10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(2):481-492. doi:10.7150/ijbs.25862 This issue Cite
Research Paper
Generation of Pigs Resistant to Highly Pathogenic-Porcine Reproductive and Respiratory Syndrome Virus through Gene Editing of CD163
1. State Key Laboratory for Agrobiotechnology, China Agricultural University, Beijing, China
2. College of animal science and technology, China Agricultural University, Beijing, China
3. Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
4. National Research Center for Veterinary Medicine, Luoyang, Henan Province, China
5. College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou, Henan Province, China
Received 2018-3-4; Accepted 2018-6-10; Published 2019-1-1
Abstract
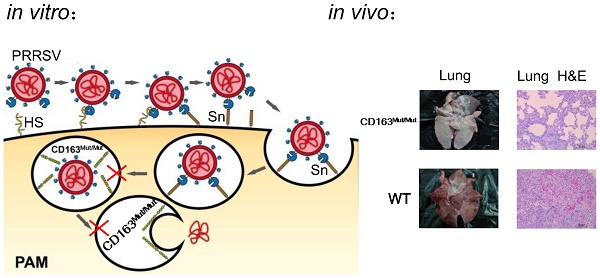
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious disease and the most economically important disease of the swine industry worldwide. Highly pathogenic-PRRS virus (HP-PRRSV) is a variant of PRRSV, which caused high morbidity and mortality. Scavenger receptor CD163, which contains nine scavenger receptor cysteine-rich (SRCR) domains, is a key entry mediator for PRRSV. A previous study demonstrated that SRCR domain 5 (SRCR5), encoded by exon 7, was essential for PRRSV infection in vitro. Here, we substituted exon 7 of porcine CD163 with the corresponding exon of human CD163-like 1 (hCD163L1) using a CRISPR/Cas9 system combined with a donor vector. In CD163Mut/Mut pigs, modifying CD163 gene had no adverse effects on hemoglobin-haptoglobin (Hb-Hp) complex clearance or erythroblast growth. In vitro infection experiments showed that the CD163 mutant strongly inhibited HP-PRRSV replication by inhibiting virus uncoating and genome release. Compared to wild-type (WT) pigs in vivo, HP-PRRSV-infected CD163Mut/Mut pigs showed a substantially decreased viral load in blood and relief from PRRSV-induced fever. While all WT pigs were dead, there of four CD163Mut/Mut pigs survived and recovered at the termination of the experiment. Our data demonstrated that modifying CD163 remarkably inhibited PRRSV replication and protected pigs from HP-PRRSV infection, thus establishing a good foundation for breeding PRRSV-resistant pigs via gene editing technology.
Keywords: HP-PRRSV, CD163, CRISPR/Cas9
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious disease and the most economically important disease of the swine industry worldwide (1, 2). PRRSV infection results in severe reproductive failure in gilts and respiratory disease in pigs of all ages and further complicates polymicrobial disease syndromes such as porcine circovirus-associated disease (3). PRRSV is a positive, single-stranded RNA virus belonging to family Arteriviridae, order Nidovirales (4). When infected by PRRSV, the pig host becomes immunologically inhibited and shows a much-delayed production of neutralizing antibodies against PRRSV (5, 6). PRRSV is highly prone to mutation, and thus, it is one of the most difficult viruses to control in pig production using traditional approaches (7, 8). In 2006, a highly pathogenic-PRRSV (HP-PRRSV) variant severely affected the swine industry of China and neighboring countries with a mortality rate of 20-100 %(9, 10).
In recent years, genetically modification technology has been widely used for anti-PRRSV research and has made good progress (11-14). Based on previous studies, we focused on the process of PRRSV entry. Three cellular factors, heparan sulfate (HS) (15), CD163 (16) and CD169 (17), have been identified as having PRRSV entry mediator activity. In a general model for cell entry, after binding to HS, PRRSV is internalized via CD169-mediated endocytosis and is uncoated by CD163 in the endosome, which subsequently releases the viral genome into the cytoplasm (18). However, HS molecules and CD169 are not strictly required for PRRSV to infect pigs (19). Pigs with defective CD163 were resistant to PRRSV (20), showing that CD163 is the most important receptor.
CD163 is a macrophage differentiation antigen belonging to a membrane protein in the scavenger receptor cysteine-rich (SRCR) family (21). CD163 was first described as an endocytic receptor that binds a complex of hemoglobin (Hb) and a plasma protein haptoglobin (Hp) (22), and further study demonstrated that the amino-terminal third of the SRCR region in CD163 was crucial for this interaction (23). CD163 was also found to function as an erythroblast adhesion receptor in erythroblastic islands and interact with its ligand via its SRCR domain 2 (24, 25). SRCR domain deletion and replacement experiments revealed that SRCR domain 5 (SRCR5) of porcine CD163 (pCD163) was essential for PRRSV infection (26). Replacing CD163 SRCR5 with the corresponding domain of human CD163L1 (hCD163L1) resulted in a loss of infectivity, and thus, it may be more advantageous to delete or modify only domain 5 than to delete the whole CD163 gene. Here, we report the efficient generation of biallelic exon replacement pigs using the CRISPR/Cas9 system combined with a donor vector. In this study, our data demonstrated that modifying CD163 remarkably inhibited PRRSV replication in vitro and in vivo. These findings suggest that modifying CD163 may provide a potential strategy for anti-PRRSV therapies.
Results
Generation and characterization of pigs with CD163-biallelic modification
To replace endogenous CD163 SRCR5 with the corresponding domain of hCD163L1, we constructed a pX330-501 vector and a donor vector. pX330-501 vector was designed to target exon 7 of CD163. T7 endonuclease 1 (T7E1) analysis of PCR products demonstrated that pX330-501 efficiently mutated the endogenous CD163 site in porcine fetal fibroblasts. The circular donor vector containing exon 11 of hCD163L1, part of pCD163 intron 6, a drug-selectable marker flanked by two loxP sites and two homologous arms was used as a template to repair a double-strand break by homologous recombination (Fig. 1A). The pX330-501 plasmid (1 μg/µl) was pooled with the linearized donor vector (1 μg/µl) and transfected into porcine fetal fibroblasts.
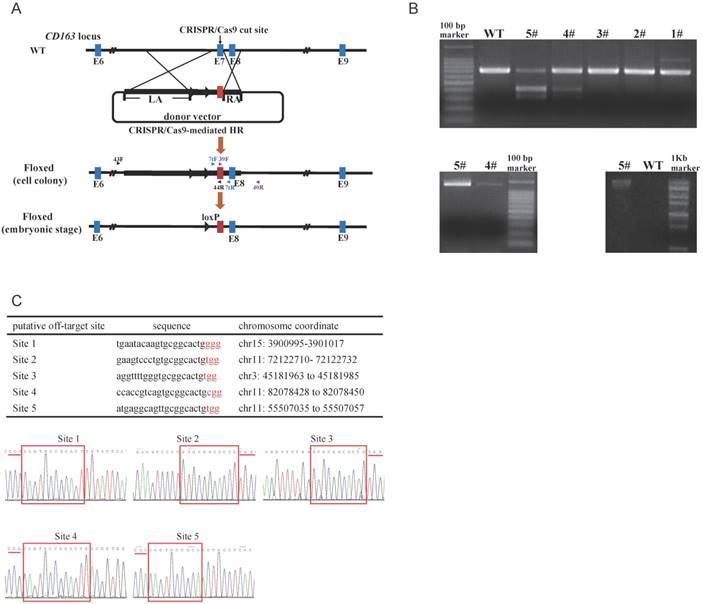
To detect targeting events, three pairs of primers were used (Fig. 1A). The amplified fragment (primers: CD7tF, CD7tR) was either exon 7 of pCD163 or exon 11 of hCD163L1, and we distinguished between the two fragments using restriction endonuclease (BbsI) reaction, such that exon 11 of hCD163L1, but not exon 7 of pCD163, could be digested by BbsI. The second amplified fragment (primers: 39F, 40R) was the right arm, while the third (primers: 43F, 44R) was the left arm and the drug-selectable marker (Fig. 1B). Interestingly, the PCR product (CD7tF, CD7R) of colony #5 was almost completely digested by BbsI, suggesting a potential biallelic modification. We tested the five potential off-target sites in colony #5. Sanger sequencing analysis of PCR products showed that colony #5 lacked off-target mutations (Fig. 1C) and thus was used as a nuclear donor for somatic cell cloning. A total of 4 cloned pigs were born, and the same detection strategy used to characterize cell colonies was performed. The results confirmed that all cloned pigs were positive for the CD163 mutant. Moreover, Sanger sequencing of the PCR products (CD7tF, CD7R) and Southern blotting indicated that the pigs were all biallelic modified (submitted for publication). Through the second batch of cell colony screening and somatic cell nuclear transfer (SCNT), we additionally created 11 biallelic modified CD163 piglets.
Expression of CD163 and CD169 in PAMs from CD163Mut/Mut pigs
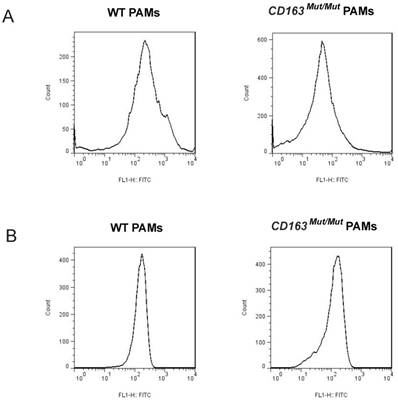
CD163 and CD169 play a pivotal role in virion entry. Thus, we examined the expression level of CD163 and CD169 in porcine alveolar macrophages (PAMs) using fluorescence-activated cell sorting (FACS). PAMs were collected by lung lavage, stained for surface expression of CD163 (clone EDHu-1) and CD169 (clone 3B11/11). As shown in Fig. 2A, in PAMs from CD163Mut/Mut pigs, the surface expression of CD163 was reduced compared to the WT PAMs, ranging from slightly detectable CD163 to a moderate level. However, CD169 expression levels were similar for both WT and CD163Mut/Mut pigs, and thus, the modified CD163 gene did not alter the expression of the PRRSV coreceptor CD169 (Fig. 2B).
Generation and characterization of pigs with CD163-biallelic modification. (A) A schematic overview of the strategy for generating a modified CD163 allele. sgRNA targeting site is shown as a black arrow. The donor vector was designed to substitute the pCD163 exon 7 with the corresponding exon of hCD163L1, and thus, the drug-selectable marker gene was flanked by two loxP sites. During the embryonic stage, Cre/loxP-mediated recombination resulted in excision of the drug-selectable marker gene, leaving one loxP site in intron 6 of pCD163. Homologous arms of the donor vector were indicated as LA (6392 bp) and RA (999 bp). (B) Identification of fibroblast colonies. (top) Identification of PCR products from colonies #1-5 by restriction endonuclease digestion. The unmodified genome PCR product could not be digested, and exon 7 substitution could result in two bands of ~307 bp and ~369 bp. (bottom left)PCR amplification of colony #4 and #5 genomes at the right flanking region and exon 11. The modified genome PCR was predicted to result in a 1317 bp product. (bottom right) PCR amplification of colony #5 genome at the left flanking region and exon 11. The modified genome PCR was predicted to result in a 12544 bp product. (C) Off-target analysis in cell colony #5. (top) Summary of putative off-target sites for the pX330-501 plasmid. (bottom) Off-target efficiency of cell colony #5 was assessed by Sanger sequencing. PAM site is underlined by a red line, and homologous sequence is shown in a red box.
CD163 gene modification does not affect red blood cell counts or circulating haptoglobin levels
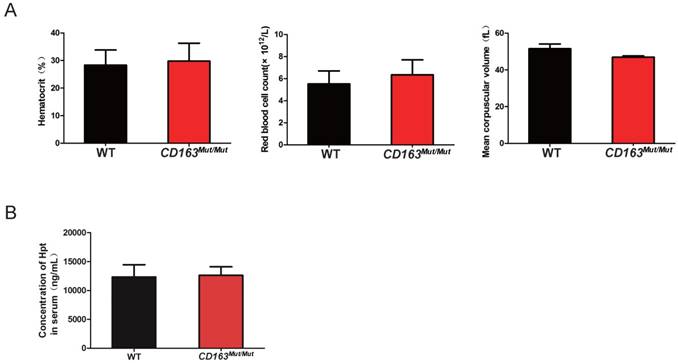
As an erythroblast adhesion receptor, CD163 is responsible for promoting erythroblast growth and survival. Thus, pigs were subjected to routine blood examinations to determine whether the CD163 modification influenced its function as an erythroblast adhesion receptor. Blood samples were collected from WT and CD163Mut/Mut pigs at four weeks of age. As shown in Fig. 3A, there was no significant difference in the blood hematocrit level, red blood cell count or mean corpuscular volume between the WT and CD163Mut/Mut groups. In addition, CD163 has been described to function as an Hb-Hp complex scavenger receptor. Upon release into circulation, free Hb binds to the plasma glycoprotein Hp, leading to exposure of an epitope that then interacts with the third SRCR domain of CD163. Hb-Hp complexes are internalized to early endosomes and degraded. Hp levels in sera from WT and CD163Mut/Mut pigs were measured at four weeks of age, and results showed that there was no significant difference in Hp levels between the WT and CD163Mut/Mut groups (Fig. 3B). All these data demonstrated that the overall functional state of CD163Mut/Mut macrophages remained normal.
CD163Mut/Mut PAMs are remarkably resistant to HP-PRRSV infection
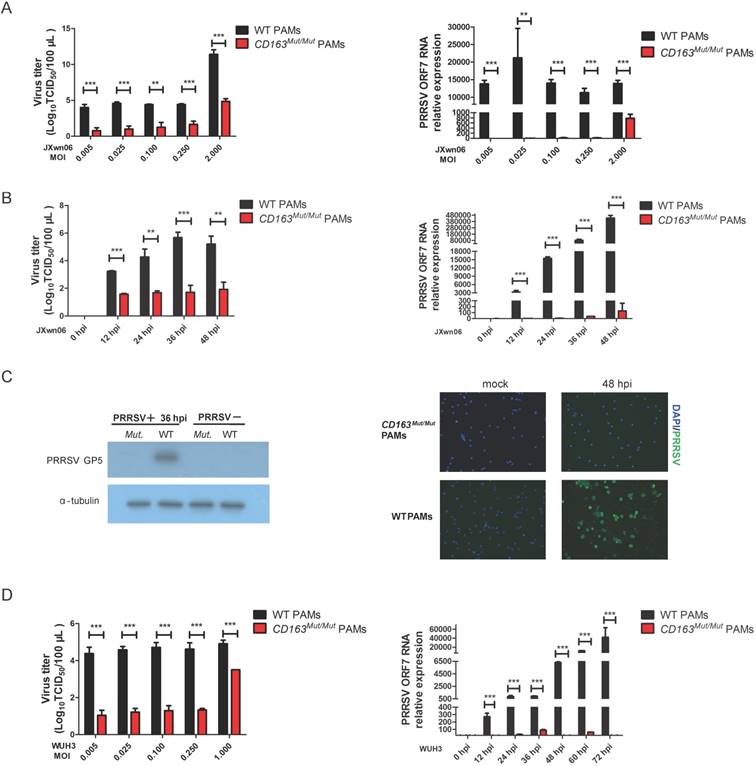
To investigate whether CD163 modification can inhibit PRRSV replication, CD163Mut/Mut and WT PAMs were infected with HP-PRRSV strain JXwn06 at several MOIs (0.005, 0.025, 0.1, 0.25 and 2.0). As expected, WT PAMs developed the cytopathic effect (CPE) after infection. However, in CD163Mut/Mut PAMs, no CPE was induced by JXwn06 at any MOI tested (Figure S1). At 36 h after infection, culture supernatants and infected cells were collected. As shown in Fig. 4A, the viral titers of the CD163Mut/Mut group were lower than those of the WT group, showing a statistically significant difference. Additionally, compared to viral gene expression in WT PAMs, expression in CD163Mut/Mut PAMs was significantly reduced (Fig. 4A).
Expression of CD163 and CD169 in PAMs from CD163Mut/Mut pigs. (A) Expression of CD163 on the surface of PAMs. PAMs were stained for CD163 (clone EDHu-1). (B) Expression of CD169 on the surface of PAMs. PAMs were stained for CD169 (clone 3B11/11). The y-axis shows the number of cells and the x-axis shows fluorescence intensity.
We next analyzed the virus-resistance activity at different time points after infection. CD163Mut/Mut and WT PAMs were seeded into 6-well plates and infected with the HP-PRRSV strain JXwn06 (MOI=0.1). At 0, 12, 24, 36 and 48 hpi, culture supernatants were measured by means of TCID50, and viral gene expression was analyzed by qPCR. Compared with WT PAMs, CD163 modification resulted in 100 to nearly 10000-fold reduction in virus yield during replication (12-48 hpi) (Fig. 4B). Consistent with viral titers, CD163 modification resulted in over 1000-fold reduction in the ORF7 gene expression of JXwn06 strain (Fig. 4B). To further confirm the effect of viral replication inhibition, we examined the expression of PRRSV GP5 and N (nucleocapsid) protein by Western blotting and immunofluorescence assay, respectively. As shown in Fig. 4C, compared to the control groups, PRRSV GP5 and N (Green) protein were not detected in CD163Mut/Mut PAMs.
Mutant CD163 protein still functions as an Hb-Hp scavenger and erythroblast adhesion receptor. (A) Effect of the modified CD163 gene on erythroblast growth. Routine blood examinations included measurements of hematocrit level (left), red blood cell count (middle) and mean corpuscular volume (right) in the venous blood of WT (n=6) and CD163-modified pigs (n=6). Data are presented as the mean±SD. (B) Serum Hp concentrations in WT (n=6) and CD163-modified pigs (n=6). Hp measurements were conducted on a single ELISA plate. Data are presented as the mean±SD.
To investigate whether CD163Mut/Mut PAMs were resistant to infection with another HP-PRRSV strain, viral infections with the HP-PRRSV WUH3 strain were also performed. Results of viral gene expression and viral titers were similar to those obtained with JXwn06 infection (Fig.4D). Taken together, these data suggested that CD163Mut/Mut PAMs were remarkably resistant to HP-PRRSV infection.
CD163 gene modification inhibits PRRSV replication but has no effect on PRRSV binding or internalization
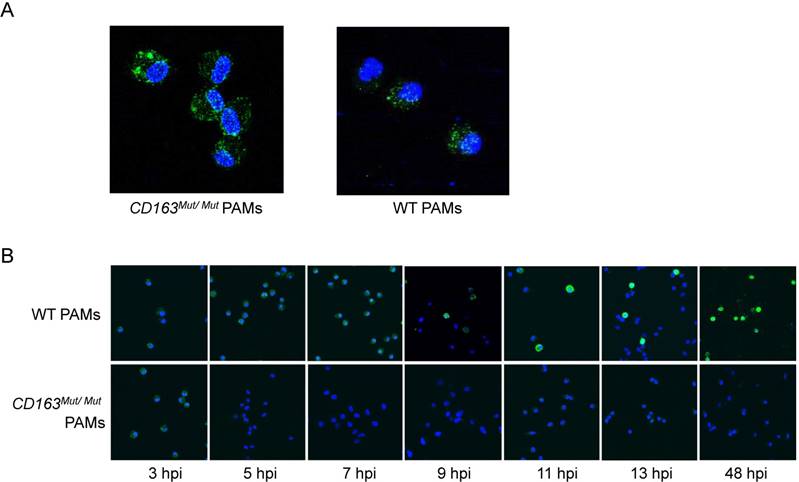
To further explore how the CD163 gene modification influences PRRSV infection, we first determined whether the mutant CD163 protein affects cell entry by PRRSV. Exploiting the fact that PRRSV can be stained with SDOW17-FITC, the process of PRRSV entry into PAMs was visualized by confocal microscopy. After PAMs were incubated with PRRSV at 37°C for 60 min, conditions under which viral attachment and internalization were reached, PAMs were stained with SDOW17-FITC and observed under the microscope. As expected, PRRSV virions bound as normal and were internalized by CD163Mut/Mut PAMs, confirming that modification of the CD163 gene has no effect on viral entry into the cells (Fig. 5A). Then, cells were incubated with PRRSV at 37°C for 5, 7, 9, 11, 13 and 48 h to allow viral internalization and replication. CD163Mut/Mut PAMs exhibited an obvious inhibitory effect on PRRSV replication, showing no viral replication in cells until 48 hpi (Fig. 5B). Taken together, these data suggest that the CD163 gene modification inhibits PRRSV replication but has no effect on PRRSV binding and internalization.
CD163Mut/Mut PAMs are remarkably resistant to HP-PRRSV infection. (A) After infection with the HP-PRRSV strain JXwn06 at the indicated MOIs (0.005, 0.025, 0.1, 0.25, 2.0), culture supernatants were collected at 36 hpi, and viral titers were analyzed by a standard TCID50 assay (left). Cells were collected to measure relative expression of viral RNA by qRT-PCR (right). GAPDH mRNA was used as an endogenous control. (B) After infection with HP-PRRSV strain JXwn06 at an MOI of 0.1, viral titers were measured by TCID50 at the indicated time points (12, 24, 36 and 48 h) (left). Relative expression of viral RNA was analyzed using qRT-PCR (right). GAPDH mRNA was used as an endogenous control. (C) PAMs were infected with JXwn06 at an MOI of 0.1, and 36 h later, levels of PRRSV protein GP5 were analyzed by Western blotting analysis (left). Expression of α-tubulin was shown as a loading control. After 48 h, cells were fixed for detection of PRRSV N protein (Green) by immunofluorescent staining(right). The nuclei (blue) were stained with DAPI. (D) The in vitro infection experiment was carried out with the HP-PRRSV strain WUH3. At the indicated MOIs (0.005, 0.025, 0.1, 0.25, 1.0), viral titers were analyzed by a standard TCID50 assay (left). After infection at an MOI of 0.025, relative expression of viral RNA was analyzed using qRT-PCR at the indicated time points (12, 24, 36, 48, 60 and 72 h). GAPDH mRNA was used as an endogenous control. Data are presented as the mean±SD, n=3. * P<0.05, ** P<0.01, *** P<0.001.
CD163 gene modification inhibits PRRSV replication but has no effect on PRRSV binding or internalization. (A) After incubating with PRRSV at 37°C for 1 h, PAMs were stained with SDOW17-FITC and observed by confocal microscopy. (B) PAMs were inoculated with PRRSV and fixed at different time points after inoculation as indicated under the images (hpi). PRRSV was visualized via SDOW17-FITC, which recognizes the viral nucleocapsid protein.
CD163 modification significantly inhibits PRRSV replication in pigs
Finally, to investigate whether CD163 mutant-mediated inhibition of PRRSV replication can be repeated in pigs, we tested the resistance of cloned CD163Mut/Mut pigs to HP-PRRSV infection by intramuscular injection. We chose 4 cloned CD163Mut/Mut pigs from the F0 generation and 6 WT pigs of the same age for the in vivo challenge experiments. After acclimation, pigs were infected with the HP-PRRSV strain JXA1 at 106.5 TCID50 per animal and followed for 21 days after infection.
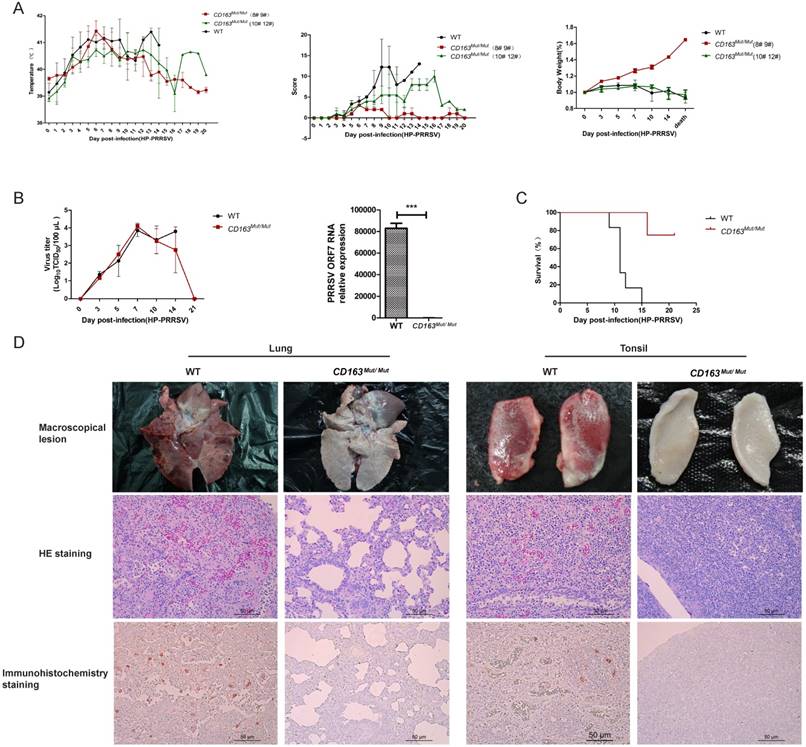
It is well known that HP-PRRSV strain infection is characterized by high fever and high mortality in pigs. Therefore, we recorded daily clinical signs, including rectal temperature, overall body condition and respiratory signs, of each pig until death. As shown in Fig. 6A, all challenged pigs exhibited a high fever (over 40°C) from 3 days post-infection (dpi), but two CD163Mut/Mut pigs (#8 and #9) recovered after 13 dpi. As expected, all WT pigs showed typical signs of PRRSV infection and weight loss (Fig. 6A). Clinical scores of the CD163Mut/Mut group were lower than the scores of the WT group. Specifically, pigs #8 and #9 showed mild clinical signs over the entire study period. The body weights of the two piglets were notably increased after infection (Fig. 6A). Because of a considerable difference between #8#9 pigs and #10#12 pigs in the CD163Mut/Mut group, the above data were analyzed separately.
PRRSV viremia during the study period was measured by TCID50 assay. These results showed that the mean viremia for WT pigs steadily increased until the pigs died. In the CD163Mut/Mut group (n=4), viremia was transient, reaching a peak at 7 dpi and rapidly declining thereafter. At the termination of the study (21 dpi), viremia in the CD163Mut/Mut pigs declined to negative levels (Fig. 6B). Notably, when died, the relative PRRSV RNA expression in the lungs of challenged CD163Mut/Mut pigs was significantly lower than that in the lungs of challenged WT pigs (P<0.001) (Fig. 6B). Challenged WT pigs showed a 0% survival rate, and three of four CD163Mut/Mut pigs survived to the end of the study (21 dpi) (Fig. 6C). Moreover, these three CD163Mut/Mut pigs recovered from the illness before euthanasia. Macroscopic lesions and hematoxylin/eosin (HE) staining of major tissues revealed that WT pigs had a severely abnormal pathology compared to CD163Mut/Mut pigs, which had no evidence of pathologic changes (Fig. 6D). Using immunohistochemical staining with an antibody specific to PRRSV, we found that a high percentage of positive viral protein signal could be observed in the tissues of WT pigs but not in CD163Mut/Mut pigs (Fig. 6D).
CD163 modification significantly inhibits PRRSV replication in pigs. (A) Rectal temperature curves (left), clinical sign score (middle) and body weight curves (right) of pigs from two challenged groups after PRRSV JXA1 infection. WT group, n=6. CD163Mut/Mut group, n=4. Scoring was based on the appearance of respiratory distress, inappetence, lethargy and fever (see 'Materials and methods'). Because of the considerable difference between #8 #9 pigs and #10 #12 pigs in the CD163Mut/Mut group, data for these pigs were analyzed separately. (B) Viral load in pigs of the two groups. (left) Analysis of viral load in the serum of pigs from the two groups at indicated time points. (right) Relative expression of viral RNA in lungs of the infected pigs. Samples were collected from the lungs of dying challenged pigs. Data in panels A-B are presented as the mean±SD. * P<0.05, ** P<0.01, *** P<0.001. (C) Survival curves for pigs from the two challenged groups after infection with PRRSV JXA1. CD163Mut/Mut pigs survived significantly longer than WT pigs. (D) Macroscopic lesion, histopathology and immunohistochemical staining of lungs and tonsils from infected pigs. (top) Diseased lungs and tonsils of pigs in different groups showed different damage extent. (middle) Representative photomicrographs of HE-stained tissues from WT and CD163Mut/Mut pigs. (bottom) Representative photomicrographs of immunohistochemically stained tissues from WT and CD163Mut/Mut pigs. PRRSV (red) was visualized via a monoclonal antibody recognizing the viral nucleocapsid protein.
Discussion
In this study, we successfully generated pigs in which the endogenous CD163 exon 7 was substituted with the corresponding exon of hCD163L1. Routine blood examinations and ELISA for Hp demonstrated that the overall functional state of CD163Mut/Mut macrophages remained normal. In vitro infection experiments demonstrated that CD163Mut/Mut PAMs were remarkably resistant to HP-PRRSV infection.. In vivo challenge experiments showed that the CD163 mutant protected pigs from HP-PRRSV infection. This study suggested that gene editing-mediated modification of key factors could play an important role in inhibiting PRRSV replication and could be used as a therapeutic tool for treating viral diseases.
Previous studies demonstrated that CD163 and CD169 were receptors for PRRSV. The efficacy of PRRSV infection in susceptible cells was significantly decreased after the receptor molecules were blocked by antibodies (27). Thus, depletion of these receptor genes through genetic modification is an attractive approach for inhibiting PRRSV infection in pigs. A previous study demonstrated that CD169-/- pigs followed the same clinical disease course and PRRSV viremia as WT pigs. However, pigs with defective CD163 were resistant to PRRSV (20). As a scavenger receptor, the biological role of CD163 mainly involves clearance of Hb and potential anti-inflammatory activity (22). CD163 protein consists of nine SRCR class B domains. Previous studies demonstrated that the SRCR domain 3 of CD163 is the critical determinant for the clearing of the Hb-Hp complexes (23). In addition, CD163 also functions as an erythroblast adhesion receptor, in which the SRCR domain 2 of CD163 displays specific erythroblast binding (24, 25). Thus, it may be more advantageous to delete or modify only domain 5, which is essential for PRRSV infection, than to delete the entire CD163 gene. Here, we generated pigs carrying biallelic modified CD163 using the CRISPR/Cas9 system combined with a circular donor vector. Challenge experiment results demonstrated that modification of CD163 remarkably inhibited PRRSV replication and had no effect on the biological role of CD163.
The in vitro infection experiments in the present study demonstrated that modification of CD163 gene remarkably inhibited PRRSV replication in PAMs. We further demonstrated that the virus-resistance effect was sustained and stable at several MOIs. In the entry process, HS and CD169 on PAMs contribute to the attachment and internalization of PRRSV virions. Subsequently, CD163 promotes uncoating and release of the viral genome from early endosomes into the cytoplasm. After PAMs were incubated with PRRSV at 37°C for 60 min, immunofluorescence showed that PRRSV virions could bind to and be internalized by CD163Mut/Mut PAMs. Longer incubation and observation showed that the PRRSV virions could not replicate after internalization. Consistent with previous studies, the modified CD163 gene inhibited PRRSV replication but had no effect on PRRSV binding and internalization.
Although CD163Mut/Mut pigs showed a transient fever and viremia after infection, we didn't detect respiratory signs, lung pathology and viral load in the lungs of CD163Mut/Mut pigs. These evidences demonstrated that these pigs were resistant against HP-PRRSV infection, especially pigs #8 and #9. Over the entire study period, pigs #8 and #9 showed mild clinical signs. The body weights of the two piglets were notably increased after infection. In addition, the PRRSV RNA relative expression in the lungs of the four cloned pigs was significantly lower than that in the lungs of challenged WT pigs (P<0.001). Fifteen days after infection, all six pigs in the WT group died, showing a 0% survival rate. However, in the CD163Mut/Mut group, only pig #10 died at 16 dpi, and the other three pigs (#8 #9 and #12) survived to the termination of the study. Although pig #10 did not survive, the PRRSV RNA relative expression in its lung showed that it effectively repressed PRRSV infection compared to the WT pigs. In the CD163Mut/Mut group, the virus-resistance effect in pigs #10 and #12 was weaker than that in pigs #8 and #9. Interestingly, the same cell colony was used to generate these four pigs. Considering that pigs #8 #9 were littermates and pigs #10 #12 were littermates in the F0 generation, we speculated that the weak virus-resistance effect in pigs #10 and #12 was due to the SCNT technology and background of the surrogate pig recipient.
In this study, we used highly pathogenic strain of PRRSV (HP-PRRSV) for challenge experiments, and this strain contains a unique 30-amino acid deletion in its nsp2 region and was isolated from diseased swine in China in 2006. Since then, HP-PRRSV has been the major epidemic viral strain in pigs in China and neighboring countries, causing enormous economic losses. In a recent publication, Burkard et al. excised exon 7 from the genome of pigs yielding a CD163 ΔSRCR5 genotype by using two CRISPR/Cas9 sgRNAs flanking exon 7 of CD163(28) , but this publication lacked in vivo challenge experiments. According to other publications, CD163-/- pigs were protected from PRRSV NVSL 97-7895(20), and CD163-HL11m pigs were protected from Type 1 PRRSV SD13-15(29). However, these PRRSV strains are not HP-PRRSV. In the present study, there of four CD163Mut/Mut pigs infected with HP-PRRSV survived and showed only slightly clinical signs compared with controls.
The results of the present study suggested that modifying CD163 remarkably inhibited HP-PRRSV replication and protected pigs from HP-PRRSV infection. Thus, the present study established a good foundation for breeding PRRSV-resistant pigs via gene editing technology.
Material and methods
Ethics statement
Experiments involving animals were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals in China. The protocol was approved by the Committee on the Ethics of Animal Experiments of China Agricultural University (Permit Numbers: SKLAB-2013-04-02, SKLAB-2013-04-03). In cages, the pigs were provided HEPA-filtered air, pig diet and tap water ad libitum. The environmental conditions included a controlled light cycle (9 h light), temperature (20-26˚C) and air humidity (40%-60%). Any animal that displayed excessive infection was immediately euthanized via CO2 asphyxiation. At the termination of the study, survived pigs were euthanized by lethal CO2 overdoses. The pigs used for obtaining PAMs were euthanized by sodium pentobarbital overdose.
Plasmids, cells and viruses
pX330 vector was purchased from Addgene. Based on exon 7 sequence of CD163, we designed gRNAs. The paired synthesized oligonucleotides were 501F (5'-CACCGGGAACTACAGTGCGGCACTG-3') and 501R (5'- AAACCAGTGCCGCACTGTAGTTCCC-3'), which were annealed and cloned into the pX330 plasmid (Addgene) following the Zhang laboratory protocol (30).
The donor vector was constructed on the basis of porcine CD163 genomic sequence and human CD163L1 cDNA sequence (NCBI Reference Sequence: NM_001297650). The 5' and 3' arms of the construct were amplified by PCR from DNA isolated from the fetal fibroblasts that would be used for later transfections. The 5' arm was 6392 bp and the 3' arm was 999 bp. The 315 bp replacement exon was CD163L1 exon 11. The inserted fragment and two arms were cloned into pPGKneo plasmid (provided by Dr. Yu Zhengquan; China angricultural University, Beijing, China).
Marc-145 cells and porcine fetal fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Cat. 11995-073) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Porcine alveolar macrophages (PAMs) were obtained using lung lavage as previously described, and maintained in RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin. The porcine fetal fibroblasts were transfected with DNA constructs using an Amaxa Nucleofector Kit (Lonza) according to the manufacturer's instructions.
Three HP-PRRSV strains were used to infect PAMs or the animals: JXwn06 (GenBank accession No. EF641008.1), WUH3 (GenBank accession No. HM853673.2) and JXA1 (GenBank accession No. EF1122445). JXwn06 was a gift from Prof. Wenhai Feng of China Agricultural University. WUH3 was a gift from Prof. Shujun Zhang of Huazhong Agricultural University. JXA1 was a gift from Prof. Kegong Tian of National Research Center For Veterinary Medicine. The PRRSV strains were grown as previously described (31). And viral titers were determined by microtitration infectivity assay and expressed as TCID50 as previously described (32).
Generation of gene-modified pigs
Porcine fetal fibroblasts (from Landrace pigs) were established as described previously (33). Approximately 1 × 106 fetal fibroblasts were transfected with 1.7 μg pX330-501 vector and 3.3 μg donor vector. After 48 h, the cells were transferred to ten 10-cm plates with selective medium containing G418 (600 μg/mL, promega). After 10 days, the resistant colonies were selected and expanded, and a small sample of every colony was subjected to genotyping. DNA was extracted from these samples and subjected to PCR analysis using the following primers: 7tF/7tR, 39F/40R and 43F/44R. Using the primers 7tF (5'-TTCTCCCTCACCGAAATGCT-3')/7tR (5'-GCAGTGACGGAACAATCTCC-3'), the amplified fragments of the positive and negative colonies were both 703 bp, but we could distinguish them by the restriction endonuclease (BbsI) reaction. The primers used for the second PCR amplification were 39F (5'-AGATGCCATATCTCTTTCTG-3') and 40R (5'-ATATCGGAGATACCCACAGT-3'). The amplified fragment was 1317 bp in length and included the 3' arm sequence. The primers used for the third PCR amplification were 43F (5'-CTAACCAGTGGCTTTACACCAGGCA-3') and 44R (5'-CCCACAGAAAGAGATATGGCATCTCC-3'). The amplified fragment was 12544 bp and contained the 5' arm, the drug-selectable marker and part of intron 6. After identifying the positive clone, somatic cell nuclear transfer (SCNT) was conducted as described (33, 34). Approximately 400 reconstructed embryos were transferred to each surrogate sow, and pregnancy was checked using abdominal ultrasound examination at 1 month after the SCNT. Cloned pigs were delivered by natural birth after approximate 115 days.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol (Invitrogen) following manufacturer's instructions. M-MLV Reverse Transcriptase was used for reverse transcription according to the manufacturer's protocol (Promega). qRT-PCR analysis was performed in 96-well plates using the Roche Light Cycler 480 System (LC 480; Roche, Basel, Switzerland). The relative expression of these genes were calculated using the 2-ΔΔct method (35) and GAPDH mRNA was set up as endogenous controls. The primers used for qPCR amplification were ORF7-F (5'-AATAACAACGGCAAGCAGCA-3') and ORF7-R (5'-GCACAGTATGATGCGTCGGC-3'). qRT-PCR was performed on each sample in triplicate.
Western blotting
PAMs were lysated using immunoprecipitation (IP) lysis buffer with protease inhibitors (Biotechnology, China). Protein concentrations of the extracts were measured with a BCA assay (Beyotime). Equal amount of each sample was separated using sodium dodecyl-sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes, and then blocked with 5% milk in TBST at room temperature for one hour. PRRSV GP5 protein is probed with a specific polyclonal antibody (produced and characterized by Pro. Wenhai Feng). Then, blots were incubated with goat anti-rabbit secondary antibody (for the GP5 polyclonal antibody) (1:10,000 dilution, ZSGB-Bio) for one hour at room temperature, followed by wash in TBST. The PVDF membranes were then processed using a Super Signal West Pico Chemiluminescent Kit (Thermo Scientific) and exposed to autoradiography film.
Immunofluorescence assay (IFA)
PAMs were fixed with cold methanol-acetone (1:1) for 10 min at 4°C, washed three times with phosphate buffered saline (PBS), and blocked with 1% BSA-PBS for 30 min at 37°C. Then, the cells were incubated with SDOW17 (1:200, mAb; Rural Technologies) against PRRSV N protein at 37°C for 1 h. After three washes with PBS, the cells were stained with (FITC)-conjugated goat anti-mouse IgG antibody (1:1000, abcam) at 37°C for 1 h. Finally, the nuclei were stained with DAPI (1:1000, Beyotime) for 5 min. Stainings were observed using fluorescence microscopy.
Flow cytometry
PAMs were blocked with 1% BSA-PBS for 30 min at 37°C. Then the cells were incubated with antibodies against CD163 (mouse anti-human CD163 antibody, AbD Serotec, MCA1853) or CD169 (FITC-conjugated mouse anti-pig CD169 antibody, BIO-RAD, MCA2316F) at 37°C for 1 h. After washing with PBS three times, PAMs for detecting CD163 were incubated with Alexa Fluor 488-labeled goat anti-mouse IgG antibody (Invitrogen) at 37°C for 1 h. After washing with PBS three times, PAMs were immediately analyzed on a BD LSRFortessa flow cytometer (BD Biosciences) with FCS Express 5 software (De Novo Software). A minimum of 10,000 cells were analyzed for each sample.
ELISA
The amount of Hp in serum was measured using an ELISA kit for Haptoglobin (Cloud-Clone Corp Inc.) according to the manufacturer's instructions. Examination was performed on each sample in triplicate.
In vitro infection of PAMs with HP-PRRSV
PAMs were obtained using lung lavage from CD163Mut/Mut pigs and wild-type pigs of the same age and gender. Approximately 106 PAMs per well were added to 6 well plates and incubated overnight at 37oC in 5% CO2, then were gently washed to remove non-adherent cells. Viral infections were performed using PRRSV JXwn06 or WUH3 strains at several MOIs respectively. The PAMs and culture medium were harvested at different time points.
In vivo infection with HP-PRRSV
In vivo studies were carried out at National Research Center for Veterinary Medicine. Two groups were established: the CD163Mut/Mut group and the WT group (n = 4 for the CD163Mut/Mut group; n = 6 for the WT group). These ten healthy, 6-week-old piglets were brought into challenge facility. Before virus challenge, all piglets were confirmed to be negative for PRRSV infection. Pigs were infected by intramuscular injection with the HP-PRRSV strain JXA1 at 106.5 TCID50 per headage and followed for 21 days after infection. The rectal temperature was recorded daily, starting 2 days prior to challenge until death. PRRSV-induced clinical signs including respiratory distress, inappetence, lethargy and fever were recorded using a scoring system (36, 37). Clinical sign scores ranged from 0: normal, to 1: mild inappetence and/or lethargy, 2: cough and/or sneeze and/or rubefaction, 4: severe respiratory distress and/or blue ear and/or depletion of appetite. Blood samples were collected at 0, 3, 5, 7, 10, 14 and 21 dpi. Serum was separated from blood sample, and stored at -80°C. After euthanasia, pigs were necropsied and tissues were fixed in 10% buffered formalin for histopathology study.
Histopathology assay and immunohistochemistry staining
To evaluate histological lesions in the tissues of the different groups, hematoxylin/eosin (HE) staining was performed. The lung and tonsil were fixed in 10% buffered formalin, routinely processed, paraffin-embedded, and stained using Harris's HE staining. For immunohistochemistry, the mouse monoclonal antibody specific to PRRSV nucleocapsid protein was used as primary antibody. All steps were carried out as described previously (38). The slides were visualized by microscope photographs.
Abbreviations
PRRS: porcine reproductive and respiratory syndrome; PRRSV: porcine reproductive and respiratory syndrome virus; HP-PRRSV: highly pathogenic- porcine reproductive and respiratory syndrome virus; HS: heparan sulfate; SRCR: scavenger receptor cysteine-rich; Hb: hemoglobin; Hp: haptoglobin; pCD163: porcine CD163; hCD163L1: human CD163-like 1; CRISPR/Cas9: clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) gene 9; WT: wild-type; T7E1: T7 endonuclease 1; SCNT: somatic cell nuclear transfer; PAMs: porcine alveolar macrophages; FACS: fluorescence-activated cell sorting; CPE: cytopathic effect; MOI: multiplicity of infection; HE: hematoxylin/eosin; DMEM: Dulbecco's modified Eagle's medium; FBS: fetal bovine serum; qRT-PCR: quantitative real-time polymerase chain reaction; IP: immunoprecipitation; SDS-PAGE: sodium dodecyl-sulphate polyacrylamide gel electrophoresis; PVDF: polyvinylidene difluoride; IFA: immunofluorescence assay; N protein: nucleocapsid protein.
Supplementary Material
Supplementary figures.
Acknowledgements
This project was supported in part by Genetically modified major projects (Project Grant No. 2014ZX08009003-006) to Professor Xiaoxiang Hu, Genetically modified major projects (Project Grant No. 2011ZX08006) to Professor Ning Li, and National Key Research and Development Program (Grant No. 2016YFD0500703) to Professor Kegong Tian.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH. et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. Journal of the American Veterinary Medical Association. 2005;227(3):385-92
2. Nieuwenhuis N, Duinhof TF, van Nes A. Economic analysis of outbreaks of porcine reproductive and respiratory syndrome virus in nine sow herds. The Veterinary record. 2012;170(9):225
3. Rossow KD. Porcine reproductive and respiratory syndrome. Veterinary pathology. 1998;35(1):1-20
4. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17-37
5. Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK. et al. Porcine Reproductive and Respiratory Syndrome Virus Nonstructural Protein 1 beta Modulates Host Innate Immune Response by Antagonizing IRF3 Activation. Journal of virology. 2010;84(3):1574-84
6. Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R. et al. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology. 2002;302(1):9-20
7. Chen J, Liu T, Zhu CG, Jin YF, Zhang YZ. Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochemical genetics. 2006;44(9-10):425-35
8. Stadejek T, Oleksiewicz MB, Potapchuk D, Podgorska K. Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in eastern Europe support the definition of new genetic subtypes. J Gen Virol. 2006;87:1835-41
9. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B. et al. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Veterinary journal (London, England: 1997). 2007;174(3):577-84
10. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C. et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PloS one. 2007;2(6):e526
11. Guo XK, Zhang Q, Gao L, Li N, Chen XX, Feng WH. Increasing Expression of MicroRNA 181 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication and Has Implications for Controlling Virus Infection. Journal of virology. 2013;87(2):1159-71
12. He YX, Hua RH, Zhou YJ, Qiu HJ, Tong GZ. Interference of porcine reproductive and respiratory syndrome virus replication on MARC-145 cells using DNA-based short interfering RNAs. Antiviral research. 2007;74(2):83-91
13. Li L, Li Q, Bao Y, Li J, Chen Z, Yu X. et al. RNAi-based inhibition of porcine reproductive and respiratory syndrome virus replication in transgenic pigs. Journal of biotechnology. 2014;171:17-24
14. Lu T, Song Z, Li Q, Li Z, Wang M, Liu L. et al. Overexpression of Histone Deacetylase 6 Enhances Resistance to Porcine Reproductive and Respiratory Syndrome Virus in Pigs. PloS one. 2017;12(1):e0169317
15. Jusa ER, Inaba Y, Kouno M, Hirose O. Effect of heparin on infection of cells by porcine reproductive and respiratory syndrome virus. Am J Vet Res. 1997;58(5):488-91
16. Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG. et al. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. Journal of virology. 2007;81(14):7371-9
17. Crocker PR, Gordon S. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. The Journal of experimental medicine. 1986;164(6):1862-75
18. Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X. et al. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. The Journal of general virology. 2010;91(Pt 7):1659-67
19. Prather RS, Rowland RR, Ewen C, Trible B, Kerrigan M, Bawa B. et al. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. Journal of virology. 2013;87(17):9538-46
20. Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG. et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nature Biotechnology. 2016;34(1):20-2
21. Van Gorp H, Delputte PL, Nauwynck HJ. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Molecular immunology. 2010;47(7-8):1650-60
22. Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SKA. et al. Identification of the haemoglobin scavenger receptor. Nature. 2001;409(6817):198-201
23. Madsen M, Moller HJ, Nielsen MJ, Jacobsen C, Graversen JH, van den Berg T. et al. Molecular characterization of the haptoglobin center dot hemoglobin receptor CD163 - Ligand binding properties of the scavenger receptor cysteine-rich domain region. J Biol Chem. 2004;279(49):51561-7
24. Barbe E, Huitinga I, Dopp EA, Bauer J, Dijkstra CD. A novel bone marrow frozen section assay for studying hematopoietic interactions in situ: the role of stromal bone marrow macrophages in erythroblast binding. Journal of cell science. 1996;109( Pt 12):2937-45
25. Fabriek BO, Polfliet MM, Vloet RP, van der Schors RC, Ligtenberg AJ, Weaver LK. et al. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood. 2007;109(12):5223-9
26. Van Gorp H, Van Breedam W, Van Doorsselaere J, Delputte PL, Nauwynck HJ. Identification of the CD163 Protein Domains Involved in Infection of the Porcine Reproductive and Respiratory Syndrome Virus. Journal of virology. 2010;84(6):3101-5
27. Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. The Journal of general virology. 2008;89(Pt 12):2943-53
28. Burkard C, Lillico SG. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. Plos pathogens. 2017;13(2):e1006206
29. Wells KD, Bardot R, Whitworth KM, Trible BR, Fang Y, Mileham A. et al. Replacement of Porcine CD163 Scavenger Receptor Cysteine-Rich Domain 5 with a CD163-Like Homolog Confers Resistance of Pigs to Genotype 1 but Not Genotype 2 Porcine Reproductive and Respiratory Syndrome Virus. Journal of virology. 2017;91(2):11
30. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8(11):2281-308
31. Zhang Q, Guo XK, Gao L, Huang C, Li N, Jia X. et al. MicroRNA-23 inhibits PRRSV replication by directly targeting PRRSV RNA and possibly by upregulating type I interferons. Virology. 2014;450-451:182-95
32. Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G. et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nature medicine. 2005;11(1):56-62
33. Zhang Y, Pan D, Sun X, Sun G, Wang X, Liu X. et al. Production of porcine cloned transgenic embryos expressing green fluorescent protein by somatic cell nuclear transfer. Science in China Series C, Life sciences. 2006;49(2):164-71
34. Wei HX, Zhang K, Ma YF, Li Y, Li QY, Dai YP. et al. Stage-dependent effect of leptin on development of porcine embryos derived from parthenogenetic activation and transgenic somatic cell nuclear transfer. The Journal of reproduction and development. 2009;55(2):99-104
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402-8
36. Halbur P, Thanawongnuwech R, Brown G, Kinyon J, Roth J, Thacker E. et al. Efficacy of antimicrobial treatments and vaccination regimens for control of porcine reproductive and respiratory syndrome virus and Streptococcus suis coinfection of nursery pigs. Journal of clinical microbiology. 2000;38(3):1156-60
37. Halbur PG, Paul PS, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J Vet Diagn Invest. 1996;8(1):11-20
38. Zhang C, Guo L, Jia X, Wang T, Wang J, Sun Z. et al. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine. 2015;33(21):2432-7
Author contact
![]() Corresponding authors: Ning Li (ninglcauedu.cn), Xiaoxiang Hu (huxxedu.cn) and Kegong Tian (tiankgnet).
Corresponding authors: Ning Li (ninglcauedu.cn), Xiaoxiang Hu (huxxedu.cn) and Kegong Tian (tiankgnet).

 Global reach, higher impact
Global reach, higher impact