10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(3):657-667. doi:10.7150/ijbs.30099 This issue Cite
Research Paper
EBV miR-BART10-3p Promotes Cell Proliferation and Migration by Targeting DKK1
Department of Biomedicine & Health Sciences, Department of Medical Lifescience, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
Received 2018-9-20; Accepted 2018-12-19; Published 2019-1-24
Abstract
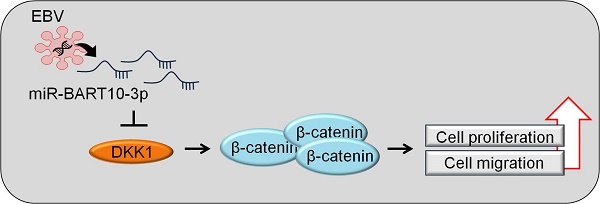
In Epstein-Barr virus (EBV)-infected epithelial cancers, BamHI A rightward transcript (BART) miRNAs are highly expressed. However, only a few target genes of BART miRNAs have been investigated. Our mRNA microarray data showed that DKK1 was markedly down-regulated in EBV-associated gastric carcinoma (EBVaGC) cells. Using luciferase reporter assay we tested whether miR-BART10-3p regulates DKK1 by directly targeting the 3'-UTR of DKK1 mRNA. We observed that miR-BART10-3p transfection decreased DKK1 expression, while an LNA inhibitor of miR-BART10-3p (LNA-miR-BART10-3p(i)) increased DKK1 expression. Furthermore, miR-BART10-3p and siDKK1 promoted cell proliferation and migration. In contrast, transfecting GC cells with LNA-miR-BART10-3p(i) or DKK1 over expression vector suppressed cell proliferation and migration. Our results suggest that miR-BART10-3p may be involved in the tumor progression of EBVaGC by targeting DKK1.
Keywords: Epstein-Barr virus, BART miRNA, DKK1, cell proliferation, cell migration
Introduction
Gastric carcinoma (GC) is the third leading cause of cancer mortality worldwide [1]. Most GC cases are diagnosed at an advanced stage, and the clinical outcomes remain unpredictable. Recently, the Cancer Genome Atlas (TCGA) classified GC into four subtypes based on molecular characteristics: microsatellite-unstable, genomically stable, chromosomally unstable, and Epstein-Barr virus (EBV)-associated [2]. As GC subtypes have distinct features, investigating potential targets in each subtype may provide guidelines for treating different GC patient populations.
EBV is a gamma herpesvirus harboring oncogenic DNA that infects more than 90% of the world's adult population. EBV is closely associated with several lymphoid and epithelial malignancies. EBV-associated GC (EBVaGC) accounts for almost 10% of GC cases, which is considerable because of the high incidence of GC. EBVaGC cells express restricted EBV latent genes, such as EBNA1, EBERs, BART microRNAs (miRNAs), and latent membrane protein 2A (LMP2A) [3-5].
MiRNAs are short, single-stranded RNAs about 22 nucleotides in length. They modulate gene expression by forming complementary duplexes with their target mRNAs, leading to translational inhibition and degradation of the target mRNAs. Single miRNA can regulate many targets, and more than one miRNA may target an individual mRNA [6-8]. Because miRNAs have the ability to inhibit gene expression, they play important roles in human cancers. For example, they regulate potential oncogenes or tumor suppressor genes [9, 10]. EBVaGC cells express high levels of BART miRNAs, which are encoded in the BamHI fragment A rightward transcript (BART) region [4, 11, 12]. By targeting cellular or viral genes, these miRNAs are involved in the regulation of multiple cellular responses such as host cell proliferation, apoptosis [12-15], and immune escape [16, 17]. Thus, EBV miRNAs are thought to contribute to the carcinogenesis of EBVaGC. Further studies are needed to elucidate the functions of most EBV-encoded miRNAs.
The Dickkopf (DKK) protein family consists of four members (DKK1~4) and a unique DKK3-related gene, Soggy (DKKL1). DKK1, the most studied member, is a soluble secreted protein involved in embryonic development. DKK1 is known as an antagonist of canonical Wnt signaling. DKK1 competitively interacts with a Wnt co-receptor (LDL receptor-related protein (LRP) 5 or LRP6), leading to the degradation of β-catenin [18-20]. DKK1 is also involved in various tumor processes such as cell proliferation, survival, migration, and invasion [21, 22]. However, the way in which DKK1 functions in EBVaGC cells has not been revealed.
In this study, we founded that DKK1 was markedly decreased in EBVaGC cell lines, and then investigated whether DKK1 was regulated by EBV BART miRNAs or not.
Materials and Methods
Cell culture and reagents
AGS is an EBV-negative gastric carcinoma cell line, while SNU-719 and AGS-EBV are EBV-positive gastric carcinoma cell lines [23, 24]. All gastric carcinoma cells were cultured in RPMI-1640 containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. AGS-EBV cells were AGS infected with a recombinant Akata virus [25]. To culture AGS-EBV cells, 400 µg/ml of G418 (Gibco, Carlsbad, CA, USA) was added to the medium. The human embryonic kidney cell line HEK293T was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. All cells were incubated at 37°C and supplemented with 5% CO2.
Target prediction
The DKK1 sequence used for miRNA target prediction was extracted from the National Center for Biotechnology Information database (NM_012242.3). To examine whether the 3′-UTR of DKK1 could be targeted by BART miRNAs, we used a publicly available RNA hybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). This tool finds the minimum free energy of hybridization needed for miRNAs to specific RNAs.
Transfection of miRNA mimics and LNA-miRNA inhibitors
All BART miRNA mimics and the scrambled control were purchased from Genolution Pharmaceuticals (Seoul, South Korea). The locked nucleic acid (LNA) inhibitor of miR-BART10-3p (LNA-miR-BART10-3p(i)) and the negative control LNA-miRNA inhibitor (control-LNA) were purchased from Exiqon (Vedbaek, Denmark). All transfection experiments were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Protein and RNA were extracted 48 h after transfection.
Plasmid constructs
The full-length 3'-UTR of DKK1 was amplified from the cDNA of AGS cells. The 3'-UTR of DKK1 was then cloned into XhoI/NotI sites located between the Renilla luciferase-coding sequence and the poly (A) site of the psiCHECK-2 plasmid (Promega, Madison, WI, USA) to produce psiC_DKK1. The primers used to amplify DKK1 were 5'-TCTAGGCGATCGCTCGAGACCAGCTATCCAAATGCAGT-3' and 5'-TTATTGCGGCCAGCGGCCGCAGGTATTATTAATTTATTGGAAACTATTTTTGA-3'. Mutations were introduced into the seed match sequence of psiC_DKK1 to produce psiC_DKK1m using an EZchange site-directed mutagenesis kit (Enzynomics, Daejeon, South Korea). The primers used for this purpose were 5'-ACAAAATTTTTGTACACATTGATTGTTATCTTGACTGA-3' and 5'-TATCAAGAGGAAAAATAGGCAGTGCAG-3'. The DKK1 coding sequences (DKK1- CDS) were amplified by using cDNA prepared from AGS cells. The primers used were 5'-TGCTAGCGGCCGCTCGAGATGATGGCTCTGGGCGCAGC-3' and 5'-CTTATCATGTCTGGATCCTTAGTGTCTCTGACAAGTGTG-3'. The amplicons were cloned into the XhoI/BamHI sites of the pCEP4 vector (Invitrogen) by using an EZ-Fusion cloning kit (Enzynomics, Daejeon, South Korea). The constructed DKK1 expression vector (pCEP4DKK1) contained a hygromycin selection marker for enrichment of transfected cells.
Preparation of DKK1 over-expressing cells
In order to establish stable cell lines, AGS-EBV cells were seeded into 100 mm cell culture dishes. After 24 h, the cells were transfected with 20 µg of empty vector (pCEP4) or pCEP4DKK1. After selection of transfected cells following 15 days of treatment with 200 µg/ml hygromycin, the cells were harvested to assess DKK1 expression.
Luciferase reporter assay for miRNA target analyses
To investigate the effect of BART miRNAs on the expression of DKK1, HEK293T cells or AGS cells were seeded in 96-well plates (5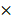 103 cells/well). After 24 h, the cells were co-transfected with 20 ng of psiC_DKK1 and 20 nM of indicated BART miRNA. To confirm sequence specific binding of miR-BART20-5p, a seed- sequence-mutated miR-BART10-3p (miR-BART10- 3pm) was also transfected. Luciferase activity was measured 48 h post-transfection using the Dual-Glo luciferase reporter assay system (Promega). For each sample, Renilla luciferase activity was normalized using firefly luciferase activity.
103 cells/well). After 24 h, the cells were co-transfected with 20 ng of psiC_DKK1 and 20 nM of indicated BART miRNA. To confirm sequence specific binding of miR-BART20-5p, a seed- sequence-mutated miR-BART10-3p (miR-BART10- 3pm) was also transfected. Luciferase activity was measured 48 h post-transfection using the Dual-Glo luciferase reporter assay system (Promega). For each sample, Renilla luciferase activity was normalized using firefly luciferase activity.
Quantitative reverse transcription PCR (qRT-PCR) for DKK1
AGS, AGS-EBV, or SNU-719 cells were harvested, and total RNA was extracted using the RNAiso Plus reagent (TaKaRa, Tokyo, Japan) according to the manufacturer's instructions. cDNA was synthesized using 3 μg total RNA, oligo(dT) primers (Macrogen, Seoul, South Korea), and Moloney murine leukemia virus reverse transcriptase (Invitrogen). Real-time PCR for the indicated genes was carried out using TOPrealTM Qpcr 2x Pre-MIX SYBR-Green (Enzynomics, Daejeon, Korea) with a real-time PCR system (CFX96, BioRad, Hercules, CA, USA). The sequences of the primers were 5'-AGCACCTTGGATGGGTATTC-3' and 5'-CACAATCCTGAGGCACAGTC-3' for DKK1, and 5'-ATGGGGAAGGTGAAGGTCG-3' and 5'-GGGGTCATTGATGGCAACAATA-3' for GAPDH. PCR conditions were 95°C for 10 min, followed by 35 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. To confirm specific amplification of the PCR product, dissociation curves were checked routinely. For this, reaction mixtures were incubated at 95°C for 60 s, and then ramped from 60°C to 95°C at a heating rate of 0.1°C/s, with fluorescence measured continuously. Relative gene expression was calculated using quantification cycle (Cq) values, with GAPDH as an internal standard.
Quantitative reverse transcription PCR for miRNA analyses
miRNA cDNA was synthesized using the Mir-X miRNA first-strand synthesis kit (Clontech, Mountain View, CA, USA) according to the manufacturer's instructions. Real-time quantitative PCR was performed using TOPrealTM Qpcr 2x Pre-MIX SYBR-Green (Enzynomics, Daejeon, Korea). The forward primer used for miR-BART10-3p was 5'-TACATAACCATGGAGTTGGCTGT-3'. All amplifications were performed in triplicate, and values were normalized to the value for an endogenous control, U6. The U6 RT primer was supplied in the kit.
Cell proliferation assay
Cell proliferation was analyzed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Amresco, Shanghai, China). AGS-EBV, SNU-719, or DKK1 over-expressing AGS-EBV cells ( cells/well) were seeded into 96-well plates. After the indicated periods following transfection, 20 µl of MTT solution (5 mg/ml) was added to each well. The absorbance at 595 nm was measured with a SoftMax apparatus (Molecular Devices, Sunnyvale, CA, USA) 4 h after adding MTT solution.
cells/well) were seeded into 96-well plates. After the indicated periods following transfection, 20 µl of MTT solution (5 mg/ml) was added to each well. The absorbance at 595 nm was measured with a SoftMax apparatus (Molecular Devices, Sunnyvale, CA, USA) 4 h after adding MTT solution.
Knocking down DKK1 with small interfering RNAs (siRNAs)
Small interfering RNAs (siRNAs) specific for DKK1 (siDKK1), and a control siRNA lacking any known target, were synthesized by Bioneer (Daejon, South Korea). The sequence of the siDKK1 was 5'-CACUAAACCAGCUAUCCAA-3'. AGS-EBV cells (1  106 cells/dish) were transfected with 30 nM siRNA using Lipofectamine 2000 (Invitrogen) in 100 mm dishes. Cells were harvested to analyze DKK1 expression 48 h after transfection.
106 cells/dish) were transfected with 30 nM siRNA using Lipofectamine 2000 (Invitrogen) in 100 mm dishes. Cells were harvested to analyze DKK1 expression 48 h after transfection.
Western blot analyses
Cell lysates in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, and 10 µg/ml aprotinin) were mixed with 5 loading buffer (Fermentas, Waltham, MA, USA) and heated at 95°C for 5 min. Samples were separated electrophoretically on 8% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Membranes were blocked and probed with the following antibodies: rabbit anti-DKK1 (1:500; Cell Signaling Technology, Danvers, MA, USA), mouse anti-β-catenin (1:500; Santa Cruz Biotechnology), and rabbit anti-β-actin (1:3,000; Cell Signaling Technology). Bound antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibodies (Santa Cruz Biotechnology) at a dilution of 1:3,000 for 45 min at room temperature. Protein bands were visualized using an enhanced chemiluminescence detection system (Amersham Bioscience, GE Healthcare, Piscataway, NJ, USA), and the membrane was exposed to X-ray film (Agfa, Mortsel, Belgium). Anti-β-actin antibody was used to confirm that loading was comparable between gel lanes. The density of each protein band was read and quantified using ImageJ software.
loading buffer (Fermentas, Waltham, MA, USA) and heated at 95°C for 5 min. Samples were separated electrophoretically on 8% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Membranes were blocked and probed with the following antibodies: rabbit anti-DKK1 (1:500; Cell Signaling Technology, Danvers, MA, USA), mouse anti-β-catenin (1:500; Santa Cruz Biotechnology), and rabbit anti-β-actin (1:3,000; Cell Signaling Technology). Bound antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibodies (Santa Cruz Biotechnology) at a dilution of 1:3,000 for 45 min at room temperature. Protein bands were visualized using an enhanced chemiluminescence detection system (Amersham Bioscience, GE Healthcare, Piscataway, NJ, USA), and the membrane was exposed to X-ray film (Agfa, Mortsel, Belgium). Anti-β-actin antibody was used to confirm that loading was comparable between gel lanes. The density of each protein band was read and quantified using ImageJ software.
Wound healing assay
To study the effect of miR-BART10-3p on cell migration, cells (3 x 105 cells/well) were seeded into 12-well plates and allowed to reach 90-95% confluence. Cells were scratched with a sterile 200 µl pipette tip through the confluent monolayer and subsequently washed with phosphate buffered saline to remove cell debris. Four hours later, the cells were transfected with miR-BART10-3p, LNA-miR-BART10-3p(i), siDKK1, or pCEP4DKK1. The cells were then cultured in RPMI-1640 medium containing 3% FBS at 37˚C in a humidified chamber supplied with 5% CO2. The scratched wounds were observed by an Axiovert 200 (Carl Zeiss, Thornwood, NY, USA) microscope just after transfection (time 0) and 48 h after transfection. Photographs were taken to assess the level of migration in each group of transfected cells, and wound areas were measured by ImageJ software. The experiments were performed in triplicate.
Statistical analyses
The data were analyzed using the Student's t-test, two-way repeated-measure analysis of variance (ANOVA), or curve fit. Analyses were performed using GraphPad Prism software (GraphPad Software, San Diego, CA, USA). P-values <0.05 were considered statistically significant. All results were expressed as the mean 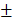 standard deviation (SD).
standard deviation (SD).
Results
DKK1 is down-regulated in AGS-EBV cells
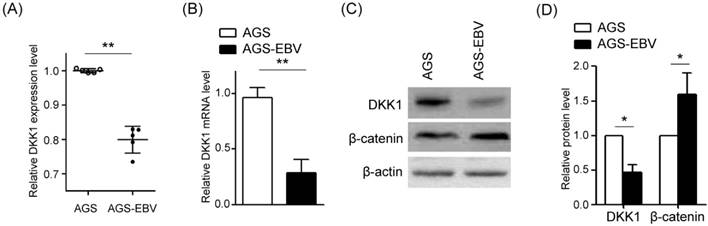
To analyze the effect of EBV infection on DKK1 expression, we compared microarray data [13] between AGS and AGS-EBV cells. Noticeably, DKK1 showed reduced expression in EBV-infected cells (Fig. 1A). To validate these data, DKK1 expressions in AGS and AGS-EBV cell lines were determined by real-time RT-PCR and Western blot. We found that DKK1 mRNA and protein levels decreased in AGS-EBV cells by 71% and 47%, respectively, compared to those in AGS cells (Fig. 1B-D). We also analyzed β-catenin expression to assess the possible relationship between DKK1 and β-catenin. The results showed that β-catenin protein levels are elevated in AGS-EBV cells than in AGS cells (Fig. 1D).
Reduction of DKK1 expression in EBV-infected GC cells. (A) Microarray analyses of AGS and AGS-EBV cells (n=5). (B) DKK1 mRNA expression was measured by real-time RT-PCR using a SYBR green qPCR kit. (C) DKK1 and β-catenin protein levels were assessed by Western blot analysis. Anti-β-actin antibody was used as a loading control. (D) Western blots were performed on two additional sets of independently cultured cells. Results were normalized using β-actin and expressed as ratios relative to values obtained from AGS cells. Error bars indicate the SD (n=3).
Screening BART miRNAs for the ability to target DKK1
To test whether EBV miRNAs down-regulate DKK1 expression in EBV-positive cells, EBV miRNAs showing a seed match to DKK1 were identified using an RNA hybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Four BART miRNAs (miR-BART5-5p, miR-BART8-3p, miR-BART10-3p, and miR-BART17-5p) were selected to further analyze their possible interaction with the 3'-UTR of DKK1 (Fig. 2A).
To investigate whether these miRNAs directly targeted DKK1, we constructed a luciferase reporter vector containing the entire 3'-UTR region of DKK1 (psiC_DKK1). The psiC_DKK1 reporter vector and each of the four BART miRNA mimics were co-transfected into HEK293T cells. Only the miR-BART10-3p mimic significantly reduced Luciferase activity when compared to the scrambled control in HEK293T cells (Fig. 2B). Because the other mimics did not inhibit luciferase activity, miR-BART10-3p was selected for further studies.
miR-BART10-3p directly targets DKK1
To identify whether miR-BART10-3p specifically targets DKK1, miR-BART10-3pm, which contains a mutation at sites 1-3 of the seed sequence of miR-BART10-3p, was used for a luciferase assay (Fig. 2C). Unlike miR-BART10-3p, the mutant mimic (miR-BART10-3pm) failed to suppress luciferase activity in HEK293T cells (Fig. 2D). We also substituted sites 2, 3, 4, and 7 of the seed match sequence for miR-BART10-3p in psiC_DKK1 to prepare psiC_DKK1m (Fig. 2C). When psiC_DKK1m was transfected into HEK293T (Fig. 2D) or AGS (Fig. 2E), luciferase activity was not reduced with co-transfected miR-BART10-3p. These results suggest that miR-BART10-3p inhibits DKK1 expression through a sequence-specific interaction with the 3'-UTR of DKK1 mRNA.
We next tested the effect of endogenously expressed miR-BART10-3p on DKK1 expression in EBV-infected GC cells using an LNA-inhibitor (LNA-miR-BART10-3p(i)). When co-transfected into ASG-EBV or SNU-719 cells, LNA-miR-BART10-3p(i) increased the luciferase activity of psiC_DKK1, but not that of psiC_DKK1m (Fig. 2F). These results suggest that the endogenously expressed level of miR-BART10-3p in ASG-EBV and SNU-719 cells was sufficient to suppress DKK1 expression by specifically targeting the 3'-UTR of DKK1. Collectively, these results support the hypothesis that miR-BART10-3p binds to the 3'-UTR of DKK1 sequence specifically.
Screening of BART miRNAs targeting DKK1. (A) Seed match results between BART miRNAs and the 3'-UTR of DKK1 mRNA. (B) Luciferase activity was measured in HEK293T cells co-transfected with BART miRNA mimics and psiC_DKK1. (C) Sequences of the seed match sequence-mutated (psiC_DKK1m) 3'-UTR of DKK1 mRNA (upper panel) and mutant miR-BART10-3p (miR-BART10-3pm) (lower panel) are shown. (D) Luciferase activity was measured in HEK293T cells co-transfected with the miR-BART10-3p mimic or miR-BART10-3pm, and psiC_DKK1 or psiC_DKK1m. (E) Luciferase activity was measured in AGS cells co-transfected with 30 nM BART miRNA mimics, and wild type or mutant luciferase reporter vector. (F) Luciferase activity was measured in AGS-EBV (left panel) or SNU-719 (right panel) cells co-transfected with the control LNA or LNA-miR-BART10-3p(i), and psiC_DKK1 or psiC_DKK1m. Luciferase activity was measured 48 h after transfection. Luciferase activity was normalized using firefly luciferase activity, and expressed as a ratio to the luciferase activity obtained from the scrambled control-transfected cells. Error bars indicate the SD (n=3).
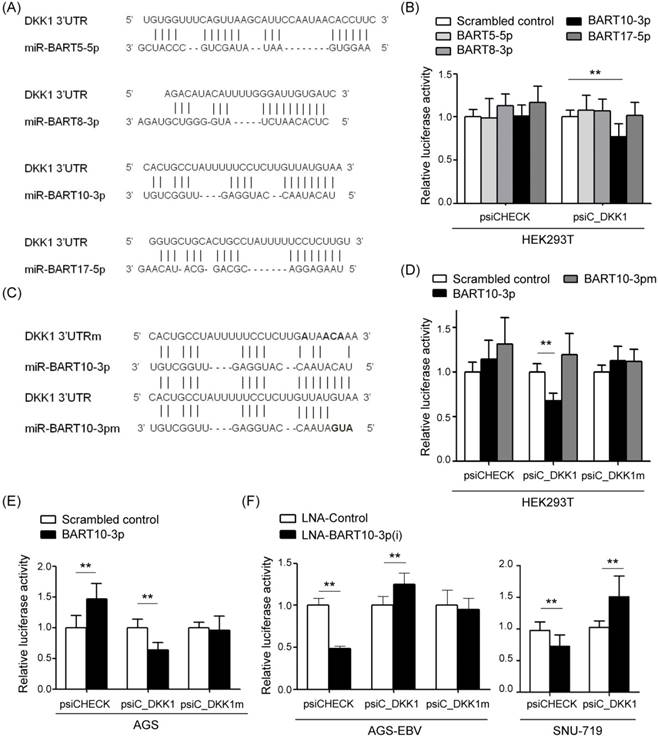
Unexpectedly, AGS cells co-transfected with the psiCHECK empty vector and miR-BART10-3p showed up-regulated luciferase activity (Fig 2E). Accordingly, both EBV infected GC cell lines (AGS-EBV and SNU-719) showed down-regulated luciferase activity when co-transfected with psiCHECK and LNA-miR-BART10-3p(i) (Fig 2F).
miR-BART10-3p regulates DKK1 expression, cell proliferation, and migration
To confirm the regulatory effect of miR-BART10-3p on DKK1, AGS cells were transfected with miR-BART10-3p, and DKK1 mRNA levels were quantified by real-time RT-PCR. Transfected miR-BART10-3p reduced DKK1 mRNA expression by 25% compared to the scrambled control in AGS cells (Fig. 3A). Likewise, a Western blot analysis revealed that DKK1 protein levels decreased approximately 60% following miR-BART10-3p transfection (Fig. 3B, C). These results support the hypothesis that miR-BART10-3p regulates DKK1 expression. We then investigated the effect of miR-BART10-3p on cell proliferation and migration. A cell proliferation assay revealed that miR-BART10-3p had a growth-enhancing effect on transfected AGS cells (Fig. 3D). A wound healing assay showed that AGS cell migration was significantly enhanced following miR-BART10-3p transfection, relative to transfection with a scrambled control (Fig. 3E, F).
An EBV miR-BART10-3p inhibitor increases DKK1 mRNA and protein expression
We next investigated whether the endogenously expressed level of miR-BART10-3p was sufficient to affect DKK1 and β-catenin expression in EBV-infected GC cells. Real-time RT-PCR and Western blot were performed to compare DKK1 levels after transfection with LNA-miR-BART10-3p(i) or with control LNA. In AGS-EBV cells, DKK1 mRNA and protein levels increased 2.9-fold and 6.5-fold, respectively, following LNA-miR-BART10-3p(i) transfection relative to cells transfected with control LNA. In SNU-719 cells, DKK1 mRNA and protein levels increased 1.3-fold and 2.9-fold, respectively, by LNA-miR-BART10-3p(i) transfection (Fig. 4A-C). These results indicate that endogenously expressed levels of miR-BART10-3p efficiently suppressed DKK1 in EBV-infected GC cells. In contrast, β-catenin protein levels decreased following by LNA-miR-BART10-3p(i) transfection in both AGS-EBV and SNU-719 cells. A cell proliferation assay showed reduced growth by LNA-miR-BART10-3p(i) transfection in both AGS-EBV and SNU-719 (Fig. 4D). Next, we examined the effect of miR-BART10-3p inhibition on cell migration. A wound healing assay showed suppressed cell migration following transfection with LNA-miR-BART10-3p(i) than control LNA (Fig. 4E, F).
Effect of miR-BART10-3p on DKK1 expression in AGS cells. AGS cells were transfected with 30 nM scrambled control or miR-BART10-3p. After 48 h, cells were harvested for RNA and protein preparation. (A) Real-time RT-PCR analysis was carried out to access DKK1 mRNA expression using a SYBR green qPCR kit. (B) DKK1 and β-catenin protein levels were analyzed by Western blot analysis. Anti-β-actin antibody was used as a loading control. (C) Western blots were performed on two additional sets of independently transfected AGS cells. Results were normalized to β-actin and are expressed as ratios. (D) At the indicated times after transfection, 20 µl of MTT solution was added to each well to assess cell proliferation. (E) A wound healing assay was performed to evaluate the effect of miR-BART10-3p on cell migration using transfected AGS cells. (F) Widths between wound edges were evaluated using ImageJ software. The percentages of wound closure from three independent experiments are shown as bar graphs. Error bars indicate the SD (n=3).
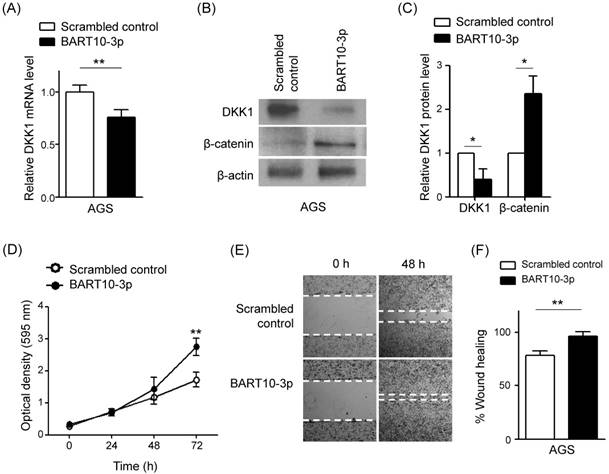
Effect of LNA-miR-BART10-3p(i) in AGS-EBV and SNU-719 cells. AGS-EBV or SNU-719 cells were transfected with 30 nM control LNA or LNA-miR-BART10-3p(i). After 48 h, cells were harvested for RNA and protein preparation. (A) Real-time RT-PCR analysis of DKK1 mRNA expression was carried out using a SYBR green qPCR kit. (B) DKK1 and β-catenin protein levels were analyzed by Western blot analysis. Anti-β-actin antibody was used as a loading control. (C) Western blots were performed on two additional sets of independently transfected AGS-EBV or SNU-719 cells. Results were normalized to β-actin and are expressed as ratios. (D) At the indicated times after transfection, 20 µl of MTT solution was added to each well to assess cell proliferation. (E) A wound healing assay was performed to evaluate the effect of LNA-miR-BART10-3p(i) on cell migration using transfected AGS-EBV cells. Widths between wound edges were evaluated using ImageJ software. The percentages of wound closure from three independent experiments are shown as bar graphs. Error bars indicate the SD (n=3).
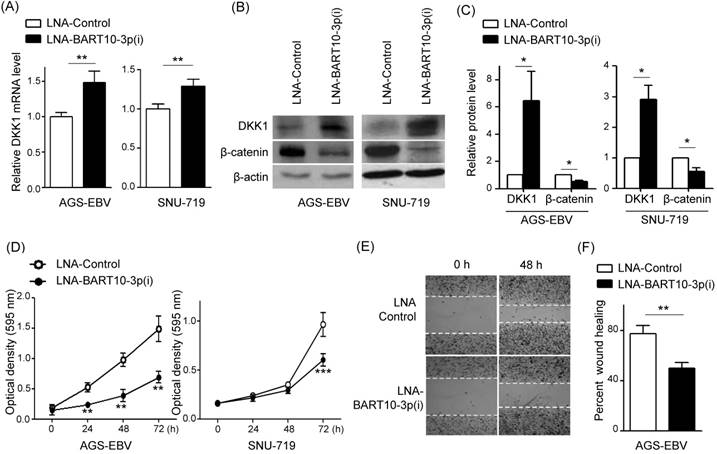
Knockdown of DKK1 induces cell proliferation and migration in AGS cells
To investigate whether siRNA targeting DKK1 (siDKK1) generates a similar phenotype to miR-BART10-3p, siDKK1 was transfected into AGS cells. After 48h, real-time RT-PCR and Western blot analyses were performed to assess DKK1 and β-catenin expression levels. The siDKK1 transfection of AGS cells efficiently knocked down DKK1 mRNA expression (by approximately 76%, Fig. 5A) and protein expression (by approximately 72%, Fig. 5B, C). In addition, β-catenin level was increased by knocking down DKK1. To further explore the role of DKK1 in AGS cells, we analyzed the effect of siDKK1 in controlling cell proliferation and migration. MTT and wound healing assays demonstrated that DKK1 knockdown by siDKK1 transfection promoted cell proliferation (Fig. 5D) and migration (Fig. 5E, F).
Knockdown of DKK1 induces cell proliferation and migration in AGS-EBV cells
The effect of DKK1 knockdown was accessed in AGS-EBV cells, too. After 48 h, real-time RT-PCR and Western blot analyses were performed to assess DKK1 and β-catenin expression levels. The siDKK1 transfection of AGS-EBV cells efficiently knocked down DKK1 mRNA expression (by approximately 65%, Fig. 6A) and protein expression (by approximately 68%, Fig. 6B, C). In addition, β-catenin level was increased by knocking down DKK1.
To further explore the role of DKK1 in AGS-EBV cells, we analyzed the effect of siDKK1 in controlling cell proliferation and migration. MTT and wound healing assays demonstrated that DKK1 knockdown by siDKK1 transfection promoted cell proliferation (Fig. 6D) and migration (Fig. 6E, F).
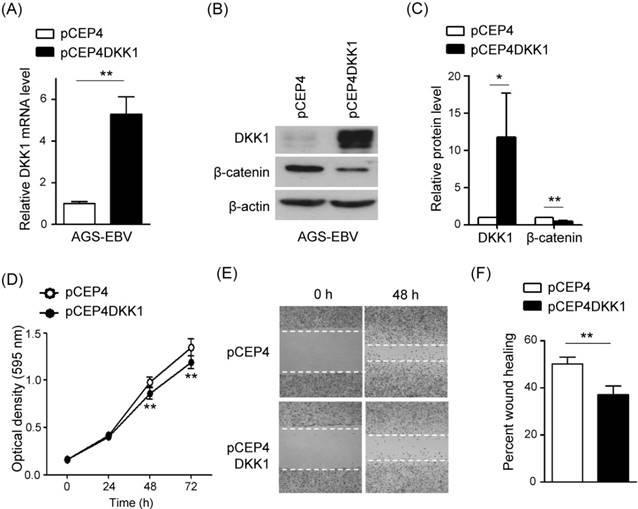
DKK1 over-expression inhibits cell proliferation and migration in AGS-EBV cells
In order to confirm the function of DKK1 in EBV-positive GC cells, we tested the effect of DKK1 over-expression on cell proliferation and migration. AGS-EBV cells were transfected with a DKK1 over-expression vector (pCEP4DKK1) or an empty control vector (pCEP4). After selecting for 15 days in 200 µg/ml hygromycin-containing media, the transfected cells were used for analyses. Real-time RT-PCR and Western blot analyses showed that DKK1 mRNA and protein levels increased 5.2-fold and 8.2-fold, respectively, in pCEP4DKK1-transfected AGS-EBV cells (Fig. 6A-C). Conversely, β-catenin level was suppressed by DKK1 overexpression. As expected, the over-expression of DKK1 in AGS-EBV cells suppressed cell proliferation and migration (Fig. 6D-F).
Effect of DKK1 knockdown using siDKK1 in AGS cells. AGS cells were transfected with 20 nM siDKK1 or control siRNA. (A) A real-time RT-PCR analysis of DKK1 mRNA expression was carried out using a SYBR green qPCR kit. (B) DKK1 and β-catenin protein levels were analyzed by Western blot analysis. Anti-β-actin antibody was used as a loading control. (C) Western blots were performed on two additional sets of independently transfected AGS cells. Results were normalized to β-actin and are expressed as ratios. (D) At the indicated times after transfection, 20 µl of MTT solution was added to each well to assess cell proliferation. (E) Widths between wound edges were evaluated using ImageJ software. The percentages of wound closure from three independent experiments are shown as bar graphs. Error bars indicate the SD (n=3).
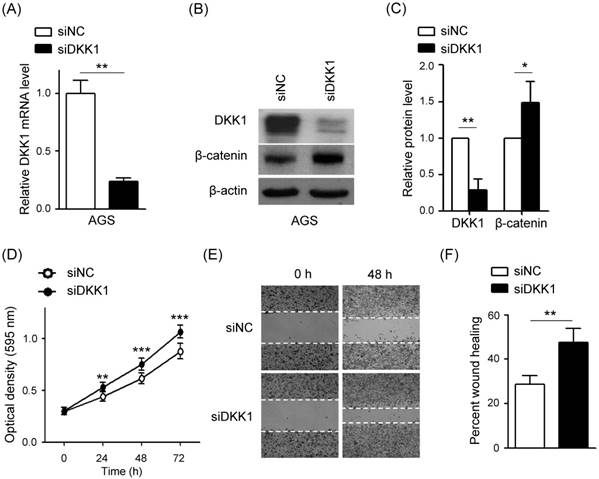
Effect of DKK1 knockdown using siDKK1 in AGS-EBV cells. AGS-EBV cells were transfected with 20 nM siDKK1 or control siRNA. (A) A real-time RT-PCR analysis of DKK1 mRNA expression was carried out using a SYBR green qPCR kit. (B) DKK1 and β-catenin protein levels were analyzed by Western blot analysis. Anti-β-actin antibody was used as a loading control. (C) Western blots were performed on two additional sets of independently transfected AGS-EBV cells. Results were normalized to β-actin and are expressed as ratios. (D) At the indicated times after transfection, 20 µl of MTT solution was added to each well to assess cell proliferation. (E) Widths between wound edges were evaluated using ImageJ software. The percentages of wound closure from three independent experiments are shown as bar graphs. Error bars indicate the SD (n=3).
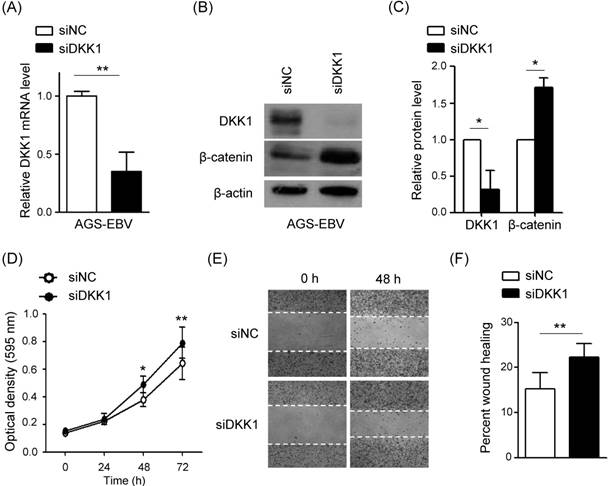
Effect of DKK1 over-expression in AGS-EBV cells. (A) A real-time RT-PCR analysis of DKK1 mRNA expression was carried out using a SYBR green qPCR kit. (B) DKK1 and β-catenin protein levels were analyzed by Western blot. Anti-β-actin antibody was used as a loading control. (C) Western blots were performed on two additional sets of independently transfected AGS-EBV cells. Results were normalized to β-actin and are expressed as ratios. (D) At the indicated times after transfection, 20 µl of MTT solution was added to each well to assess cell proliferation. (E) Widths between wound edges were evaluated using ImageJ software. The percentages of wound closure from three independent experiments are shown as bar graphs. Error bars indicate the SD (n=3).
Discussion
In this study, we demonstrated a role for miR-BART10-3p and DKK1 in the cell proliferation and migration of EBV-infected cells. Data from microarrays, real-time RT-PCR, and Western blot assays revealed that DKK1 expression was suppressed in EBV-infected cells. We therefore hypothesized that DKK1 may function as a tumor suppressor in EBV-infected GC cells. miR-BART10-3p reduced the expression of DKK1 by directly targeting the 3'-UTR of DKK1. Accordingly, the inhibitor of miR-BART10-3p rescued DKK1 expression in AGS-EBV and SNU-719 cells. In addition, miR-BART10-3p induced cell proliferation and migration, which was mimicked by siDKK1. In contrast, the inhibitor to miR-BART10-3p reduced cell proliferation and migration, as did over-expressed DKK1.
Unexpectedly, the luciferase activity of the psiCHECK control vector was enhanced by co-transfected miR-BART10-3p mimic (Fig. 2D, E) and suppressed by a co-transfected LNA-BART10-3p(i) inhibitor (Fig. 2F). To understand these observations, we have checked coincidental seed match between miR-BART10-3p and psiCHECK control vector, but could not find the possibility. Further investigations are warranted to clarify the underlying mechanisms of these observations.
In AGS and AGS-EBV cells, miR-BART10-3p enhanced cell proliferation and migration suggesting that BART10-3p may contribute to EBVaGC metastasis. Consistent with our findings, Yan et al. [26] demonstrated that miR-BART10-3p promotes the invasion and migration capabilities of nasopharyngeal carcinoma (NPC) cells. They showed that miR-BART10-3p targeted the beta-transducin repeat-containing E3 ubiquitin protein ligase (BTRC) in NPC. Taken together, miR-BART10-3p may induce metastasis in EBV-associated carcinomas such as NPC and GC by promoting cell growth and migration.
It is well known that DKK1 acts as an important negative regulator of Wnt/β-catenin signaling by directly binding to LRP5/6. In the absence of Wnt stimulation, β-catenin is phosphorylated by CK1α and GSK3β, and then degraded by β-TrCP-mediated ubiquitination. On the other hand, Wnt binding to the Frizzled (FZD) receptor and a co-receptor (LRP5/6) leads to the accumulation of β-catenin in the cytoplasm, allowing localization to the nucleus. There, β-catenin interacts with TCF/LEF to transactivate downstream genes including cyclin D1 and c-myc. Wnt target genes are diverse, and cell- and context-dependent [27, 28]. DKK1 has been shown to be regulated by TCF/β-catenin [21]. Accordingly, previous studies showed increase in c-myc, cyclin D1, and β-catenin expression in EBVaGC cells [29-32]. We also observed that AGS-EBV cells expressed increased levels of β-catenin, along with decreased levels of DKK1, relative to AGS cells.
Even though miR-BART10-3p transfected AGS-EBV cells showed decreased DKK1 expression at both mRNA and protein levels, there might be some other EBV factor(s) down regulating DKK1. For example, DKK1 promoter has 5' CpG islands, and aberrant methylation of this region has been implicated in DKK1 silencing observed in GC [33], colorectal cancer [34], cervical cancer [35], and multiple myeloma [36]. Notably, DNA methylation of both host and viral genomes is one of the features of EBVaGC [2, 37, 38].
DKK1 has been reported to be down- or up-regulated in various cancers and the role of DKK1 in cancer is controversial. DKK1 is elevated in multiple myeloma [39], breast cancer [40, 41], hepatocellular carcinoma [42], and prostate cancer [43, 44]. DKK1 has been shown to be a tumor suppressor as well. For example, DKK1 over-expression restored the epithelial phenotype and decreased expression of the EMT transcription factor in colon cancer [45]. DKK1 is down-regulated in metastatic LM-MCF-7 breast cancer cells, and involved in cell proliferation and dysregulation of the Wnt/β-catenin signaling pathway [46].
In GC, increased levels of serum DKK1 have been reported [47]. However, Sato et al. [33] have demonstrated that DKK family genes are frequent targets of epigenetic silencing in gastrointestinal tumors. Jia et al. [48] showed that DKK1 is decreased in the serum and tissues of GC patients. The same authors demonstrated that miR-493 promotes GC progression by suppressing DKK1 expression. We observed that DKK1 knockdown by siDKK1 transfection exerted similar effect with miR-BART10-3p. These results indicated that DKK1 play an important role in promoting gastric cancer cell migration and invasion. Furthermore, slightly bigger changes were observed in AGS cells than in AGS-EBV cells following DKK1 depletion. These may be due to the fact that expression of DKK1 is somewhat suppressed in AGS-EBV cells than in AGS by endogenously expressed miR-BART10-3p. Moreover, we showed reduced expression of DKK1 by miR-BART10-3p causing enhanced cell proliferation and migration in EBVaGC cells. Our data support the hypothesis that DKK1 functions as a tumor suppressor in EBV-associated tumors.
β-Catenin is not only a key protein in Wnt/β-catenin signaling, but also interacts with E-cadherin to play an important role in cell junctions. This binding prevents nuclear translocation of β-catenin, while loss of E-cadherin by such as tumor likely promotes β-catenin release from membrane, and promotes epithelial mesenchymal transition [49]. Here we found that DKK1 expression is inversely associated with β-catenin expression in EBVaGC. Our results support that β-catenin is regulated by DKK1.
Taken together, our findings demonstrate that miR-BART10-3p regulates EBVaGC cell proliferation and migration by directly targeting DKK1, which functions as a tumor suppressor. Thus, miR-BART10- 3p may serve as a potential therapeutic target for patients with EBVaGC.
Abbreviations
EBV: Epstein-Barr virus; BART: BamHI A rightward transcript; EBVaGC: EBV-associated gastric carcinoma; GC: gastric carcinoma; TCGA: The cancer genome atlas; DKK1: Dickkopf 1; miRNAs: microRNAs; UTR: Untranslated region; RT-PCR: reverse transcription polymerase chain reaction; LNA: locked nucleic acid; siNC: siRNA negative control.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2018R1D1A1B07041763).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359-86
2. The Cancer Genome Atlas Research N, Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-9
3. Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N. et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9131-5
4. Kim DN, Chae HS, Oh ST, Kang JH, Park CH, Park WS. et al. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. Journal of virology. 2007;81:1033-6
5. Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH. et al. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World journal of gastroenterology. 2005;11:629-33
6. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Current genomics. 2010;11:537-61
7. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduction And Targeted Therapy. 2016;1:15004
8. Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2010;1803:1231-43
9. Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer research. 2012;72:5576-87
10. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257-61
11. Barth S, Meister G, Grasser FA. EBV-encoded miRNAs. Biochimica et biophysica acta. 2011;1809:631-40
12. Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF. et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. The Journal of experimental medicine. 2008;205:2551-60
13. Kim H, Choi H, Lee SK. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer letters. 2015;356:733-42
14. Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. Journal of virology. 2013;87:8135-44
15. Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival. PLoS pathogens. 2015;11:e1004979
16. Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J. et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E6467-e75
17. Zhou L, Bu Y, Liang Y, Zhang F, Zhang H, Li S. Epstein-Barr Virus (EBV)-BamHI-A Rightward Transcript (BART)-6 and Cellular MicroRNA-142 Synergistically Compromise Immune Defense of Host Cells in EBV-Positive Burkitt Lymphoma. Medical science monitor: international medical journal of experimental and clinical research. 2016;22:4114-20
18. Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nature cell biology. 2001;3:683-6
19. Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A. et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321-5
20. Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Current biology: CB. 2001;11:951-61
21. Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y. et al. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520-6
22. Menezes ME, Devine DJ, Shevde LA, Samant RS. Dickkopf1: a tumor suppressor or metastasis promoter? International journal of cancer. 2012;130:1477-83
23. Oh ST, Seo JS, Moon UY, Kang KH, Shin DJ, Yoon SK. et al. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology. 2004;320:330-6
24. Park JG, Yang HK, Kim WH, Chung JK, Kang MS, Lee JH. et al. Establishment and characterization of human gastric carcinoma cell lines. International journal of cancer. 1997;70:443-9
25. Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nature medicine. 2002;8:594-9
26. Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766-82
27. Vlad A, Rohrs S, Klein-Hitpass L, Muller O. The first five years of the Wnt targetome. Cellular signalling. 2008;20:795-802
28. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781-810
29. Pajic A, Polack A, Staege MS, Spitkovsky D, Baier B, Bornkamm GW. et al. Elevated expression of c-myc in lymphoblastoid cells does not support an Epstein-Barr virus latency III-to-I switch. The Journal of general virology. 2001;82:3051-5
30. Zhu S, Sun P, Zhang Y, Yan L, Luo B. Expression of c-myc and PCNA in Epstein-Barr virus-associated gastric carcinoma. Experimental and therapeutic medicine. 2013;5:1030-4
31. Wiech T, Nikolopoulos E, Lassman S, Heidt T, Schopflin A, Sarbia M. et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Archiv: an international journal of pathology. 2008;452:621-7
32. Koriyama C, Akiba S, Itoh T, Sueyoshi K, Minakami Y, Corvalan A. et al. E-cadherin and beta-catenin expression in Epstein-Barr virus-associated gastric carcinoma and their prognostic significance. World journal of gastroenterology. 2007;13:3925-31
33. Sato H, Suzuki H, Toyota M, Nojima M, Maruyama R, Sasaki S. et al. Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis. 2007;28:2459-66
34. Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J. et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116-21
35. Lee J, Yoon YS, Chung JH. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecologic oncology. 2008;109:270-4
36. Kocemba KA, Groen RW, van Andel H, Kersten MJ, Mahtouk K, Spaargaren M. et al. Transcriptional silencing of the Wnt-antagonist DKK1 by promoter methylation is associated with enhanced Wnt signaling in advanced multiple myeloma. PloS one. 2012;7:e30359
37. Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ. et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Laboratory investigation; a journal of technical methods and pathology. 2008;88:161-70
38. Shinozaki-Ushiku A, Kunita A, Fukayama M. Update on Epstein-Barr virus and gastric cancer (review). International journal of oncology. 2015;46:1421-34
39. Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B. et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. The New England journal of medicine. 2003;349:2483-94
40. Bu G, Lu W, Liu CC, Selander K, Yoneda T, Hall C. et al. Breast cancer-derived Dickkopf1 inhibits osteoblast differentiation and osteoprotegerin expression: implication for breast cancer osteolytic bone metastases. International journal of cancer. 2008;123:1034-42
41. Xu WH, Liu ZB, Yang C, Qin W, Shao ZM. Expression of dickkopf-1 and beta-catenin related to the prognosis of breast cancer patients with triple negative phenotype. PloS one. 2012;7:e37624
42. Chen L, Li M, Li Q, Wang CJ, Xie SQ. DKK1 promotes hepatocellular carcinoma cell migration and invasion through beta-catenin/MMP7 signaling pathway. Molecular cancer. 2013;12:157
43. Hall CL, Daignault SD, Shah RB, Pienta KJ, Keller ET. Dickkopf-1 expression increases early in prostate cancer development and decreases during progression from primary tumor to metastasis. The Prostate. 2008;68:1396-404
44. Rachner TD, Thiele S, Gobel A, Browne A, Fuessel S, Erdmann K. et al. High serum levels of Dickkopf-1 are associated with a poor prognosis in prostate cancer patients. BMC cancer. 2014;14:649
45. Qi L, Sun B, Liu Z, Li H, Gao J, Leng X. Dickkopf-1 inhibits epithelial-mesenchymal transition of colon cancer cells and contributes to colon cancer suppression. Cancer science. 2012;103:828-35
46. Zhou XL, Qin XR, Zhang XD, Ye LH. Downregulation of Dickkopf-1 is responsible for high proliferation of breast cancer cells via losing control of Wnt/beta-catenin signaling. Acta pharmacologica Sinica. 2010;31:202-10
47. Gao C, Xie R, Ren C, Yang X. Dickkopf-1 expression is a novel prognostic marker for gastric cancer. Journal of biomedicine & biotechnology. 2012;2012:804592
48. Jia X, Li N, Peng C, Deng Y, Wang J, Deng M. et al. miR-493 mediated DKK1 down-regulation confers proliferation, invasion and chemo-resistance in gastric cancer cells. Oncotarget. 2016;7:7044-54
49. Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR. et al. E-cadherin/beta-catenin complex and the epithelial barrier. Journal of biomedicine & biotechnology. 2011;2011:567305
Author contact
![]() Corresponding author: Suk Kyeong Lee, Mailing address: Department of Biomedicine & Health Sciences, Department of Medical Lifescience, College of Medicine, The Catholic University of Korea, 222 Banpo-daero, Seocho-gu, Seoul, 06591, Republic of Korea. Tel: + 82-2-2258-7480; Fax: + 82-2-535-7481; E-mail: sukkleeac.kr
Corresponding author: Suk Kyeong Lee, Mailing address: Department of Biomedicine & Health Sciences, Department of Medical Lifescience, College of Medicine, The Catholic University of Korea, 222 Banpo-daero, Seocho-gu, Seoul, 06591, Republic of Korea. Tel: + 82-2-2258-7480; Fax: + 82-2-535-7481; E-mail: sukkleeac.kr

 Global reach, higher impact
Global reach, higher impact