CNS functioning.
We further examined the expressional profiles of X9 in various regions of brain. It was found that the levels of X9 differed in distinct regions (Fig. 3B-D). Of note, X9 transcript was sharply decreased as the development progressed. Specifically, X9 was highly accumulated at PND 0, showing a robust dominance over EZH2-WT, a tendency subsequently reversed at PND 14 and 28. All the brain regions complied with the similar alteration profiles. The results here suggested that X9 was likely implicated in the early development of mammal brains.
Besides, the protein levels of X9 were determined pertaining to four brain regions, namely olfactory bulb, hippocampus, S and V region, at PND 0. An EZH2 antibody, which recognize the consensus domain (695-746 aa) of EZH2 variants was utilized. Fig. 3E showed that the olfactory bulb possessed less X9 than the other regions tested, and the proportion of X9 exhibited a similar tendency with its mRNA transcript. Overall, EZH2-X9 was expressed in a spatiotemporal-specific way in rat brains.
Ezh2-X9 was functionally associated with proliferation and differentiation of PC-12 cells
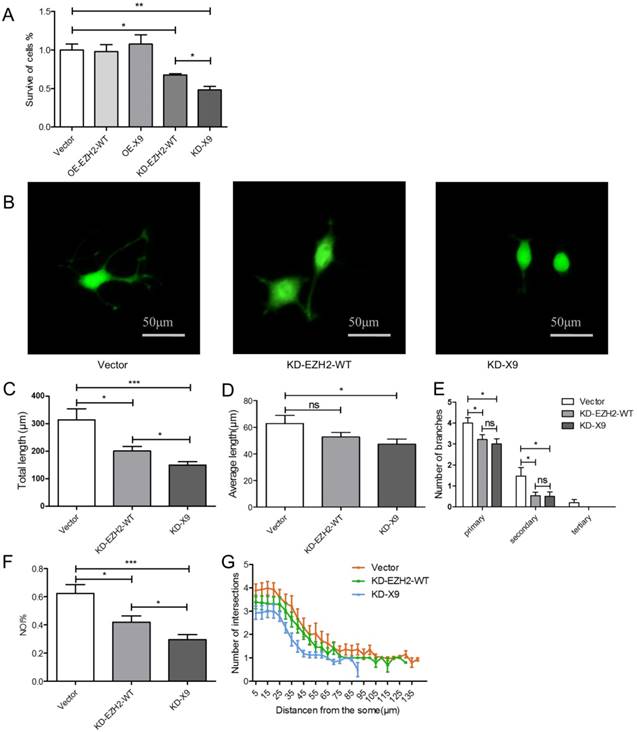
EZH2 is an important factor to influence the balance of self-renewal and differentiation of neural stem cells [4]. Thus the functional consequence of Ezh2-X9 in the cultured neurons was next inspected. PC12 cells were transfected with four respective plasmids to knockdown or overexpress either variant, and cell proliferation was first determined by MTT assay. According to the results (Fig. 4A), the depletion of EZH2-WT or X9 led to the compromised proliferation capacity, probably indicating that no functional redundancy was involved in their relations. Moreover, when compared to the “EZH2-WT-depleting” event, the proliferation of X9-KD cells was weakened to a larger extent, showing a relatively close relationship with PC-12 cell proliferation. No obvious changes were detected in the EZH2-overexpressed cells.
The impact of X9 on the neural differentiation was subsequently investigated. The representative images of the neurite outgrowth profiles were evidenced in Fig. 4B. Consequently, as manifested by total and average lengths, number of branches, NOI and sholl analysis (Fig. 4C-G), the depletion of X9 significantly decreased the neurite outgrowth, a similar case with EZH2-WT. In the event of total length, NOI and sholl analysis, the X9-KD cells showed a more dramatic reduction in comparison to EZH2-WT cells, suggesting that X9 is likely closely-related to PC-12 cell differentiation. Therefore, X9 is an important factor in regulating the proliferation and differentiation of nervous cells.
Ezh2-X9 was functionally associated with proliferation and differentiation of PC-12 cells. (A) MTT assay of the PC-12 cells with the treatment of KD or OE EZH2 variant, respectively (n = 4). (B) Representative graphs of PC-12 cells induced by NGF with the treatment of KD EZH2-WT or K9, respectively. Bars represent 50 μM. (C-G) Neurite outgrowth profiles of PC-12 cells with the treatment of KD EZH2-WT or K9, respectively. Total (C) and average (D) neurite length, number of branches (E), NOI value (F) and sholl analysis (G) were calculated (n > 150). The data are represented as Mean ± SEM; *P<0.05, **P<0.01, ***P< 0.001, ns, not significant.
EZH2-X9 lost the ability to introduce H3K27me3
Ezh2 is a key catalytic unit of PRC2 complex (namely, Polycomb repressive complex 2), which recruits H3K27me3 mark to specific genetic loci to control the expression pattern of the structural gene [7]. Thus, in order to explore the exact function of Ezh2-X9, its relation with H3K27me3 is warranted to be determined. To this end, the KD- and OE-K9/ EZH2-WT plasmids were used to transfect PC-12 cells. The knockdown efficiency was first validated through the measurement of the mRNA level (Fig. 5A, B). Interestingly, the transcription levels of Ezh2-WT and X9 were not entirely independent, but interconnected to a certain degree. When the mRNA level of Ezh2-WT was decreased, the expression of X9 was surprisingly stimulated (Fig. 5A). However, the counteracting relations could not be copied under the opposite circumstance, as blocking X9 did not promote the expression of EZH2-WT (Fig. 5B).
Considering the tri-methylation of H3K27, it's discovered that while blocking EZH2-WT reduced the presence of H3K27me3, the epigenetic mark remains unaffected in the X9-depleted cells (Fig. 5C). The results might conclude that X9 lost the capacity to introduce H3K27me3, which is consistent with the previous finding that CXC domain is indispensible for the methyltransferase activity of EZH2 [17].
EZH2-X9 lost the ability to introduce H3K27me3. (A-B) Relative mRNA levels of EZH2-WT (A) and X9 (B) in PC-12 cells in the treatment of KD EZH2-WT or X9, respectively (n = 4). (C) Immunoblots and quantification of H3K27me3 in PC-12 cells in the treatment of OE or KD EZH2 variants, respectively (n = 4). (D) Co-immunoprecipitation (Co-IP) of Myc in PC-12 cells in the treatment of OE EZH2-WT or X9, respectively. IgG represents a control antibody used for IPs. Antibodies used for IP and Western blotting (WB) were anti-Myc and anti-EED, respectively. Prior to carrying out the IP experiments, one tenth of total lysates were subjected to the respective WB as input controls. The data are represented as Mean ± SEM; *P<0.05, **P<0.01, ***P< 0.001, ns, not significant.
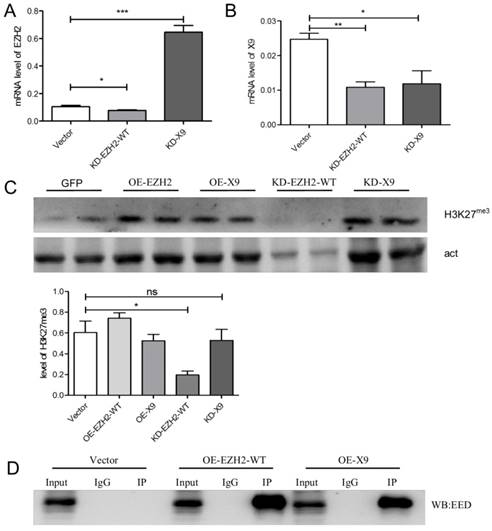
To further investigate if X9 entered the PRC2 complex under the studied neural context, a Co-IP trial was carried out. Either EZH2 variant was fused with Myc-tag, and the protein samples pulled-down by Myc antibody were subjected to immunoblotting of EED, a constituting protein of PRC2 complex. As evidenced by Fig. 5D, X9 interacted with EED, suggesting that it retained the ability to be incorporated into PRC2 complex. This data explains the fact that X9 possessed the entire domains accounting for EED-interaction, and implies that X9 might play its roles in a methylase-independent fashion.
Ezh2-X9 displayed a distinct mode of action as a transcriptional regulator
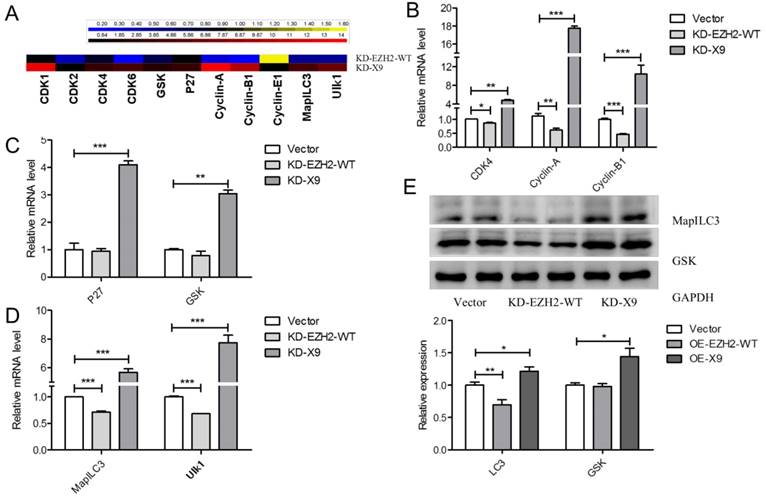
Since EZH2-X9 did not introduce H3K27me3, whether it could still regulate gene expression was subsequently examined. A series of genes responsible for cell cycle regulation, apoptosis and autophagy were subjected to transcript profiling, in the PC-12 cells blocking the expression of X9. It was seen from Fig. 6A, that most of the tested genes displayed a promoted mRNA abundance upon the knockdown of X9, demonstrating that X9 retained its ability to repress gene expression. Of note, great discrepancies occurred when the knockdown of EZH2-WT was used for comparisons. It suggested that X6 is a distinct transcriptional regulator from EZH2-WT.
In terms of CDK4, Cyclin-A and Cylin-B1, the blockade of X9 considerably stimulated their expression (Fig. 6B), showing that the presence of X9 was closely related to cell cycle regulation. The similar occasion occurred pertaining to the expression of apoptosis- (Fig. 6C) and autophagy-related genes (Fig. 6D). Interestingly, under the studied context, X9 seems potent in repressing gene expression, whereas EZH2-WT did not display pronounced repressive patterns. The role of X9 as a transcriptional repressor was further verified through measuring the protein levels of LC3 and GSK (Fig. 6E). Overall, X9 was still a transcriptional repressor and it did not share the regulatory targets with EZH2-WT.
Discussion
Alternative splicing expands protein diversity and augments the coding potential by means of complex regulation of gene expression [9]. This is particularly essential in the central nervous system, which requires a large protein repertoire to generate its intricate neural circuits [11, 20]. Deregulation of splicing is regularly associated with the functional abnormalities of CNS. For instance, the disruption of alternative splicing of Dcc led to an impaired neuronal migration and axon guidance [21]; a series of neuronal microexons were misregulated in autistic brains [22]. Of note, the functions of the vast majority of AS events detected to date are not known, and only a small proportion of AS variants were genuinely expressed and implicated in the cellular processes. As a molecule playing key roles in various developmental stages of CNS, no specific alternative splicing of EZH2 is yet characterized in the mammal neural cells. Among all the variants predicted, only EZH2-WT, as well as a novel X9, were found present in the nervous system, where their expressional profiles, intracellular location and molecular functions were characterized in succession. Based on it, it's rational to hypothesize that the previously-proposed function of EZH2 was actually an integrated manifestation of its variants, with their precise roles required to be specifically assigned.
35 % of known and 17 % new cassette exons in rodents have conserved splicing patterns in human [20]. While no homologue of X9 was found by sequence alignment of nucleotide sequences, it's very intriguing that an EZH2 variant, called EZH2△14, was recently discovered in the human renal cancer, with architectural identity as X9 [17]. Thus, the splicing pattern of X9 is not restrained to the rodent species, but probably conserved in mammals. In light of distinct roles in rat CNS, the evidence that X9 contributes to the human nervous function, as well as the development of psychiatric disorders, is required to be further provided. On the other side, despite that EZH2△14 shares the same organization of variable exons with X9, they considerably differed in the regulation of target genes and psychological roles: EZH2△14 had the opposite effect in promoting the migration and invasion of cancer cells with EZH2-WT [17], whereas X9 and EZH2-WT probably showed the similar tendency in regulating proliferation and differentiation of PC-12 cells (Fig. 4). Moreover, X9 retained its capacity to directly regulate gene expression in an H3K27me3-independent fashion, a characteristic that EZH2△14 did not possess. The discrepancies of analogues might reflect that the precise cellular roles of X9 are dependent on the physiological or pathological context it resides. In addition, the promoter methylation promoted by EZH2 might confound the ultimate expressional profiles of the target genes, due that EZH2 directly controlled DNA methylation [23]. It seems that the relationship between the transcription factors and the bona fide transcript levels of target genes was rather complex and intricate, enabling impact of X9 on CpG methylation an interesting topic for future investigations.
EZH2 is responsible for the balance of proliferation and differentiation of neural stem cells [4, 5]. According to the evidence here (Fig. 4), both of the neuronal processes were mediated by the cooperated action of EZH2-WT and X9. Comparatively, X9 seems more potent in maintaining the normal cellular status, as its depletion caused more severe injuries of neural-like cells, as manifested by the cell proliferation and total length of the outgrown neurites. The function of either splicing variant could not be considered redundant, due that no complementation was observed in the event of removal of EZH2-WT or X9. But why should the CNS keep two sets of EZH2 proteins? One possibility is that either of them played predominant roles at alternative developmental stages. For instance, alternative splicing of LRRTM3 and neurexin varied depending on the excitory status of hippocampal synapses [24, 25], thus a similar case might be applied to EZH2 variants, that is, their roles were dependent on various periods of life. At very early life, X9 was transcribed substantially across the brain (Fig. 3B), suggesting that X9 is likely the original form of EZH2 as neuronal development initiates. The hypothesis that functions of EZH2 variants were temporally assigned is warranted to be further validated in vivo.
The skipped exon of X9 belongs to the “CXC domain” of EZH2, a ~65 residue cys-rich region preceding the SET domain. Because of its evolutionary conservation, the CXC domain is uniquely present in Ez-related proteins, as well as a few proteins with limited diversity, such as MLS2 and TSO1 [15, 26, 27]. The exact function of the CXC domain still remains elusive, but as a pre-SET domain, it's thought to have relevance with HMT activity of EZH2 [17]. Consistent with it, in the present investigation, the mutation of CXC led to the loss of its ability to catalyze tri-methylation of lysine 27 of histone H3, as X9 did not seem to cause significantly reduced levels of H3K27me3 (Fig. 3). However, the argument that CXC domain is indispensible for the HMT activity of EZH2 was challenging, as Joshi et al. [28], stated that the CXC mutant retained catalytic activity, Lys-27 specificity and trimethylation capacity. The conflicting results might be due that a substantive knockdown occurred in X9, instead of a point mutation in the constructed “CXC mutant”. In terms of precise functions of CXC, another possibility could be raised that it's responsible for the recognition of target DNAs, because EZH2-WT and X9 displayed distinct regulatory aspects regarding specific genetic substrates. The bridging effect of CXC could be a consequence of either a direct interaction with DNA [27] or an indirect effect through assembly with PHO [28].
The relationship within splicing variants attracts much attention. EZH2α and EZH2β displayed redundancy in serving as the transcriptional regulators, as evidenced by the target genes occupied by either isoform [10]. However, this is not the case for EZH2-WT and X9, whereas they controlled the expression of distinct set of genes relating to cellular proliferation, differentiation and survival. Meanwhile, in contrast with situation in renal cancer [17], the overexpression of X9 did not show opposite effect in regulating cell proliferation with EZH2-WT (Fig. 4A). Therefore, the actions of EZH2-WT and X9 are likely neither redundant nor counteractive, but directed to a similar consequence via alternative pathways. Noteworthy, the expression of X9 was negatively affected by the induced overexpression of EZH2-WT, implying that there existed an interaction between EZH2 variants with respect to transcript regulation. No previous studies suggested that the EZH2 level could be regulated by itself or the resultant H3K27me3, and that makes the precise mechanisms governing the expression of X9 an issue to be addressed in future investigations.
In conclusion, in this study, a new form of EZH2 variant, namely X9, was first identified and characterized in the CNS of rats. X9 was enriched in the early developmental period, and was an important factor in maintaining the normal proliferation and differentiation of PC-12 cells. X9 lost the capability to recruit H3K27me3, however still retained the repressive roles in modulating gene expression. Considering the conserved splicing program of EZH2, the discovery of X9 in mammal CNS will enable a better understanding of the specified roles of EZH2 in neurogenesis and activity, as well as the relevance of its misregulation with etiology of psychiatric disorders.
Ezh2-X9 displayed a distinct mode of action as a transcriptional regulator. (A) Heatmap illustration of the expressional changes of genes regulated by EZH2 variants. qPCR was performed in the PC-12 cells transfected with KD-EZH2-WT or KD-X9 plasmids, respectively (n = 4). (B-D) Relative mRNA levels of cell cycle- (B), apoptosis- (C) and autophagy (D) -related genes in PC-12 cells in the treatment of KD EZH2-WT or X9, respectively (n = 4). (E) Immunoblots and quantification of LC3 and GSK in PC-12 cells in the treatment of KD EZH2-WT or X9, respectively (n = 4). The data are represented as Mean ± SEM; *P<0.05, **P<0.01, ***P< 0.001.
Materials and Methods
Antibodies and reagents
The following antibodies were used in this study: Anti-Myc: anti-Ezh2 (Proteintech, Wuhan, China, 21800-1-AP), Anti-Myc (Proteintech, Wuhan, China, 16286-1-AP), Anti-H3K27me3 (Millipore, MA, USA, ABE44-S), Anti-actin (Abcam, Cambridge, UK, ab16039), Anti-EED (Proteintech, Wuhan, China, 16811-1-AP), Anti-GAPDH (Abcam, Cambridge, UK, ab9484), Anti-GSK (CST, Massachusetts, USA, #12456), Anti-MapILC3 (Novus, Littleton, USA, NB100-2220).
Animals
Sprague-Dawley (SD) rats were obtained from the Laboratory Animal Center of Anhui Medical University. All animal procedures were carried out in accordance with National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Hefei University of Technology, China. The new-borne male rats were fed indirectly through their mothers and directly after weaning. Various brain regions (as indicated in [29]) were removed from the decapitated rats within 1 min at PND 0, 14 or 28, respectively, and then subjected to qPCR analysis as described above.
Cell culture
PC12 cells (undifferentiated and differentiated lines, CAS, Shanghai, China) were cultured at 37℃, 5 % CO2 in humidified atmosphere with RPMI1640 medium, supplemented with 10 % FBS (Fetal Bovine Serum) and 1% penicillin-streptomycin for undifferentiated and 10 % FBS for differentiated PC12 cells. Cells were plated onto 6-well tissue culture plates coated with poly-L-lysine (sigma P-2636) at 500μg/ml in 0.1 M Borate buffer. Subsequently, cells were allowed to continue its growth for 24 h. For undifferentiated cell lines, the medium was replaced with fresh medium containing 0.5 % FBS, 1 % HS and 1 % penicillin-streptomycin with NGF (50 ng/ml), at 24 h of growth. Plasmids transfection was performed as cells reach about 80% confluent and then allowed for growth for additional 48 h, respectively. Neuronal differentiation analysis was performed according to images from fluorescence microscopy (Nikon), and manifested by NOI (Neurite Outgrowth Index) [30], neurite length, branching and Sholl (Image J software) analysis.
Quantitative RT-PCR analysis
The transcriptional levels of genes were measured through quantitative real-time RT-PCR. First, the total RNAs (5 ng, equivalent for all samples) were extracted from cell samples or animal tissues using the AxyPrep Multisource Total RNA Miniprep kit (Axygen, Suzhou, China). Subsequently, the reverse transcription reaction was completed according to the manufacturer's instruction (Trans Gen, Beijing, China), resulting in the first strand of total cDNA.
Real-time PCR was performed on cDNA using the Roche LightCycler 96 (Shanghai, China). The 20 μl reaction pool of qPCR was composed of: 10 μl of SYBR premix Extaq; 0.4 μl of forward and reverse primer each (0.2 nM for final concentrations); 1 μl of cDNA template (10 times dilution) and 8.2 μl of deionized water. The transcriptional levels were first calculated as the -ΔCt (threshold cycle) value's difference from that of 18s rRNA under the same conditions, and normalized with -ΔCt (untreated group set as “0”) or 2-ΔΔCt (untreated group set as “1”) for gene disturbance assays, respectively. In transcriptional profiling in different regions, the qPCR results were normalized with their relative abundance with 18s rRNA, and the values of 4 weeks were multiplied with 10 to have data easily manifested. The absolute quantitative PCR was performed as described previously [31]. The primer efficiencies were shown in Fig. S1, and sequences of primers and probes used in this study were listed in Table 1.
Cell proliferation assay
Cell proliferation was assessed by the MTT reduction assay as described previously with some modifications [18]: PC 12 cells were seeded into 96-well plates at a concentration of 2×104 cells/well. 24 h later, plasmids transfection was carried out and incubated for 24 h. 50 μl of MTT/PBS solution (5 mg/ml) was added to the culture and incubated in the dark for 4 h at 37℃. The supernatants were removed and the formazan crystals in each well were dissolved in 50 μl of DMSO. The relative amount was determined based on the absorbance at 570 nm using a plate reader. The cell proliferation index of the vector group was then normalized as 100 %.
Plasmid construction
The vector pRNAT-U6.1-Neo (Genescript, Beijing, China) was used to construct the Ezh2-shRNA-expression vector and X9-shRNA-expression vector. The annealed shRNA fragment was ligated into the BamH I/Hind III restriction sites of the vector, resulting in the pRNAT-sh Ezh2. The target sequence of shEzh2 and shX9 were 5'-GCTCCTCTAACCATGTTTACA-3' and 5'-GGGTCAAAACCGCTTTCCTGG-3', respectively. A vector targeting the scrambled sequence was also established as a negative control.
The vector pEASY-blunt M2 (TransGen, Beijing, China) was used to construct the Ezh2-overexpression vector and X9-overexpression vector. The ORF of rat Ezh2 and X9 was amplified and ligated to the blunt ends downstream of CMV promoter, resulting in the pEASY-Ezh2 and pEASY-X9. The resultant constructs were subjected to nucleotide sequencing to ensure the fidelity of genetic manipulation. A vector with a scrambled sequence was also constructed as a negative control. All transfections were carried out using Lipofectamine 3000 (Thermo, Shanghai, China) with the referred amount of total plasmids for differentiated and undifferentiated PC 12 cells in the instruction manual, respectively.
Primers and probes used in this study
| Primers/probes | Sequences (5'-3') | Methods |
|---|---|---|
| F1 | ACGGCTCCTCTAACCATG | qPCR |
| R1 | AGGGCACGAACTGTCACA | qPCR |
| F2 | TCCAGTATCATAGCACCTGTTCC | qPCR |
| R2 | GCGGTTTTGACCCTTTTTCA | qPCR |
| F3 | ATGGGCCAGACTGGGAAGA | Over-expression |
| R3 | TCAAGGGATTTCCATTTCTCGTTC | Over-expression |
| sh-EZH2 | GCTCCTCTAACCATGTTTACA | KD-EZH2-WT |
| sh-X9 | GGGTCAAAACCGCTTTCCTGG | KD-X9 |
| Cyclin A-F | AGACTGAGTGGTTGGATGGCA | qPCR |
| Cyclin A-R | TGTCCACAGTCAGCAATGGTG | qPCR |
| Cyclin B1-F | AAAGGCGTAACTCGAATGGA | qPCR |
| Cyclin B1-R | CCGACCTTTTATTGAAGAGCA | qPCR |
| Cyclin E1-F | GGATTATTGCACCATCCAGAGGCT | qPCR |
| Cyclin E1-R | CTTGTGTCGCCATATACCGGTCAA | qPCR |
| CDK-1-F | TCCGCAACAGGGAAGAAC | qPCR |
| CDK-1-R | GAGCCTTTTTAGATGGCTGCT | qPCR |
| CDK-2-F | CTTTGGAGTCCCTGTCCGTA | qPCR |
| CDK-2-R | CGAAAGATCCGGAAGAGTTG | qPCR |
| CDK-4-F | TGCACAGTGTCACGAACAGA | qPCR |
| CDK-4-R | ACCTCGGAGAAGCTGAAACA | qPCR |
| CDK-6-F | CATCGTTCACCGAGATCTGA | qPCR |
| CDK-6-R | CCAACACTCCACATGTCCAC | qPCR |
| GSK-3β-F | TCCATTCCTTTGGGATCTGCC | qPCR |
| GSK-3β-R | ATCAGCTCTGGTGCCCTGTAGTAC | qPCR |
| p27-F | AGCGACCTGCTGCAGAAGAT | qPCR |
| p27-R | TTACGTCTGGCGTCGAAGGC | qPCR |
| MaplLC3-F | TGAGTGTCACAGTGGGCTCCA | qPCR |
| MaplLC3-R | ACAGTCTTTGTAAGGGCGGTTCT | qPCR |
| Ulk1-F | AGCCCACAGTAAATACCACAG | qPCR |
| Ulk1-R | ACTDACAGCCTACAGGAGAAA | qPCR |
| X9-probe | AAGCGGTTTTGACCCTTTTTCAGTTGGA | FISH (FITC) |
| EZH2-WT-probe | TGCCGTGGATGGTCACAGGGTTGATAGTT | FISH (Cy3) |
Western blot analysis
PC12 cells were washed with PBS and lysed in Laemmli lysis buffer. For brain samples, samples were dissected at PND0 or PND28, homogenized, washed and lysed in Laemmli lysis buffer. Proteins were loaded onto a 12 % SDS-PAGE for electrophoresis. The separated proteins were then transferred to PDVF membrane (Millipore, MA, USA). For immunodetection, the blots were blocked for 1 h in PBST (0.01M, PH7.2-7.4, 0.1% Tween 20) containing 5% nonfat dry milk at room temperature, followed by incubation with the primary antibodies overnight at 4℃. After incubation with the second antibody and extensive washing, immunoreactivity was detected by EasySee Western Blot Kit (TransGen, Beijing, China). The band intensity was normalized to the internal control (β-actin for whole) for comparisons.
Immunostaining
Immunostaining was performed according to the method of Stansfield et al [32]. Briefly, PC 12 cells grown on coverslips were rinsed in PBS and fixed in 4 % paraformaldehyde, followed by additional fixation in ice-cold methanol. 0.2 % of Triton X-100 was added to permeabilize the membranes and subsequently 10 % of normal horse serum was added to block the process. Fixed cells were incubated in anti-Myc antibody overnight at 4℃ with the dilution of 1:1000. Subsequently, coverslips were incubated in the second antibody diluted in block solution. Coverslips were mounted onto slides in Prolong Gold mounting media with DAPI (Invitrogen, Shanghai, China). Immunofluorescence-labeled cells were imaged at 40× magnification using fluorescence microscopy (Nikon, Tokyo, Japan).
Coimmunoprecipitation
Co-IP assay was performed using the Nuclear Complex Co-IP Kit (Active Motif, Shanghai, China) according to manufacturer's instructions. The PC-12 cells were transfected with vector, EZH2-WT-OE and X9-OE plasmids, respectively. Anti-Myc antibody was used to perform the immunoprecipitation assay and immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with anti-EED antibody.
FISH
PC 12 cells grown on coverslips were rinsed in PBS and fixed in 4 % paraformaldehyde/PBS with 0.1 % DEPC. 3 % H2O2 was added for 10 minutes to inactivate the endogenous peroxidase at room temperature. Pepsins resolved in citric acid were pipetted on the coverslips to expose mRNA fragments, followed by repeated washes of 0.5 M PBS. DIG-probe was then added and incubated at 37℃ overnight, after being treated with pre-hybridization solution. The subsequent washes were performed with 2× SSC, 0.5× SSC and 0.2× SSC successively. The mixture was then subjected to blocking, biotinylated binding, detection and washing. Coverslips were mounted onto slides in Prolong Gold mounting media with DAPI (Invitrogen, Shanghai, China). Fluorescence-labeled cells were imaged at 40× magnification using fluorescence microscopy (Nikon, Tokyo, Japan).
Statistical analysis
Graph data are presented as means ± SEM. Statistical analysis was performed using SPSS software. Unpaired, two-tailed t test was used to perform two group comparisons. The number of samples examined in each analysis was shown in the legends.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Science Foundation of China (No. 81773475, 21477031, 31401671), the National Key Basic Research Program of China (973 Program, No. 2012CB525003), the Fundamental Research Funds for the Central Universities (PA2017GDQT0018) and Key Laboratory of Xin'an Medicine Ministry of Education, Anhui University of Chinese Medicine (No. 2018xayx01).
Competing Interests
The authors have declared that no competing interest exists.
References
1. T Muto, G Sashida, M Oshima. et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. Journal of Experimental Medicine. 2013;210(12):2627-2639
2. Gan L, Yang Y, Li Q. et al. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. 2018;6:10
3. Prokopuk L, Hogg K, Western PS. Pharmacological inhibition of EZH2 disrupts the female germline epigenome. Clin Epigenetics. 2018;10:33
4. Pereira JD, Sansom SN, Smith J. et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107(36):15957-62
5. Zhang J, Ji F, Liu Y. et al. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014;34(15):5184-99
6. Sher F, Rossler R, Brouwer N. et al. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875-83
7. Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343-9
8. Guzman RE, Miranda-Laferte E, Franzen A. et al. Neuronal ClC-3 Splice Variants Differ in Subcellular Localizations, but Mediate Identical Transport Functions. J Biol Chem. 2015;290(43):25851-62
9. Yang WJ, Xu KK, Cong L. et al. Identification, mRNA expression, and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in oriental fruit fly, Bactrocera dorsalis. Int J Biol Sci. 2013;9(4):331-42
10. Grzenda A, Lomberk G, Svingen P. et al. Functional characterization of EZH2beta reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics Chromatin. 2013;6(1):3
11. Zhu H, Hasman R.A, Barron VA. et al. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell. 2006;17(12):5105-14
12. Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749-61
13. Pan Q, Shai O, Lee LJ. et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413-5
14. Jin N, Yin X, Gu J. et al. Truncation and Activation of Dual Specificity Tyrosine Phosphorylation-regulated Kinase 1A by Calpain I: A MOLECULAR MECHANISM LINKED TO TAU PATHOLOGY IN ALZHEIMER DISEASE. J Biol Chem. 2015;290(24):15219-37
15. Kim E, Ilagan JO, Liang Y. et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27(5):617-30
16. Shirahata-Adachi M, Iriyama C, Tomita A. et al. Altered EZH2 splicing and expression is associated with impaired histone H3 lysine 27 tri-Methylation in myelodysplastic syndrome. Leuk Res. 2017;63:90-97
17. Chen K, Xiao H, Zeng J. et al. Alternative Splicing of EZH2 pre-mRNA by SF3B3 Contributes to the Tumorigenic Potential of Renal Cancer. Clin Cancer Res. 2017;23(13):3428-3441
18. Arifuzzaman S, Das A, Kim SH. et al. Selective inhibition of EZH2 by a small molecule inhibitor regulates microglial gene expression essential for inflammation. Biochem Pharmacol. 2017;137:61-80
19. Su IH, Dobenecker MW, Dickinson E. et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121(3):425-36
20. Yan Q, Weyn-Vanhentenryck SM, Wu J. et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A. 2015;112(11):3445-50
21. Leggere JC, Saito Y, Darnell RB. et al. NOVA regulates Dcc alternative splicing during neuronal migration and axon guidance in the spinal cord. Elife. 2016:5
22. Irimia M, Weatheritt RJ, Ellis JD. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159(7):1511-23
23. Vire E, Brenner C, Deplus R. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871-4
24. Um JW, Choi TY, Kang H. et al. LRRTM3 Regulates Excitatory Synapse Development through Alternative Splicing and Neurexin Binding. Cell Rep. 2016;14(4):808-822
25. Aoto J, Martinelli DC, Malenka RC. et al. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154(1):75-88
26. Zheng S, Villa R, Wang J. et al. Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation. Genes Dev. 2014;28(23):2652-62
27. Zheng S, Wang J, Feng Y. et al. Solution structure of MSL2 CXC domain reveals an unusual Zn3Cys9 cluster and similarity to pre-SET domains of histone lysine methyltransferases. PLoS One. 2012;7(9e):45437
28. Joshi P, Carrington EA, Wang L. et al. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J Biol Chem. 2008;283(41):27757-66
29. Keith GP, Franklin BJ. The mouse brain in stereotaxic coordinates, second edition. 2001: A Harcourt Science and Technology Company.
30. Hashimoto K. and Ishima T. Neurite outgrowth mediated by translation elongation factor eEF1A1: a target for antiplatelet agent cilostazol. PLoS One. 2011;6(3e):17431
31. Walker AW, Martin JC, Scott P. et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26
32. Stansfield KH, Pilsner JR, Lu Q. et al. Dysregulation of BDNF-TrkB signaling in developing hippocampal neurons by Pb(2+): implications for an environmental basis of neurodevelopmental disorders. Toxicol Sci. 2012;127(1):277-95
Author contact
![]() Corresponding author: xuyiedu.cn
Corresponding author: xuyiedu.cn

 Global reach, higher impact
Global reach, higher impact