10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(4):895-908. doi:10.7150/ijbs.29744 This issue Cite
Research Paper
Systematic Analysis of tRNA-Derived Small RNAs Reveals Novel Potential Therapeutic Targets of Traditional Chinese Medicine (Buyang-Huanwu-Decoction) on Intracerebral Hemorrhage
1. Institute of Integrative Medicine, Xiangya Hospital, Central South University, Changsha, China
2. Department of Gerontology, Traditional Chinese Medicine Hospital Affiliated to Xinjiang Medical University, Urumqi, China
3. Department of Neurology, Henan Province People's Hospital, Zhengzhou, China
4. Department of Clinical Laboratory, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Received 2018-9-6; Accepted 2019-1-28; Published 2019-3-1
Abstract
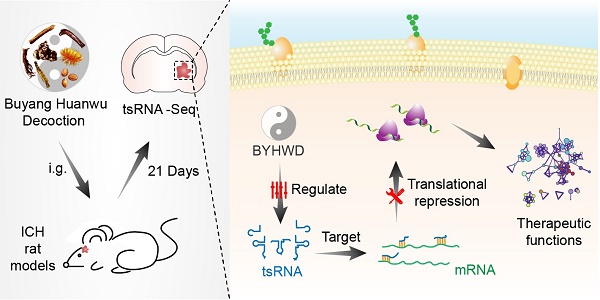
Although Buyang-Huanwu-Decoction (BYHWD), a famous traditional Chinese medicine, has been utilized to promote the recovery of neurological function in intracerebral hemorrhage (ICH) for centuries, its therapeutic mechanisms remain unclear. tRNA-derived small RNA (tsRNA) is a novel class of short non-coding RNA, possessing potential regulating functions. In the current study, we explored the novel therapeutic targets of BYHWD by tsRNA-sequencing. Rats were randomly divided into three groups: sham, ICH, and BYHWD-treated groups. The modified neurological severity score, corner turn test, foot-fault test, and weight change were used to assess neurological injury. After BYHWD treatment, these behavioral tests were obviously meliorated compared with ICH group in the recovery phase. In the rat brain tissues surrounding the hemorrhagic region, a total of 350 tsRNAs for exact match were identified. 12 of tRNAs (fold change >1.3 and P-value <0.05) were significantly changed in ICH group compared to sham group. Among them, 3 of tRNAs (rno-tRFi-Ser-25a, rno-tRF5-Ala-16a and rno-tRF5-Glu-29a) were markedly regulated by BYHWD treatment and validated with quantitative real-time PCR. Additionally, target prediction and bioinformatics analyses revealed that these tsRNAs could play therapeutic roles through FoxO signaling pathway, positive regulation of long term synaptic depression, autophagy - animal, IL-17 signaling pathway and regulation of cytoskeleton and transforming growth factor beta. In conclusion, tsRNAs are the potential therapeutic targets of BYHWD on ICH treatment. The present study provides novel insights for future investigations to explore the mechanisms, by which BYHWD promotes neurological function recovery after ICH.
Keywords: traditional Chinese medicine, Buyang-Huanwu-Decoction, intracerebral hemorrhage, transcriptomics, non-coding RNA, tsRNA
Introduction
Stroke is the second leading cause of death and disability worldwide [1]. Intracerebral hemorrhage (ICH), occurring in over 2 million people each year globally, is a devastating subtype of stroke with a high mortality rate of 32-50% [2, 3]. Moreover, fewer than 20% of survivors have good functional outcomes in the chronic stage of ICH [4]. Although the understanding of ICH-induced primary and secondary brain damage has improved [5, 6], no proven treatments are available for functional recovery after ICH [7]. The development of new effective therapies for ICH is urgently required. Fortunately, traditional Chinese medicine (TCM), with more than 2,000-year history, represents one of the oldest herbal medicine in the world [8]. Based on a multi-component and multi-target approach, TCM has become an important strategy for new drug development to treat stroke [9, 10]. World Health Organization encourages that herbal remedies should be advocated to incorporate into main stream medical systems [11].
Buyang-Huanwu-Decoction (BYHWD), a famous TCM formula, is by far the most frequently used for the treatment of stroke (40.32%) [12]. Accumulating evidence suggests that BYHWD improves the outcome of ischemic stroke through neuroprotection and neurogenesis effects, an improvement in synaptic plasticity, etc. [13-15]. Moreover, our previous study confirms that BYHWD tends to improve the recovery of kinetic function and promote angiogenesis after ICH by enhancing VEGFR2 phosphorylation through the PI3K/Akt signaling pathway [16, 17]. However, the multi-faceted regulatory mechanisms of BYHWD acting on ICH have not been fully elucidated due to the lack of appropriate methods.
Recent studies have investigated the pharmacological mechanisms and potential therapeutic targets of TCM through transcriptomics technologies [18-20]. Novel high-throughput technologies such as deep-transcriptome sequencing, demonstrate that most of the genome is transcribed into RNAs. Non-coding RNAs (ncRNAs) accounting for 98% of total RNAs are particularly abundant in the central nervous system [21]. As the important regulators of various molecular processes, ncRNAs are divided into two subclasses: small ncRNAs (<200 nucleotides, nt) such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) with >200 nt in length [22]. Within the past decade, ncRNAs have emerged as key mediators of posttranscriptional gene expression in pathological aspects of stroke [23]. It has recently been shown that BYHWD could exposure augmented angiogenetic miRNAs in ischemic stroke [24]. Additionally, we first found that lncRNAs were possible therapeutic targets of BYHWD in ICH treatment [25, 26]. Nevertheless, considering a huge amount of ncRNAs, more functional ncRNAs are needed to reveal the therapeutic mechanisms of BYHWD in ICH.
The tRNA-derived small RNA (tsRNA) is a novel class of small ncRNA (<50nt) [27]. There are at least six types of tsRNAs: tRNA-derived fragments 1 (tRF-1), tRF-3, tRF-5, internal tRF (i-tRF), stress-induced tRNA fragments 3 (tiRNA-3) and tiRNA-5, which differ in the cleavage position of the precursor or mature tRNA transcripts [27, 28]. tsRNAs are not simply the random products of tRNA degradation, but play important roles in biological processes [29]. Increasing studies have demonstrated that tsRNAs can silence their target mRNAs in an RNA interference manner [30-32]. In addition, tsRNAs are abundant in most organisms and have been implicated in stress responses, cancer, viral infection responses, and neurological disorders [33-37]. However, the biological roles are still not well understood. Particularly, a recent study showed that tRNAVal- and tRNAGly-derived fragments could modulate angiogenesis in stroke brain tissues [38]. Consequently, in view of the angiogenic function of BYHWD [17, 24], we hypothesize that BYHWD probably performs therapeutic actions by tsRNAs.
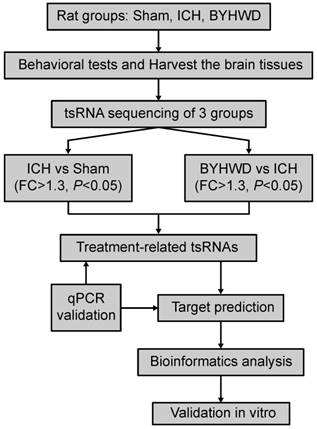
For that purpose, we aimed to utilize RNA-sequencing technologies to evaluate the tsRNA expression levels in sham, ICH model, and BYHWD-treated groups. Then ICH-induced changed tsRNAs (ICH vs sham) and BYHWD-regulated altered tsRNAs (BYHWD vs ICH) were first identified. Finally, using bioinformatics, we evaluated their biological functions to reveal the therapeutic mechanisms of BYHWD (Figure 1). These new findings may provide a novel perspective to illustrate the molecular mechanisms of BYHWD treatment for long-term neurological function recovery after ICH.
Study design illustration. ICH, intracerebral hemorrhage; BYHWD, Buyang-Huanwu-Decoction; FC, fold change; qPCR, quantitative real-time PCR.
Materials and Methods
Preparation of BYHWD
The ingredients of BYHWD including seven medicinal herbs: Huang Qi (Radix astragali), Chi Shao (Radix paeoniae Rubra), Dang Gui (Radix angelicae Sinensis), Chuan Xiong (Rhizoma Ligustici Chuan xiong), Hong Hua (Flos Carthami), Tao Ren (Semen persicae), and Di Long (Pheretima Aspergillum) at a ratio of 60:6:4.5:3:3:3 (dry weight). All herbs were obtained from Xiangya Hospital, Central South University (CSU) (Changsha, China). Furthermore, each herb was authenticated by the herbal medicinal botanist, Professor Suiyu Hu, at the Institute of Integrative Medicine of Xiangya Hospital, CSU. The decoction was prepared and subjected to quality control as previously described [26]. Finally, the powder was dissolved in distilled water at a concentration of 0.13 g/ml for intragastric administration [26].
Animals and Experimental Groups
Adult male Sprague-Dawley rats weighing 180-220g were obtained from the Laboratory Animal Centre of CSU. All the animals were reared under identical conditions (room temperature at 25 °C and 12-hour light-dark cycle) and had free access to standard rodent water and food. All animal protocols were approved by the Committee on the Use and Care of Animals of CSU and conformed to the Guidelines for the Care and Use of Laboratory Animals.
Rats were randomly divided into three groups: the sham-operated group, ICH group, and BYHWD- treated group [39]. Collagenase-induced ICH rat models were important tools for studying ICH and had been used by many researchers [40, 41]. ICH was conducted as described in detail previously and sham rats were injected with the same volume of saline [26, 42]. For the BYHWD-treated group, ICH rats were administered once daily from 1 to 21 days by intragastric administration. According to our previous study [17], we chose 2.05 g/kg as the dosage of BYHWD in the present study. The distilled water with equal volumes was administered to the sham and ICH groups for 21 days.
Assessment of Neurological Function
The modified neurological severity score (mNSS), corner turn test, foot-fault test, and weight change were used to assess the neurological injury. These tests were detailly performed in our previous studies [17, 43]. Firstly, the mNSS was an integrative test to evaluate the motor (muscle status, abnormal movement), sensory (visual, tactile, and proprioceptive), and reflex abilities (Supplementary Table S1). And a higher score meant a more severe injury (normal score: 0; maximal deficit score: 18). Secondly, the corner turn test could assess the motor function. Rats were made to proceed into a corner with the angle of 30 degrees. The individual rats could turn either left or right when exiting the corner, and then the direction was recorded. The test was repeated 10 times for each animal and the percentage of right turns was calculated. The percentage of right turn in normal rats was about 50%, while the injured ones would have a higher percentage. Thirdly, the placement dysfunction of forelimbs was tested by the foot-fault test. Rats were put on an elevated grid surface and placed their paws on the wire while moving along the grid. The total number of steps (movement of each forelimb) which the rat used to cross the grid was counted, and the total number of foot-faults for left forelimb was recorded. Data were presented as the percentage of foot faults per the total number of steps (normal score: 0%; maximal deficit score: 100%). Lastly, weight change was used for general condition evaluation. All of the tests were carried out on the day of surgery but before anesthesia (0 day), and 1st and 21st days after ICH (n=6, each group).
tsRNA Sequencing (tsRNA-Seq)
On the 21st day, rats were deeply anesthetized by intraperitoneal injection of pentobarbital and perfused with ice-cold saline. Then, brain tissues surrounding the hemorrhagic region were harvested for following detections. Total RNAs were extracted from sham, ICH and BYHWD groups (n=3 each group) according to the manufacturer's instruction (Qiagen, USA). Subsequently, using rtStar tRF& tiRNA Pretreatment Kit protocols (Arraystar, USA), some RNA modifications which might interfere with small RNA-sequencing library construction were removed. cDNA was then synthesized and amplified using Illumina's proprietary reverse transcription primers and amplification primers. Afterwards, 135-170 nt PCR amplified fragments (corresponding to 15-50 nt small RNA) were extracted and purified from the PAGE gel using an automated gel cutter. The libraries are qualified and absolutely quantified using Agilent BioAnalyzer 2100. Finally, the sequencing run was performed on Illumina NextSeq 500 system using NextSeq 500/550 V2 kit (Illumina, USA). Sequencing was carried out by running 50 cycles.
Data Analysis
The raw sequencing data that passed the Illumina chastity filter were used for the following analysis. After Illumina quality control, the sequencing reads were 5', 3'-adaptor trimmed, filtered for over 15nt by Cutadapt software. And then using NovoAlign software (v2.07.11), trimmed reads were aligned to mature-tRNA and pre-tRNA sequences from GtRNAdb (http://gtrnadb.ucsc.edu/). The exactly matched reads were thought as tsRNAs. Moreover, tsRNA expression levels could be measured and normalized as tag counts per million of total aligned tRNA reads (TPM). tsRNA expression profiling and differential expression analysis were calculated by the average TPM. Fold changes (FC; ICH versus sham or BYHWD versus ICH) were used for comparing two groups of profile differences. In a ddition, FC>1.3 and P-value <0.05 were considered significantly different expression and these tsRNAs were chosen for further analysis.
In the standard of FC>1.3 and P<0.05, we could identify ICH-induced tsRNAs (ICH vs sham) and BYHWD-induced tsRNAs (BYHWD vs ICH). Furthermore, tsRNAs that were both ICH-induced upregulated and BYHWD-induced downregulated, were considered as treatment-related ones. Similarly, the genes downregulated after ICH and meanwhile upregulated after BYHWD treatment were also treatment-related tsRNAs.
Target Prediction of treatment-related tsRNAs
tsRNAs contained some seed sequences which might match the crosslink-centered regions of target mRNAs [44, 45]. Besides increasing studies strongly suggested that tsRNAs have a miRNA-like function, leading to mRNA silence [31, 46, 47]. In this study, we used four common algorithms to predict tsRNA targets, namely, TargetScan (http://www.targetscan.org), miRanda (http://www.microrna.org), miRDB (http://www.mirdb.org) and RNAhybrid (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid). Additionally, to reduce false-positive results, only genes predicted by all four software were considered as targets of tsRNAs. The network illustration was visualized with Cytoscape software (version 3.5.1, the Cytoscape Consortium, San Diego, CA, USA).
Validation by Quantitative Real-Time PCR (qPCR)
To validate the changes of treatment-related tsRNAs detected by RNA-seq, all tsRNAs were selected and their expression was examined by qPCR. Firstly, total RNA was reverse-transcribed into cDNA using rtStar™ First-Strand cDNA Synthesis Kit (3' and 5' adaptor; Arraystar) according to the manufacturer's protocol. Then, qPCR amplification was performed using ViiA 7 Real-time PCR System (Applied Biosystems) and 2×PCR master mix (Arraystar). The cycling conditions were as follows: incubation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 60 s and 95 °C for 15 s. The relative tsRNA expression levels were calculated using the 2-ΔΔCt method and were normalized to U6, as an endogenous. The specific primers for each gene were listed in Table 1. All reactions were performed in triplicate.
Furthermore, the target mRNAs of treatment- related tsRNAs were validated by qPCR as well. One target of each tsRNA was randomly selected. The qPCR protocols were described as above and β-Actin was used as internal controls to normalize the data.
Sequences of primers for qPCR validation of candidate genes and internal control.
| Gene name | Primer sequence | Productlength |
|---|---|---|
| U6 | F: 5' GCTTCGGCAGCACATATACTAAAAT 3'R: 5' CGCTTCACGAATTTGCGTGTCAT 3' | 89 |
| rno-tRF5-Ala-16a | F: 5' GTTCAGAGTTCTACAGTCCGACG 3'R: 5' CCGATCTACTGAGCTACATCCC 3' | 49 |
| rno-tRF5-Glu-29a | F: 5' ACAGTCCGACGATCTCCCATA 3'R: 5' TGCTCTTCCGATCTAGGAATCC 3' | 57 |
| rno-tRFi-Ser-25a | F: 5' CCGACGATCATGGACTGCTA 3'R: 5' CTTCCGATCTAGAGCACAATGG 3' | 44 |
| rno-tRFi-Gln-16a | F: 5' GCGCGCGTCTGGACTCT 3'R: 5' AGTGCAGGGTCCGAGGTATT 3' | 60 |
| β-Actin | F: 5' ACATCCGTAAAGACCTCTATGCC 3'R: 5' TACTCCTGCTTGCTGATCCAC 3' | 223 |
| Clec11a | F: 5' CGGTACTTACGAGCTGCTCTTGC 3'R: 5' CGGTGCCAGGCGAAGAATGAC 3' | 125 |
| Itga8 | F: 5' TCGGCTACTCGCTGGACTTCTAC 3'R: 5' TCTACGATATCCGGCTGGCTAGTG 3' | 97 |
| Map3k7 | F: 5' CTCGTCCTCCTCCTCGTCTTCTG 3'R: 5' ACCTCTTCCACCTCGATCTCCTTG 3' | 102 |
| Foxo1 | F: 5' GTACGCCGACCTCATCACCAAG 3'R: 5' GGCACACTCTTCACCATCCACTC 3' | 93 |
| Mapk1 | F: 5' TGAAGACACAGCACCTCAGCAATG 3'R: 5' GGTGTTCAGCAGGAGGTTGGAAG 3' | 131 |
| Smad3 | F: 5' CTTCACAGCCGTCCATGACAGTAG 3'R: 5' CCAATGTAGTAGAGCCGCACACC 3' | 151 |
| Sos1 | F: 5' GCTCGCCATTACATCTCCAACCTC 3'R: 5' CTTCCTGTGTCAGTGGTGGTGATG 3' | 162 |
| Cdkn2b | F: 5' CCAACGCCGTCAACCGCTTC 3'R: 5' TGCCTTGTGCAGCACCATTAGC 3' | 188 |
| Crebbp | F: 5' GAGAGCAAGCGAATGGAGAGGTTC 3'R: 5' AGTCATGTCGTGTGCAGTTCTTCC 3' | 171 |
| Ikbkb | F: 5' CTGGTAGAACGGATGATGGCACTG 3'R: 5' TCCTGGCTGTCACCTTCTGTCC 3' | 170 |
| Pik3ca | F: 5' AACTCGCCTCATAGCAGAGCAATG 3'R: 5' TGGCACGCAGTCATGGTTGATC 3' | 183 |
| Prkaa1 | F: 5' TTGCGTGTGCGAAGGAAGAACC 3'R: 5' CCGATCTCTGTGGAGTAGCAGTCC 3' | 157 |
| Slc2a4 | F: 5' AGCCAGCCTACGCCACCATAG 3'R: 5' CAGCAGAGCCACCGTCATCAAG 3' | 155 |
F: forward primer. R: reverse primer.
Bioinformatics Analysisue
To assign the biological annotation of the targets, pathway and process enrichment analysis were applied by c. The analysis result was carried out with the following ontology sources: Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway, Gene Ontology (GO) Biological Processes, and Reactome Gene Sets. Enriched terms with P<0.01, minimum count 3, and enrichment factor >1.5 were collected and grouped into clusters based on their membership similarities. More specifically, P-values were calculated based on accumulative hypergeometric distribution and enrichment factor was the ratio between observed count and the count expected by chance.
Cell Culture and Transfection
PC12 cells, which were generally used as neuron cell line were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) containing 5% heat-inactivated horse serum (Beyotime, China) and 10% of fetal bovine serum (PAN, Germany). Cells were incubated in 5 % carbon dioxide incubators at 37 °C. During the exponential phase of growth, PC12 cells were cultured in 12-well plates for transfection. The rno-tRF5-Glu-29a mimic (UCCCAUAUGGUCUAGCGGUUAGGAUUCCU), rno-tRFi-Ser-25a mimic (AUGGACUGCUAAUCCAUUGUGCUCU), rno-tRF5-Ala-16a mimic (GGGGAUGUAGCUCAGU) and the negative control (NC; UUUGUACUACACAAAAGUACUG) were obtained from RiboBio (Guangzhou, China) and Sangon Biotech (Shanghai, China). The transfection of mimics and NC was performed using Lipofectamine 3000 (Invitrogen, USA) at a final concentration of 100 nmol, according to the manufacturer's instructions. All groups were performed in triplicate. After 48 hours of transfection, the transfected cells were harvested for total RNA isolation. The tsRNA-targeted genes were then measured by qPCR. The specific primers were listed in Table 1 and the protocols were described as above.
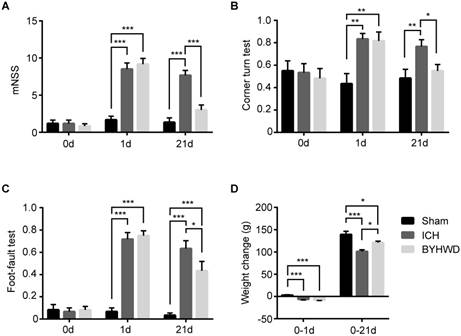
Effects of BYHWD on neurological recovery after ICH. On 1 day, the mNSS (A), corner turn test (B), foot-fault test (C) and weight change (D) of ICH and BYHWD group were significantly different with sham group, which indicated the successful ICH models. On 21 day, the mNSS (A), corner turn test (B), foot-fault test (C) and weight change (D) of sham group and BYHWD group showed significantly different values than ICH group, which suggested the therapeutic effects of BYHWD for functional recovery after ICH. Data were presented as mean ± SEM (n=6 each group), *𝑃<0.05, **𝑃<0.01, ***𝑃<0.001. mNSS, modified neurological severity score; 0d, the day of surgery but before anesthesia; 1d and 21d, the 1st and 21st day after ICH; weight change, the difference between weight values of 1&21 day and 0 day.
Statistical Analysis
Results were presented as the mean ± SEM. Statistical analysis was conducted by GraphPad Prism (version 7.00; GraphPad Software, Inc., La Jolla, CA, USA). Differences between two groups were carried out using Student's t-test. One-way analysis of variance (ANOVA) was employed for three groups. P<0.05 was defined as statistically significant.
Results
Effects of BYHWD on Neurological Recovery after ICH
Behavioral tests and changes of body weight were carried out to generally evaluate whether BYWHD treatment could improve neural functional recovery after ICH (Figure 2). On 0 day, these tests were performed to show the base line levels of normal rats and there was no statistically significance between the three groups (all P>0.05) (Figure 2A-C).
The scores of the 1st day were used to prove the successful models. On the first day after surgery, the collagenase-induced rats (ICH and BYHWD groups) showed significantly higher scores of mNSS, respectively (P<0.001, P<0.001) (Figure 2A). Similarly, the percentages of ICH and BYHWD groups were notably higher than sham in the corner turn test (P<0.01, P<0.01) and the foot-fault test (P<0.001, P<0.001) (Figure 2B,C). Furthermore, the body weight of ICH and BYHWD rats decreased, while the sham ones increased. The weight change of ICH and BYHWD were significantly lower than sham (P<0.001, P<0.001) (Figure 2D). In addition, the results between have no significance between ICH and BYHWD (all P>0.05).
On the 21st day, the scores of ICH rats were still higher than sham in mNSS (P<0.001), the corner turn test (P<0.01) and the foot-fault test (P<0.001) (Figure 2A-C). However, after long-term treatment of BYHWD, the scores of BYHWD rats significantly declined in mNSS (about 60%, P<0.001) and the corner turn test (about 30%, P<0.05). Moreover, the results of BYHW in mNSS and the corner turn test were very close to sham (without statistically significance) (Figure 2A,B). Although the percentage of BYHWD was higher than sham (P<0.001), it decreased about 30% than ICH (P<0.05) (Figure 2C). Additionally, for weight change, ICH worsened the weight growth than sham (P<0.001). And BYHWD treatment could improve about 20% rise over ICH (P<0.05), even though it could not reach sham level (P<0.05) (Figure 2D).
Altered Expression Profiles of tsRNAs with BYHWD Treatment
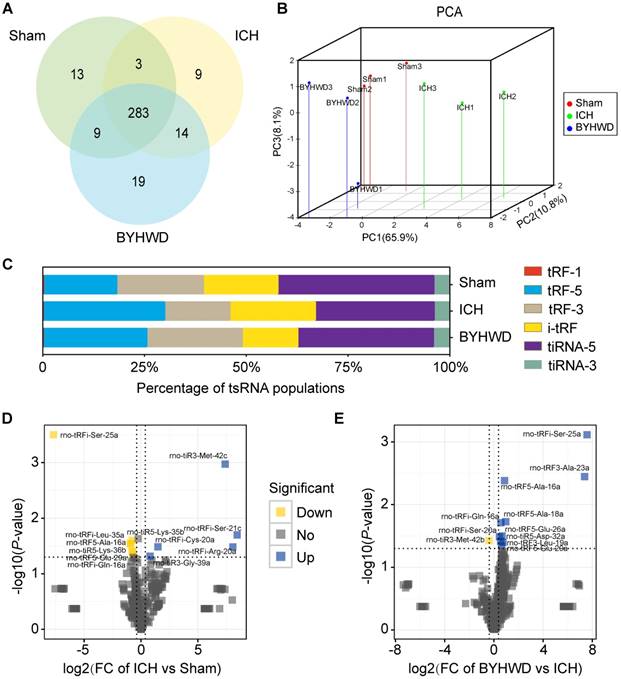
The tsRNA-Seq analysis was used to assess the expression levels of tsRNAs in sham, ICH and BYHWD rats. The raw data of tsRNA-Seq had been submitted to Gene Expression Omnibus (GEO accession: GSE109697). A total of 350 tsRNAs for exact match were identified in the right globus pallidus region of rats (308 in sham, 309 in ICH and 325 in BYHWD) (Figure 3A,B and Supplementary Table S2). More than 99% of tsRNAs were generated from mature tRNAs (tRF-5, tRF-3, i-tRF, tiRNA-5 and tiRNA-3). In those tsRNAs, expression levels of each subtype tsRNA was quite different. Moreover, in general view, tRF-5s and i-tRFs were increased while tRF-3s and tiRNA-5s were decreased after ICH over sham. Interestingly, BYHWD treatment could reverse these changes of ICH and made expression profiles close to sham levels (Figure 3C). Additionally, we identified 12 significantly dysregulated tsRNAs in ICH rats: 5 were upregulated, while 7 were downregulated (ICH vs sham; FC>1.3 and P<0.05) (Figure 3D and Supplementary Table S3). And after BYHWD treatment, 11 tsRNAs were obviously changed: 10 were upregulated, while 1 was down- regulated (BYHWD vs ICH; FC>1.3 and P<0.05) (Figure 3E).
Altered expression profiles of tsRNAs with BYHWD treatment. (A) Venn plot to show the total number of identified tsRNAs in the brain tissues of sham, ICH and BYHWD groups. (B) PCA plot to illustrate the clustering of the 3 replicates of each group and assess the variation and reproducibility (FC>1.3 and P<0.05). The red points represent the sham group; green, the ICH group; blue, the BYHWD group. (C) Histogram to show the expression levels of each subtype tsRNA in 3 groups. BYHWD treatment could reverse these changes of ICH group and made expression profiles close to sham levels. Volcano plot to exhibit 12 significantly changed tsRNAs between ICH and sham gropus (FC>1.3 and P<0.05) (D), and 11 significantly changed tsRNAs between BYHWD and ICH groups (FC>1.3 and P<0.05) (E). Yellow denotes downregulated tsRNAs and blue denotes upregulated ones. The names of the dots with significant P-value were annotated. TPM, tag counts per million of total aligned tRNA reads, representing tsRNA expression levels.
Identification of BYHWD Treatment-Related tsRNAs and qPCR Confirmation
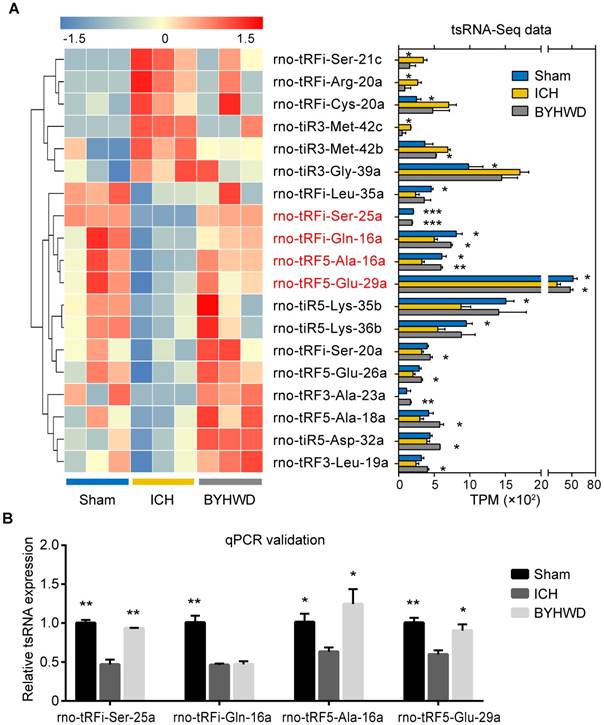
Of those significantly altered tsRNAs, 4 BYHWD treatment-related tsRNAs were identified (rno-tRFi-Ser-25a, rno-tRF5-Ala-16a, rno-tRFi-Gln-16a and rno-tRF5-Glu-29a) (Figure 4A). All of the 4 tsRNAs were significantly downregulated after ICH and meanwhile were significantly upregulated after BYHWD treatment (Supplementary Figure S2). While none of tsRNAs was both ICH-induced upregulated and BYHWD-induced downregulated (Supplementary Figure S2). Although some tsRNAs such as rno-tRFi-Ser-21c increased significantly in ICH, and the decrease after BYHWD treatment showed no statistical significance (Figure 4A and Supplementary Table S3).
To validate the tsRNA-Seq results, we used qPCR to confirm the expression changes for the 4 tsRNAs. As shown in Figure 4B, all the tsRNA transcripts were detected and exhibited significantly decreased after ICH (all P<0.05). Furthermore, rno-tRFi-Ser-25a, rno-tRF5-Ala-16a and rno-tRF5-Glu- 29a were significantly upregulated after BYHWD treatment (P<0.01, P<0.05, P<0.05) except for rno-tRF5-Gln-16a. The qPCR results of rno-tRFi-Ser- 25a, rno-tRF5-Ala-16a and rno-tRF5-Glu-29a were consistent with tsRNA-Seq data. Thus, the 3 tsRNAs were confirmed as BYHWD treatment-related tsRNAs and used for further analysis.
BYHWD treatment-related tsRNAs and qPCR confirmation. The significantly changed tsRNAs were shown in the heat map (A). Among those, rno-tRFi-Ser-25a, rno-tRF5-Ala-16a, rno-tRFi-Gln-16a and rno-tRF5-Glu-29a (in red fonts) were significantly changed both after ICH and after BYHWD treatment. (B) The qPCR results showed that rno-tRFi-Ser-25a, rno-tRF5-Ala-16a and rno-tRF5-Glu-29a were consistent with tsRNA-Seq data. Thus, the 3 tsRNAs were confirmed as BYHWD treatment-related tsRNAs and used for further analysis. Data were presented as mean ± SEM (n=3 each group), *𝑃<0.05, **𝑃<0.01, ***𝑃<0.001 indicate significant difference compared with ICH group.
Targets of BYHWD treatment-related tsRNAs and validation. (A) Venn plot to indicate that 383 mRNA targets of 3 treatment-related tsRNAs were predicted by 4 prediction software simultaneously. The targets of each tsRNA were shown respectively (B). Three mRNA transcripts (Clec11a, Map3k7, Itga8; each one corresponding to one tsRNA) were randomly selected to validate the accuracy of target prediction through qPCR. (C) The binding region of and seed sequence were exhibited. Consistent with the supposition that tsRNAs could silence the mRNAs, all the selected mRNA targets were observed to be upregulated in ICH and downregulated in BYHWD significantly (D-F). Data were presented as mean ± SEM (n=3 each group), *𝑃<0.05, **𝑃<0.01, indicate significant difference compared with ICH group.
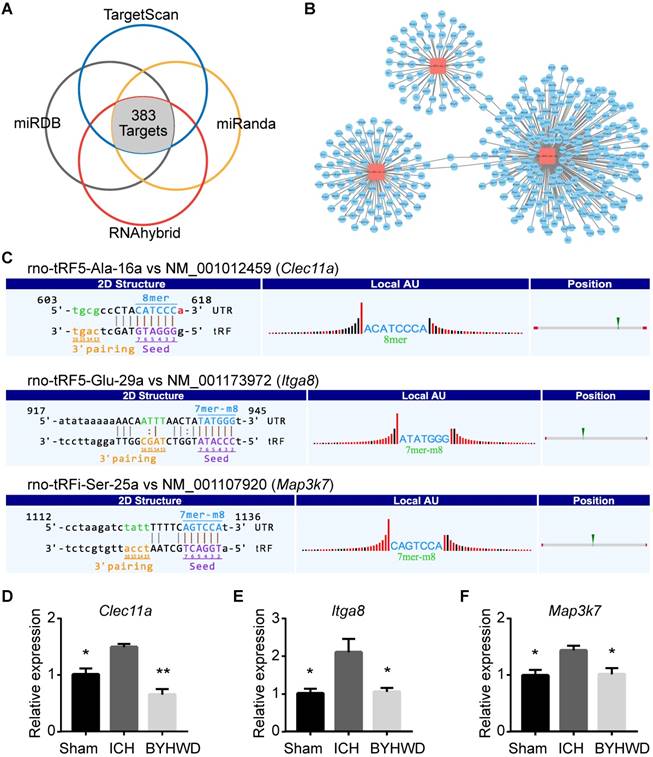
Targets of BYHWD Treatment-Related tsRNAs and Validation
Although the mechanism of tsRNA action is yet to be elucidated, there is recent evidence suggesting a miRNA-like mode of action [34]. miRNAs could recognize their mRNA targets using their seed sequence (positions 2-7nt at their 5' ends) and suppress the global mRNA translational activities [47]. Based on the rule, we used four kinds of algorithms named TargetScan, miRanda, miRDB, and RNAhybrid to predict tsRNA targets. In total, 383 mRNA targets were predicted simultaneously for the 3 BYHWD treatment-related tsRNAs (Figure 5A). rno-tRFi-Ser-25a, rno-tRF5-Glu-29a, and rno-tRF5- Ala-16a could target 59, 87, and 241 transcripts respectively (Figure 5B and Supplementary Table S4). In the present study, the BYHWD treatment-related tsRNAs were decreased after ICH and increased after BYHWD. Therefore, their target transcripts would be increased after ICH and decreased after BYHWD.
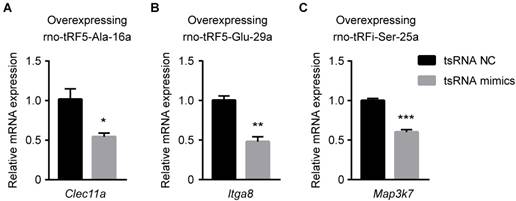
To enhance data quality, three mRNA transcripts (Clec11a, Map3k7, Itga8; each one corresponding to one tsRNA) were randomly selected to validate the accuracy of target prediction through qPCR. The binding region of and seed sequence were shown in Figure 5C. Consistent with the supposition above, all the selected mRNA targets were observed to be upregulated in ICH and downregulated in BYHWD significantly (Figure 5D-F). Additionally, to further validate the targets of tsRNAs, we overexpressed the tsRNA in PC12 cell lines to investigate the changes of targets of the corresponding tsRNA. Clec11a was significantly downregulated when transfected with rno-tRF5-Ala-16a. So were Itga8 and Map3k7 when transfected with corresponding tsRNA (Figure 6). These results in vitro were consistent with the qPCR results in rats, indicating that the target prediction results were credible for further analysis.
The relative mRNA levels determined by qPCR in PC12 cells transfected with rno-tRF5-Ala-16a (A), rno-tRF5-Glu-29a (B), rno-tRFi-Ser-25a (C) mimics. Clec11a was downregulated when transfected with rno-tRF5-Ala-16a. So were Itga8 and Map3k7 when transfected with corresponding tsRNA. Data were presented as mean ± SEM (n=3 each group), *𝑃<0.05, **𝑃<0.01, ***𝑃<0.001 indicate significant difference compared to NC group.
Biological Function Analysis Revealing Potential Therapeutic Effects of tsRNAs
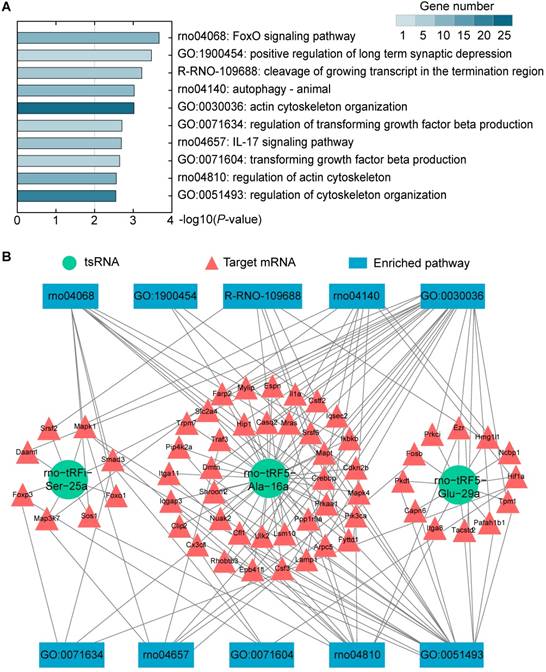
The tsRNAs could regulate mRNA translational activities, and hence, to understand their biological functions, we conducted a bioinformatics analysis of the functions of the target mRNAs. KEGG pathway, GO biological processes, and Reactome Gene Sets were executed to explore the functions of 383 target mRNAs. According to KEGG enrichment analysis, FoxO signaling pathway (rno04068; 10 genes enriched), autophagy - animal (rno04140; 9 genes enriched), IL-17 signaling pathway (rno04657; 7 genes enriched), and regulation of actin cytoskeleton (rno04810; 11 genes enriched) were significantly detected. The major biological processes surveyed by GO were positive regulation of long term synaptic depression (GO:1900454; 3 genes enriched), actin cytoskeleton organization (GO: 0030036; 24 genes enriched), regulation of transforming growth factor beta production (GO: 0071634; 4 genes enriched), etc. Moreover, cleavage of growing transcript in the termination region (conducted; 6 genes enriched) was the main term conducted by Reactome Gene Sets. Ranked by P-values, top 10 enriched terms were shown in Figure 7A and Supplementary Table S5. Among those, FoxO signaling was the most significant pathway.
Alteration of tsRNA-Targeted Genes Enriched in FoxO Signaling Pathway after Transfection
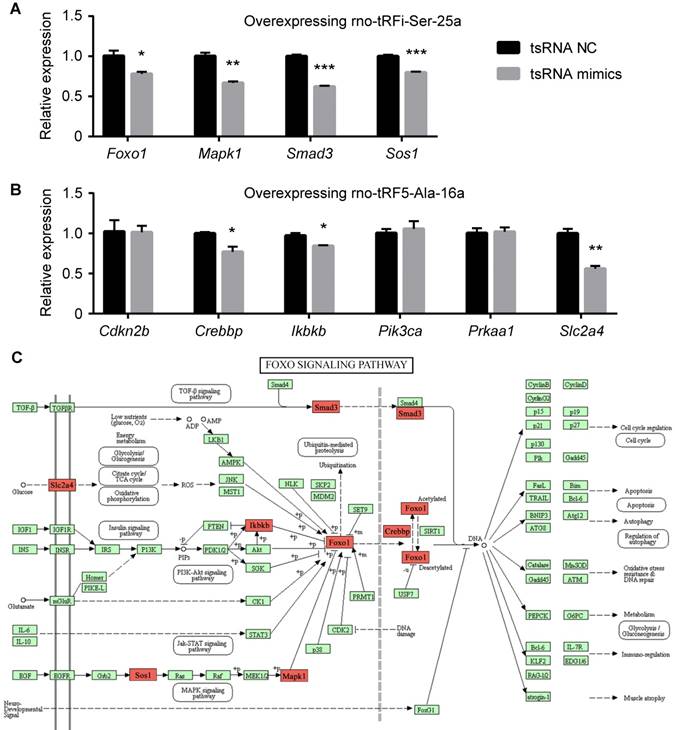
There were 10 genes enriched in FoxO signaling pathway (Cdkn2b, Crebbp, Ikbkb, Pik3ca, Prkaa1, Slc2a4, Foxo1, Mapk1, Smad3, and Sos1). These genes were regulated by 2 tsRNAs (Figure 7B). Foxo1, Mapk1, Smad3, and Sos1 were regulated by rno-tRFi-Ser-25a, and rno-tRF5-Ala-16a targeted Cdkn2b, Crebbp, Ikbkb, Pik3ca, Prkaa1, and Slc2a4. After overexpressing rno-tRFi-Ser-25a in PC12 cells, Foxo1, Mapk1, Smad3, and Sos1 were significantly downregulated (P<0.05, P<0.01, P<0.001, P<0.001) (Figure 8A). When transfected with rno-tRF5-Ala-16a mimics, Crebbp, Ikbkb and Slc2a4 were significantly downregulated (P<0.05, P<0.05, P<0.01), and Cdkn2b, Pik3ca and Prkaa1 had no change (all P>0.05) (Figure 8B). All the tsRNA-targeted genes were shown in Figure 8C.
Discussion
This is the first study to investigate the expression profiles of tsRNAs, as potential molecules regulating mRNA translational activities, in experimental ICH treated with TCM formula. After BYHWD treatment, 3 tsRNAs were significantly reversed compared with ICH. And bioinformatics analysis prompted that the 3 treatment-related tsRNAs could play therapeutic roles through regulating FoxO signaling pathway, long term synaptic depression, actin cytoskeleton organization transforming growth factor beta production, etc. These findings provide new insights into the better understanding of therapeutic targets and pharmacological mechanisms on ICH treatment by BYHWD.
BYHWD has been utilized for centuries to promote the recovery of neurological function in stroke-induced disabilities in China [14]. Evidence- based investigations have indicated that BYHWD improves neurological deficiencies in both experimental studies and clinical trials [15, 49]. And the present study has yet again indicated the therapeutic effects by the behavioral tests. Moreover, increasing studies have tried to reveal the therapeutic mechanisms, for instance, promoting angiogenesis through VEGF and angiopoietin and the regeneration of peripheral nerves [12]. Furthermore, the pharmacological networks of BYHWD were built for drug target prediction according to a structure-based method [15]. However, these studies cannot interpret why the molecular protein targets are modulated. In our previous study, we attempted to answer the question in the respect of mRNA translational regulation, and found that BYHWD could target lncRNAs to affect the mRNA levels and exhibit therapeutic biological functions in ICH rats. Nevertheless, it is still limited to reveal the therapeutic mechanisms of BYHWD in ICH treatment. More functional ncRNAs are required to elucidate the problem. And one study indicated tsRNAs were involved in ischemia stroke, and tRNAVal- and tRNAGly-derived tsRNAs could modulate angiogenesis in stroke brain tissues [38]. All these evidences suggest that tsRNAs may be potential therapeutic targets of stroke.
The tsRNA-Seq results of 3 groups indicated that tsRNAs were correlative with BYHWD treatment. Moreover, based on tsRNA-Seq data, 4 tsRNAs were identified as the targets of BYHWD, which were significantly downregulated after ICH and meanwhile were significantly upregulated after BYHWD treatment. Additionally, there were several tsRNAs (e.g. Cys-20a and Gly-39a; Figure 4A) which were significantly upregulated after ICH compared to Sham, but there was no statistical significance between ICH and BYHWD groups (P>0.05). Therefore, all the BYHWD treatment-related tsRNAs were downregulated on the ICH when compared to BYHWD and Sham groups. Furthermore, in consideration of reliability of sequencing data, we validated the 4 tsRNAs by qPCR. One tsRNA was not consistent with the sequencing result, and then the remaining 3 tsRNAs were confirmed for further analysis. Subsequently, we use bioinformatics methods to show the biological functions of the 3 BYHWD treatment-related tsRNAs and further reveal their pharmacological mechanisms.
Biological function analysis. (A) Top 10 enriched terms were shown ranked by P-value and colored by the number of enriched genes. (B) tsRNA-mRNA-pathway interaction networks. tsRNAs can mediate the global translational regulation of gene expression in a miRNA-like silencing manner. Then, the altered mRNAs affect the corresponding pathways to exhibit biological functions.
The relative mRNA levels determined by qPCR in PC12 cells transfected with rno-tRFi-Ser-25a (A) and rno-tRF5-Ala-16a (B) mimics. Foxo1, Mapk1, Smad3, Sos1, Crebbp, Ikbkb and Slc2a4 were significantly downregulated after transfection. (C) The 7 tsRNA-targeted genes were signed in red on FoxO signaling pathway map. Data were presented as mean ± SEM (n=3 each group), *𝑃<0.05, **𝑃<0.01, ***𝑃<0.001 indicate significant difference compared to NC group.
It has been demonstrated that the leading function of tsRNAs was to mediate the global translational regulation of gene expression through diverse mechanisms [29]. Thereinto, the complementarity between some tsRNAs and their target mRNAs is indispensable for efficient silencing [29]. Recent studies have demonstrated the regulatory function of these tsRNAs by postulating that they can bind and repress mRNAs acting as miRNAs [31, 50-52]. tsRNAs are found associated with Argonaute (AGO) proteins which are essential for target recognition in a miRNA-like silencing manner. The AGO-tsRNA complexes then interact with complementary sequences on target mRNAs to regulate the expression of these targets. Kumar et al. found that tRF-5s and tRF-3s associate with AGO1, AGO3, and AGO4, but not with AGO2, while the mechanisms of tsRNA-AGO associations were still unclear [31]. The interaction regions were based on the complementarity of the 5' seed sequence of a tsRNA to a short sequence within a 3' UTR of a transcript [45]. Thus, based on the miRNA-like manner, the targets of tsRNAs can be predicted by some algorithms. As the tsRNAs are newly discovered recently, the database for tsRNA target prediction is still absent. A recent study used RNAhybrid to scan the transcripts with the requirement: perfect pairing between the nucleotides 2-11 and at most four mismatches between nucleotides 12-21 of the tsRNAs and target sites [45]. Another study used RNA22 software to predict target mRNAs with 3' UTR bearing complementarity to at least the 6-mer seed sequence of the tsRNA [46]. In general, there is no unified algorithm for tsRNA target prediction, and the main principle is based on the miRNA-like manner after all. Therefore, in the present study, we used 4 kinds of common miRNA algorithms to predict the targets of BYHWD treatment-related tsRNAs. And the biological functions of target mRNAs represent the effects of tsRNAs.
To predict the functions of the tsRNA targets, we employed functional annotation tools of Metascape for KEGG pathway, GO biological processes, and Reactome Gene Sets analyses. Most of the enriched pathways were ICH-associated pathways and terms, such as FoxO signaling pathway, positive regulation of long term synaptic depression, autophagy - animal, IL-17 signaling pathway, regulation of transforming growth factor beta. Previous studies have confirmed that these pathways participate in the pathogenesis of hemorrhagic stroke. For instance, the long-term synaptic depression is associated with the memory deficits after hemorrhagic stroke [53], autophagy may contribute to the inflammatory injury and neuronal death in ICH [54], and IL-17 produced by γδT cells can increase neuroinflammation and secondary injury after ICH [55]. Furthermore, transforming growth factor beta impairs the blood-brain barrier integrity after hemorrhage and acts as potential inflammatory markers of stroke patients [55, 56]. Therefore, BYHWD are likely to exhibit therapeutic functions via tsRNAs by regulating the ICH-related pathways.
Additionally, FoxO signaling pathway is most significantly enriched among these pathways. Emerging evidence has implicated FoxO family in cognitive function, neurogenesis, axon growth and regeneration [57]. Moreover, FoxO signaling pathway may be involved to regulate the permeability of the blood brain barrier and inflammatory injury in rats after ICH [58, 59]. In this study, we found 7 targets enriched in FOXO signaling pathway were associated with rno-tRFi-Ser-25a and rno-tRF5-Ala-16a involving Foxo1, Mapk1, Sos1, Ikbkb etc. The transfection experiment showed the overexpression of rno-tRFi-Ser-25a could reduce the level of Foxo1. Conversely, when rno-tRFi-Ser-25a was downregulated, the level of Foxo1 would increase. As we know the Foxo1 is known upregulated after ICH [59], and rno-tRFi-Ser-25a was downregulated in our study. Hence, the increased expression level of Foxo1 may be caused by the tsRNA. Furthermore, BYHWD could upregulate rno-tRFi-Ser-25a to affect this signaling pathway and play a therapeutic role on ICH. tsRNAs are potential therapeutic targets in ICH treatment.
Unlike lncRNAs and miRNAs, tsRNAs are special therapeutic targets. In our previous study, we found that BYHWD could regulate biological functions including hemoglobin complex, oxygen transport, oxygen transporter activity and pyruvate metabolism via lncRNAs, to protect against long-term neurological deficiencies after ICH [26]. BYHWD exposure augmented angiogenetic miRNA (miR-26, miR-221 and miR-222) and VEGF expression to elevate angiogenesis in rat brain after stroke [24]. Compared with lncRNAs and miRNAs, the therapeutic mechanisms of tsRNAs are quite distinct. The functions of tsRNAs indicate that tsRNAs are novel and special therapeutic targets of BYHWD on ICH treatment.
Conclusion
In summary, this study initiatively revealed the changed expression patterns of tsRNAs in ICH rats after BYHWD treatment. tsRNAs might be novel potential therapeutic targets for BYHWD to regulate ICH-induced biological pathways. The present study provides the basis and direction for future investigations to explore the mechanisms by which BYHWD protects against long-term neurological deficiencies after ICH. Yet there are also limitations in this study. More sample-sizes and clinical specimens should be encouraged for the confirmation of the results. The interaction process between tsRNAs and mRNAs are needed to be clarified clearly by more studies.
Supplementary Material
Supplementary figures and table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary table S4.
Supplementary table S5.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81874425, 81673719 and 81603414) and China Postdoctoral Science Foundation (Nos. 2017T100614 and 2016M600639). We also thank Aksomics Co. (Shanghai, China) for technical support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524-37
2. Baharoglu MI, Cordonnier C, Al-Shahi Salman R, de Gans K, Koopman MM, Brand A. et al. Platelet transfusion versus standard care after acute stroke due to spontaneous cerebral haemorrhage associated with antiplatelet therapy (PATCH): a randomised, open-label, phase 3 trial. Lancet. 2016;387:2605-13
3. Adeoye O, Broderick JP. Advances in the management of intracerebral hemorrhage. Nat Rev Neurol. 2010;6:593-601
4. Hanley DF, Lane K, McBee N, Ziai W, Tuhrim S, Lees KR. et al. Thrombolytic removal of intraventricular haemorrhage in treatment of severe stroke: results of the randomised, multicentre, multiregion, placebo-controlled CLEAR III trial. Lancet. 2017;389:603-11
5. Lan X, Han X, Li Q, Yang QW, Wang J. Modulators of microglial activation and polarization after intracerebral haemorrhage. Nat Rev Neurol. 2017;13:420-33
6. Wang J. Preclinical and clinical research on inflammation after intracerebral hemorrhage. Prog Neurobiol. 2010;92:463-77
7. Hemphill JC 3rd, Greenberg SM, Anderson CS, Becker K, Bendok BR, Cushman M. et al. Guidelines for the management of spontaneous intracerebral hemorrhage: A guideline for healthcare professionals from the American heart association/American stroke association. Stroke. 2015;46:2032-60
8. Zhou G, Chen Z. Translating the power of TCM into patients' hope. Front Med. 2014;8:265-7
9. Kim HU, Ryu JY, Lee JO, Lee SY. A systems approach to traditional oriental medicine. Nat Biotechnol. 2015;33:264-8
10. Zhang X, Zheng W, Wang T, Ren P, Wang F, Ma X. et al. Danshen-Chuanxiong-Honghua ameliorates cerebral impairment and improves spatial cognitive deficits after transient focal ischemia and identification of active compounds. Front Pharmacol. 2017;8:452
11. He SM, Li CG, Liu JP, Chan E, Duan W, Zhou SF. Disposition pathways and pharmacokinetics of herbal medicines in humans. Curr Med Chem. 2010;17:4072-113
12. Hung IL, Hung YC, Wang LY, Hsu SF, Chen HJ, Tseng YJ. et al. Chinese herbal products for ischemic stroke. Am J Chin Med. 2015;43:1365-79
13. Luo L, Deng S, Yi J, Zhou S, She Y, Liu B. Buyang Huanwu decoction ameliorates poststroke depression via promoting neurotrophic pathway mediated neuroprotection and neurogenesis. Evid Based Complement Alternat Med. 2017;2017:4072658
14. Pan R, Cai J, Zhan L, Guo Y, Huang RY, Li X. et al. Buyang Huanwu decoction facilitates neurorehabilitation through an improvement of synaptic plasticity in cerebral ischemic rats. BMC Complement Altern Med. 2017;17:173
15. Guo Q, Zhong M, Xu H, Mao X, Zhang Y, Lin N. A systems biology perspective on the molecular mechanisms underlying the therapeutic effects of Buyang Huanwu decoction on ischemic stroke. Rejuvenation Res. 2015;18:313-25
16. Zhang ZQ, Tang T, Luo JK, Huang JF, Yang QD, Li XQ. et al. Effect of qi-tonifying and stasis-eliminating therapy on expression of vascular endothelial growth factor and its receptors Flt-1, Flk-1 in the brain of intracerebral hemorrhagic rats. Chin J Integr Med. 2007;13:285-90
17. Cui HJ, Yang AL, Zhou HJ, Wang C, Luo JK, Lin Y. et al. Buyang huanwu decoction promotes angiogenesis via vascular endothelial growth factor receptor-2 activation through the PI3K/Akt pathway in a mouse model of intracerebral hemorrhage. BMC Complement Altern Med. 2015;15:91
18. Li X, Zhang Y, Hong Z, Gong S, Liu W, Zhou X. et al. Transcriptome profiling analysis reveals the potential mechanisms of three bioactive ingredients of Fufang E'jiao Jiang during chemotherapy-induced myelosuppression in mice. Front Pharmacol. 2018;9:616
19. Zhang S, Zhu D, Li H, Zhang H, Feng C, Zhang W. Analyses of mRNA profiling through RNA sequencing on a SAMP8 mouse model in response to ginsenoside Rg1 and Rb1 treatment. Front Pharmacol. 2017;8:88
20. Xu S. Transcriptome profiling in systems vascular medicine. Front Pharmacol. 2017;8:563
21. Salta E, De Strooper B. Noncoding RNAs in neurodegeneration. Nat Rev Neurosci. 2017;18:627-40
22. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297-325
23. Zhang X, Hamblin MH, Yin KJ. Noncoding RNAs and stroke. Neuroscientist. 2018:1073858418769556
24. Yang J, Gao F, Zhang Y, Liu Y, Zhang D. Buyang Huanwu decoction (BYHWD) enhances angiogenic effect of mesenchymal stem cell by upregulating VEGF expression after focal cerebral ischemia. J Mol Neurosci. 2015;56:898-906
25. Hanjin C, Tao L, Pengfei L, Ali Y, Huajun Z, Jiekun L. et al. Altered long noncoding RNA and messenger RNA expression in experimental intracerebral hemorrhage - a preliminary study. Cell Physiol Biochem. 2018;45:1284-301
26. Cui H, Liu T, Li P, Yang A, Zhou H, Luo J. et al. An intersectional study of LncRNAs and mRNAs reveals the potential therapeutic targets of Buyang Huanwu decoction in experimental intracerebral hemorrhage. Cell Physiol Biochem. 2018;46:2173-86
27. Loher P, Telonis AG, Rigoutsos I. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci Rep. 2017;7:41184
28. Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141-5
29. Luo S, He F, Luo J, Dou S, Wang Y, Guo A. et al. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 2018;46:5250-68
30. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673-95
31. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
32. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404-9
33. Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci. 2016;41:679-89
34. Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res. 2016:5
35. Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T. et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636-50
36. Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790-802
37. Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397-400
38. Li Q, Hu B, Hu G-W, Chen C-Y, Niu X, Liu J. et al. tRNA-derived small non-coding RNAs in response to ischemia inhibit angiogenesis. Sci Rep. 2016;6:20850
39. Li Q, Han X, Lan X, Hong X, Li Q, Gao Y. et al. Inhibition of tPA-induced hemorrhagic transformation involves adenosine A2b receptor activation after cerebral ischemia. Neurobiol Dis. 2017;108:173-82
40. Chang CF, Cai L, Wang J. Translational intracerebral hemorrhage: a need for transparent descriptions of fresh tissue sampling and preclinical model quality. Transl Stroke Res. 2015;6:384-9
41. Wang M, Hong X, Chang CF, Li Q, Ma B, Zhang H. et al. Simultaneous detection and separation of hyperacute intracerebral hemorrhage and cerebral ischemia using amide proton transfer MRI. Magn Reson Med. 2015;74:42-50
42. Wang Y, Peng F, Xie G, Chen ZQ, Li HG, Tang T. et al. Rhubarb attenuates blood-brain barrier disruption via increased zonula occludens-1 expression in a rat model of intracerebral hemorrhage. Exp Ther Med. 2016;12:250-6
43. Xing Z, Xia Z, Peng W, Li J, Zhang C, Fu C. et al. Xuefu Zhuyu decoction, a traditional Chinese medicine, provides neuroprotection in a rat model of traumatic brain injury via an anti-inflammatory pathway. Sci Rep. 2016;6:20040
44. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57-62
45. Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct. 2015;10:51
46. Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093-105
47. Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. Global gene repression by Dicer-independent tRNA fragments. bioRxiv. 2017
48. Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA. et al. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723-35
49. Wei RL, Teng HJ, Yin B, Xu Y, Du Y, He FP. et al. A systematic review and meta-analysis of buyang huanwu decoction in animal model of focal cerebral ischemia. Evid Based Complement Alternat Med. 2013;2013:138484
50. Li Y, Luo J, Zhou H, Liao J-Y, Ma L-M, Chen Y-Q. et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048-55
51. Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909
52. Rosa Garcia-Silva M, Cabrera-Cabrera F, Catalina Gueida M, Cayota A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes (Basel). 2012;3:603-14
53. Provencio JJ, Swank V, Lu H, Brunet S, Baltan S, Khapre RV. et al. Neutrophil depletion after subarachnoid hemorrhage improves memory via NMDA receptors. Brain Behav Immun. 2016;54:233-42
54. Li Q, Weiland A, Chen X, Lan X, Han X, Durham F. et al. Ultrastructural characteristics of neuronal death and white matter injury in mouse brain tissues after intracerebral hemorrhage: coexistence of ferroptosis, autophagy, and necrosis. Front Neurol. 2018;9:581
55. Zhong Q, Zhou K, Liang QL, Lin S, Wang YC, Xiong XY. et al. Interleukin-23 secreted by activated macrophages drives gammadeltaT Cell Production of Interleukin-17 to aggravate secondary injury after intracerebral hemorrhage. J Am Heart Assoc. 2016;5:e004340
56. Jiang C, Kong W, Wang Y, Ziai W, Yang Q, Zuo F. et al. Changes in the cellular immune system and circulating inflammatory markers of stroke patients. Oncotarget. 2017;8:3553-67
57. Kim SY, Webb AE. Neuronal functions of FOXO/DAF-16. Nutr Healthy Aging. 2017;4:113-26
58. Li Z, Liang G, Ma T, Li J, Wang P, Liu L. et al. Blood-brain barrier permeability change and regulation mechanism after subarachnoid hemorrhage. Metab Brain Dis. 2015;30:597-603
59. Li Z, He Q, Zhai X, You Y, Li L, Hou Y. et al. Foxo1-mediated inflammatory response after cerebral hemorrhage in rats. Neurosci Lett. 2016;629:131-6
Author contact
![]() Corresponding author: Prof. Yang Wang. Institute of Integrative Medicine, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha 410008, China. E-mail: wangyang_xy87edu.cn. Phone: 086-731-84327213. Fax: 086-731-84327332.
Corresponding author: Prof. Yang Wang. Institute of Integrative Medicine, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha 410008, China. E-mail: wangyang_xy87edu.cn. Phone: 086-731-84327213. Fax: 086-731-84327332.

 Global reach, higher impact
Global reach, higher impact