10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(7):1336-1344. doi:10.7150/ijbs.33329 This issue Cite
Research Paper
Prognostic Score Predicts Survival in HPV-Negative Head and Neck Squamous Cell Cancer Patients
1. Division of Molecular Diagnostics, Department of Laboratory Medicine, Zhejiang Cancer Hospital, Hangzhou 310022, P.R. China
2. Department of Otorhinolaryngology, Head and Neck Surgery, Berlin Institute of Health, Charité- Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, Campus Benjamin Franklin, Berlin 12200, Germany
3. Key Laboratory of Laboratory Medicine, Ministry of Education of China, Zhejiang Provincial Key Laboratory of Medical Genetics, Wenzhou Medical University, Wenzhou 325035, P.R. China
4. Houston Methodist Research Institute, Houston 77030, United States
5. Department of Craniomaxillofacial, Oral and Plastic Surgery, Klinikum Ernst von Bergmann, Potsdam 14467, Germany
6. Clinic for Gynecology, Berlin Institute of Health, Charité- Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, Campus Benjamin Franklin, Berlin 12200, Germany
7. Department of Head and Neck Surgery, The University of Texas M. D. Anderson Cancer Center, Houston 77030, United States
*Equal contribution
Received 2019-1-19; Accepted 2019-3-17; Published 2019-5-12
Abstract
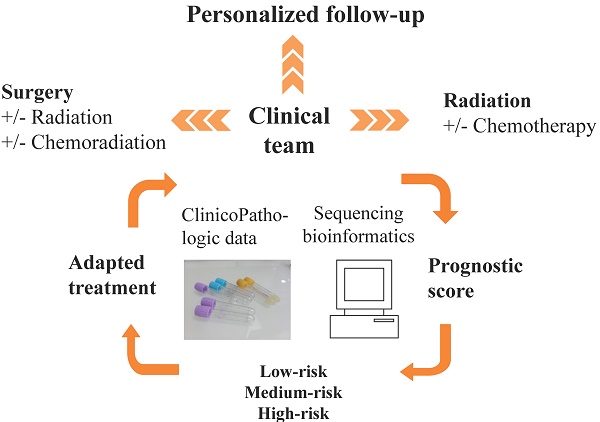
Although patients having head and neck squamous cell carcinoma (HNSCC) have high mortality, standardized prognostic tools are unavailable. As such, having a validated simple prognostic scoring system to help predict mortality in these high-risk patients is urgently needed. The current study aimed to develop and internally validate a prognostic scoring system for overall mortality in human papillomavirus (HPV)-independent HNSCC patients. Data on 400 consecutive patients from the Cancer Genome Atlas database with a known HPV-RNA negative status were analyzed. A prognostic model to predict patient overall mortality was developed using the logistic regression beta coefficients and a simple risk score was created. The model was internally validated using bootstrap validation with 2000 replications. Five covariates (age, pT, pN, perineural invasion, and EAp53 score) were used in the development of the mortality risk score in the final model. Three risk groups were stratified based on the prognostic scores: low-risk (<96 points), medium-risk (96-121 points), and high-risk (≥122 points) with a survival of 76%, 62% and 35%, respectively. The proposed model presented good discrimination in both the development (AUC = 0.76; 95% CI 0.70, 0.81) and bootstrap validation (AUC = 0.76; 95% CI 0.70, 0.81) with a non-significant Hosmer-Lemeshow chi-square of 6.17 (p = 0.63). The proposed prognostic scoring system is easy to use to predict patient overall mortality and could also help in the appropriate allocation of medical resources while managing HNSCC patients. External validation (including re-calibration if needed) should be conducted to test the model's generalizability in different populations.
Keywords: head and neck cancer, EAp53, predictive model, survival, next generation sequencing
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide. Among the estimated 500,000 new HNSCC cases reported globally every year, 63,000 are from the United States and 139,000 from the European countries [1, 2]. Despite advances in surgical technique, chemo- and radiotherapy, and more recently, immune checkpoint inhibitors, the overall survival rates of HNSCC patients remain approximately 50% over the last three decades and the treatment response for locally advanced disease remains unpredictable [3]. Meanwhile, standardized prognostic tools for mortality in this high-risk population are not available. Thus, there is a pressing need to stratify HNSCC patients into risk-categories and to adapt therapeutic and follow-up decisions acknowledging heterogeneous treatment responses seen in the same disease stage and with the same treatment and to focus care on patients with a predicted low five-year survival [4, 5].
Several predictive risk models for HNSCC have been developed [6-8]. In the mathematical predictive model developed by Emerick et al. using the Survey, Epidemiology and End-Results 2009 dataset, anatomical site, age at diagnosis, race, tumor extension, N stage, and extracapsular spread were found to contribute to mortality [6]. The authors also found tumor diameter increased along with the additional chance of death across all subsites, but not for tumors less than 1 cm and around 5 cm. The number of positive nodes, up to 5 nodes, was found to be associated with an additional ~14% risk of death [6]. However, Emerick's model did not include human papillomavirus (HPV) status. It is now clear that an oncogenic subtype, HPV-associated oropharyngeal cancer (OPSCC), is distinct from the HPV-negative tumors in genetic alterations and a better prognosis. It was recently added with separate TNM-stages to the 8th edition of AJCC toward a de-escalation treatment protocol [9]. Recently, a nomogram model has also been developed for patients with locoregionally advanced (LA) OPSCC treated with primary radiation-based therapy [7]. Factors including: age at diagnosis, smoking, Zubrod performance status, education, anemia, tumor p16 status, T stage, and N stage were selected for the final model which can predict 2-year and 5-year overall survival (OS) and progression-free survival (PFS) [7]. The strength of this nomogram model is the analysis of sociodemographic factors and HPV status in addition to clinicopathologic factors in both the development and external validation cohorts. However, Fakhry's model has been applied for LA-OPSCC patients who received primary radiotherapy without including genomic data [7].
Knowledge of genome alterations proposes that there are a more complex biologic feature for HNSCC [3]. The mutational profile of HNSCC and mutated gene-related pathways have the potential to stratify patients, aid in initial treatment decisions and based on the probability of recurrence provide a more rationally tailored after-care prognosis with clinical follow-up and imaging [3]. For example, Rebeiro et al reported that a genomic model based on chromosomal alterations has good performance in predicting HNSCC recurrence and metastasis [8]. In oral cancer, the hybrid model of ReliefF-GA-ANFIS has shown that clinicopathologic data together with alcohol consumption, tumor invasion, and p63 gene expression achieved the best accuracy for prognosis models in a total of 31 oral SCC (OSCC) cases [10]. However, these models also did not stratify patients with HPV status.
Given by the distinct tumor biology and clinical outcome of HPV-associated HNSCC, analyzing tumor HPV status should increase the accuracy in outcome predictions. Additionally, there is a need that genomics be translated and interpreted to answer questions relevant for geneticists, clinicians and patients. To date, Next-Generation Sequencing is becoming affordable and is thus moving to more frequent clinical application. However, at this stage molecular testing for alterations in HNSCC has not become a meaningful tool in clinical practice yet. Moreover, the identified specific mutations in cancer are challenged by the mutational heterogeneity, a phenomenon seen in most tumors. To stimulate these findings from discovery to clinical practice, incorporation of clinical and demographic information with genomic data is necessary. Thus, the current study aimed to develop and validate a prognostic system for HPV-negative HNSCC in cooperation with genomic and clinicopathologic factors toward an implementation of a clinically useful tool. The proposed mortality risk score is simple and easy to use in quantifying the mortality risk for HNSCC patients and could provide the appropriate guidance in post-therapy follow-up.
Materials and Methods
Study population
The study used retrospective de-identified genomic and clinical data of HPV-negative HNSCC patients from The Cancer Genome Atlas (TCGA). The TCGA raw datasets for 530 tumor samples from HNSCC patients were obtained from cBioPortal (http://www.cbioportal.org assessed on 2018/09/20) [11, 12]. The inclusion criteria for analysis was: (1) having a primary tumor in the oral cavity, oropharynx, hypopharynx, or larynx; (2) age<80 years; (3) having no prior history of malignancies nor previous treatments for cancer; (4) have available HPV E6/E7 RNA status; and (5) having documented outcome as either living or dead (“died”).
Variable definitions
Excessive alcohol consumption was identified as: >4 drinks/day for man or >3 drinks/day for woman [13]. Lifelong non-smoker was defined as negative primary tobacco exposure and current and reformed smokers as positive. PCR-based RNA-detection of viral E6/E7 oncogenes was used to identify HPV-negative tumor tissue samples and the HPV data were retrieved from a previously published article [14]. The p16 status was identified by immunohistochemistry. Affymetrix SNP6 copy number measurements were used and CDKN2A copy number loss was defined as having a Logarithm (Log 2) less than -0.7. Gene mutations of CCND1, CDKN2A, FAT1, NOTCH1, NSD1, PIK3CA, AJUBA, EGFR, PTEN, KMT2D and TP53 were defined as positive.
To estimate clinical prognosis, a computational approach named “evolutionary action p53” (EAp53) was developed to stratify TP53 missense mutations into high and low risk [15]. An online tool, EAp53 server, was used to predict the impact of P53 missense mutation and then a score between 0-100 was given according to a previous publication [15]. The threshold of EAp53 was established as previously introduced where a rank-based procedure was analyzed based on the Cox proportional hazard ratios [15]. The threshold of EAp53 was 78 in our study. Next, patients with missense mutation were subsequently divided into high-risk (≥78) and low-risk (<78) group. Other mutations such as nonsense mutation and frameshift mutation were grouped as other.
Statistical analyses
Univariate logistic regression for each variable was performed to select potential factors of overall mortality. Further, variables having a p-value of <0.2 in the univariate analysis or being considered as clinically important were investigated using the multivariate modeling. The Bayesian model averaging method was used to select variables for the multiple logistic regression model [16]. To reduce the model subsets, the Likelihood Ratio test was performed. The best model was further selected based on the smallest Bayesian information criterion. The development of the risk score has been described elsewhere [16]. Briefly, each significant variable was assigned a weighted-point based on its β coefficient value. Patient risk scores were calculated and stratified into three risk groups which were significantly distinct in predictive mortality risk (low-, medium- and high-risk). Area under the receiver operating characteristic curve (AUC) was used to determine model discrimination. Hosmer-Lemeshow's goodness of fit test was performed to evaluate the model calibration. Overall performance of the final model and risk score was evaluated using the Brier score. Internal validation was performed using the bootstrap resampling method with 2000 replications. All the analyses were performed using Stata version 15.1 (StataCorp LLC, College Station, TX, USA). A p value of <0.05 was considered statistically significant.
Results
Characteristics of the study sample
From a cohort of 528 cases, 400 consecutive cases met the inclusion criteria and were used for the univariate analysis. A total of 128 patients were excluded from the analysis due to the following reasons (patients may meet more than one exclusion criteria): 1) age>80 years old [26 cases (4.92%)]; 2) having a history of cancer [33 cases (6.25%)]; and 3) patients that were HPV RNA-positive [72 cases (13.64%)] or without HPV status data [8 cases (1.52%)].
Development of a prognostic score system for HNSCC mortality
Table 1 presents the crude association between the potential risk factors and overall mortality. Ten variables (age, pT, pN, extracapsular spread, argiolymphatic invasion, perineural invasion, lymph node dissection, CDKN2A copy number loss, NSD1 mutation and EAp53) which were found to have prognostic significance and were examined for the development of a final multiple logistic regression model. In the final model, five variables (age, pT, pN, perineural invasion, and EAp53) were significant and were used for the development of the mortality risk score (Table 2). We assigned weighted points for each of the final five risk factors by the linear transformation of the corresponding regression coefficient [(divided by the smallest β-coefficient (0.03, age), and rounded to the nearest integer, (Table 2)]. The following formula was used to calculate the prognostic score for individual patients:
Risk score=1*[Age] + 21*[pT] + 31*[pN] + [Perineural invasion: No, 0 point; Yes, 37 points; Unknown, 38 points] + [EAp53: Wild type & Low risk, 0 point; High risk & Other, 20 points;].
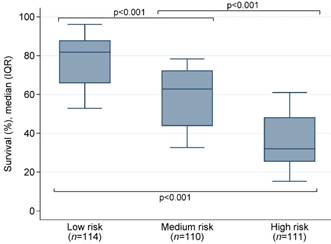
All variables were binary with “No” = 0 and “Yes” = 1. Patients were then stratified into three groups that can significantly differentiate the predictive mortality-risk: low-risk group (<96points), medium-risk group (96-121 points), and high-risk group (≥122 points). As shown in Table 3, the mortality was 24% in the low-risk group, 38% in the medium-group, and 65% in the high-risk group. The median survival probability by risk group was presented in Figure 1. The intercept (-4.358669) of the final model and corresponding regression coefficients of each variable included in the risk score were used to calculate the probability of death:
Predicted probability of death= -4.358669 + 0.0322484*[Age] + 0.6661356*[pT] + 0.9999758*[pN] + [Perineural invasion: No, *0; Yes, *1.202005; Unknown, *1.223973] + 0.633055*[EAp53].
Internal validation
The final model presented good discrimination in both the development (AUC = 0.76; 95% CI 0.70, 0.81) and bootstrap validation (AUC = 0.76; 95% CI 0.70, 0.81) (Table 3, Figure S1) models. The evaluation using the risk score alone also obtained a good discrimination in both the development and bootstrap validation (AUC = 0.72; 95% CI 0.66, 0.77 and AUC = 0.70; 95% CI 0.65, 0.76, respectively) models. A good calibration with a non-significant Hosmer-Lemeshow chi-square of 6.17 (p = 0.63) and an excellent overall performance with a Brier score of 0.198 were presented in the final model (Table 3). Patients with medium- and high-risk had significantly higher odds of mortality compared to patients with low-risk (Table 4).
The median survival probability by risk group.
Crude associations between potential risk factors and mortality.
| Variable | All | Alive | Dead | Unadjusted OR | P-value |
|---|---|---|---|---|---|
| (n=400) | (n=222) | (n=178) | (95% CI) | ||
| Age (years), median (IQR) | 61.0 (53-68) | 60.0 (53-67) | 62.0 (53-69) | 1.01(1.00, 1.03) | 0.11 |
| Gender | |||||
| Male | 293 (73.3) | 168 (75.7) | 125 (70.2) | (reference) | |
| Female | 107 (26.7) | 54 (24.3) | 53 (23.8) | 1.32 (0.85, 2.06) | 0.22 |
| Race | |||||
| White | 333 (83.2) | 186 (83.8) | 147 (82.6) | (reference) | |
| Asian | 11 (2.8) | 7 (3.2) | 4 (2.2) | 0.72 (0.21, 2.52) | 0.61 |
| Black or African American | 42 (10.5) | 21 (9.4) | 21 (11.8) | 1.27 (0.67, 2.41) | 0.47 |
| American Indian or Alaska native | 2 (0.5) | 1 (0.4) | 1 (0.6) | 1.27 (0.08, 20.40) | 0.87 |
| Unknown | 12 (3.0) | 7 (3.2) | 5 (2.8) | 0.91 (0.28, 2.91) | 0.87 |
| Tobacco use | |||||
| No | 80 (20.0) | 51 (23.0) | 29 (16.3) | (reference) | |
| Yes | 311 (77.8) | 168 (75.7) | 143 (80.3) | 1.50 (0.90, 2.49) | 0.12 |
| Unknown | 9 (2.2) | 3 (1.3) | 6 (3.4) | 3.52 (0.82, 15.13) | 0.09 |
| Excessive alcohol | |||||
| No | 242 (60.5) | 143 (64.4) | 99 (55.6) | (reference) | |
| Yes | 45 (11.3) | 26 (11.7) | 19 (10.7) | 1.06 (0.55, 2.01) | 0.87 |
| Unknown | 113 (28.2) | 53 (23.9) | 60 (33.7) | 1.64 (1.04, 2.56) | 0.03 |
| Laterality | |||||
| Left | 108 (27.0) | 66 (29.7) | 42 (23.6) | (reference) | |
| Right | 106 (26.5) | 69 (31.1) | 37 (20.8) | 0.84 (0.48, 1.47) | 0.54 |
| Midline | 31 (7.8) | 19 (8.6) | 12 (6.7) | 0.99 (0.44, 2.25) | 0.99 |
| Unknown | 155 (38.7) | 68 (30.6) | 87 (48.9) | 2.01 (1.22, 3.32) | 0.006 |
| *OPSCC | |||||
| No | 264 (66.0) | 141 (63.5) | 123 (69.1) | (reference) | |
| Yes | 136 (34.0) | 81 (36.5) | 55 (30.9) | 0.78 (0.51, 1.18) | 0.24 |
| **LSCC | |||||
| No | 295 (73.8) | 163 (73.4) | 132 (74.2) | (reference) | |
| Yes | 105 (26.2) | 59 (26.6) | 46 (25.8) | 0.96 (0.61, 1.51) | 0.87 |
| Histology | |||||
| G1+G2 | 304 (76.0) | 172 (77.5) | 132 (74.2) | (reference) | |
| G3+G4 | 86 (21.5) | 46 (20.7) | 40 (22.5) | 1.13 (0.70, 1.83) | 0.61 |
| Others | 10 (2.5) | 4 (1.8) | 6 (3.3) | 1.95 (0.54, 7.07) | 0.31 |
| pT | |||||
| T1+T2 | 133 (33.3) | 88 (39.6) | 45 (25.3) | (reference) | |
| T3+T4 | 237 (59.2) | 117 (52.7) | 120 (67.4) | 2.01 (1.29, 3.12) | 0.002 |
| Others | 30 (7.5) | 17 (7.7) | 13 (7.3) | 1.50 (0.67, 3.35) | 0.33 |
| pN | |||||
| N0+N1 | 201 (50.3) | 141 (63.5) | 60 (33.7) | (reference) | |
| N2+N3 | 144 (36.0) | 59 (26.6) | 85 (47.8) | 3.39 (2.16, 5.30) | <0.001 |
| Others | 55 (13.7) | 22 (9.9) | 33 (18.5) | 3.53 (1.90, 6.54) | <0.001 |
| Positive surgical margins | |||||
| Negative | 289 (72.2) | 163 (73.4) | 126 (70.8) | (reference) | |
| Positive & Close | 79 (19.8) | 39 (17.6) | 40 (22.5) | 1.33 (0.81, 2.18) | 0.27 |
| Unknown | 32 (8.0) | 20 (9.0) | 12 (6.7) | 0.78 (0.37, 1.65) | 0.51 |
| Extracapsular spread | |||||
| No | 197 (49.3) | 130 (58.6) | 67 (37.6) | (reference) | |
| Yes | 90 (22.5) | 30 (13.5) | 60 (33.7) | 3.88 (2.29, 6.58) | <0.001 |
| Unknown | 113 (28.2) | 62 (27.9) | 51 (28.7) | 1.60 (0.99, 2.56) | 0.049 |
| Angiolymphatin invasion | |||||
| No | 181 (45.3) | 122 (55.0) | 59 (33.1) | (reference) | |
| Yes | 100 (25.0) | 45 (20.3) | 55 (30.9) | 2.53 (1.53, 4.17) | <0.001 |
| Unknown | 119 (29.7) | 55 (24.7) | 64 (36.0) | 2.41 (1.50, 3.87) | <0.001 |
| Perineural invasion | |||||
| No | 147 (36.8) | 107 (48.2) | 40 (22.5) | (reference) | |
| Yes | 148 (37.0) | 66 (29.7) | 82 (46.0) | 3.32 (2.04, 5.41) | <0.001 |
| Unknown | 105 (26.2) | 49 (22.1) | 56 (31.5) | 3.06 (1.80, 5.18) | <0.001 |
| Lymph Node dissection | |||||
| No lymph node dissection obtained | 50 (12.5) | 21 (9.5) | 29 (16.3) | (reference) | |
| Lymph node dissection obtained | 347 (86.8) | 199 (89.6) | 148 (83.1) | 0.54 (0.30, 0.98) | 0.04 |
| Unknown | 3 (0.7) | 2 (0.9) | 1 (0.6) | 0.36 (0.03, 4.26) | 0.42 |
| CDKN2A copy number loss | |||||
| No | 264 (66.0) | 160 (72.1) | 104 (58.5) | (reference) | |
| Yes | 130 (32.5) | 60 (27.0) | 70 (39.3) | 1.79 (1.17, 2.74) | 0.01 |
| Unknown | 6 (1.5) | 2 (0.9) | 4 (2.2) | 3.08 (0.55, 17.10) | 0.20 |
| CCND1 mutation | |||||
| No | 387 (96.8) | 215 (96.8) | 172 (96.7) | (reference) | |
| Yes | 3 (0.7) | 1 (0.5) | 2 (1.1) | 2.50 (0.22, 27.80) | 0.46 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.83 (0.23, 3.00) | 0.78 |
| CDKN2A mutation | |||||
| No | 292 (73.0) | 161 (72.5) | 131 (73.6) | (reference) | |
| Yes | 98 (24.5) | 55 (24.8) | 43 (24.2) | 0.96 (0.61, 1.52) | 0.87 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.82 (0.23, 2.96) | 0.76 |
| FAT1 mutation | |||||
| No | 293 (73.2) | 161 (72.5) | 132 (74.2) | (reference) | |
| Yes | 97 (24.3) | 55 (24.8) | 42 (23.6) | 0.93 (0.59, 1.48) | 0.76 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.81 (0.22, 2.94) | 0.75 |
| NOTCH1 mutation | |||||
| No | 317 (79.3) | 178 (80.2) | 139 (78.1) | (reference) | |
| Yes | 73 (18.2) | 38 (17.1) | 35 (19.7) | 1.18 (0.71, 1.96) | 0.53 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.85 (0.24, 3.08) | 0.81 |
| NSD1 mutation | |||||
| No | 338 (84.5) | 180 (81.1) | 158 (88.8) | (reference) | |
| Yes | 52 (13.0) | 36 (16.2) | 16 (9.0) | 0.51 (0.27, 0.95) | 0.03 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.76 (0.21, 2.74) | 0.67 |
| PIK3CA mutation | |||||
| No | 325 (81.3) | 182 (82.0) | 143 (80.4) | (reference) | |
| Yes | 65 (16.2) | 34 (15.3) | 31 (17.4) | 1.16 (0.68, 1.98) | 0.59 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.85 (0.23, 3.06) | 0.80 |
| AJUBA mutation | |||||
| No | 367 (91.8) | 206 (92.8) | 161 (90.5) | (reference) | |
| Yes | 23 (5.7) | 10 (4.5) | 13 (7.3) | 1.66 (0.71, 3.89) | 0.24 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.85 (0.24, 3.07) | 0.81 |
| EGFR mutation | |||||
| No | 376 (94.0) | 212 (95.5) | 164 (92.2) | (reference) | |
| Yes | 14 (3.5) | 4 (1.8) | 10 (5.6) | 3.23 (1.00, 10.49) | 0.05 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.86 (0.24, 3.10) | 0.82 |
| PTEN mutation | |||||
| No | 381 (95.3) | 208 (93.7) | 173 (97.2) | (reference) | |
| Yes | 9 (2.2) | 8 (3.6) | 1 (0.6) | 0.15 (0.02, 1.21) | 0.08 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.80 (0.22, 2.89) | 0.74 |
| KMT2D mutation | |||||
| No | 329 (82.3) | 187 (84.2) | 142 (79.8) | (reference) | |
| Yes | 61 (15.2) | 29 (13.1) | 32 (18.0) | 1.45 (0.84, 2.51) | 0.18 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 0.88 (0.24, 3.17) | 0.84 |
| p16 | |||||
| Negative | 63 (15.8) | 43 (19.4) | 20 (11.2) | (reference) | |
| Positive | 6 (1.5) | 5 (2.2) | 1 (0.6) | 0.43 (0.05, 3.93) | 0.45 |
| Unknown | 331 (82.7) | 174 (78.4) | 157 (88.2) | 1.94 (1.09, 3.44) | 0.02 |
| ***EAp53 | |||||
| Wild type, Low risk | 133 (33.3) | 85 (38.3) | 48 (27.0) | (reference) | |
| High risk, Other | 257 (64.2) | 131 (59.0) | 126 (70.8) | 1.70 (1.11, 2.62) | 0.02 |
| Unknown | 10 (2.5) | 6 (2.7) | 4 (2.2) | 1.18 (0.32, 4.39) | 0.80 |
*OPSCC: Oropharyngeal squamous cell carcinoma; **LSCC: Laryngeal squamous cell carcinoma; ***Low risk: EAp53 score ≤78; high risk: EAp53 score >78.
Multiple logistic regression model and weighted point assignment (n=335).
| Variable | β coefficient | Adjusted OR (95% CI) | P-value | *Weight point |
|---|---|---|---|---|
| Age (years) | 0.03 | 1.03 (1.01, 1.06) | 0.01 | 1 |
| pT | 0.67 | 1.95 (1.13, 3.35) | 0.02 | 21 |
| pN | 1.00 | 2.72 (1.62, 4.57) | <0.001 | 31 |
| Perineural invasion | ||||
| No | (reference) | |||
| Yes | 1.20 | 3.33 (1.88, 5.90) | <0.001 | 37 |
| Unknown | 1.22 | 3.40 (1.76, 6.59) | <0.001 | 38 |
| EAp53 | 0.63 | 1.88 (1.12, 3.16) | 0.02 | 20 |
C-statistic = 0.76; * Using a linear transformation of the corresponding β coefficient, weighted points of each risk factor were calculated [was divided by the smallest β coefficient (0.03, age), and rounded to the nearest integer]; Intercept = -4.358669.
Risk score performance for all variables of the multiple logistic regression model in patients who have complete data.
| Risk Group | n (%) | Mean Score (±SD) | Mortality, n (%) | *P-value |
|---|---|---|---|---|
| Complete data for all variables of multiple logistic regression model | 335 (100%) | 107 (±28) | 141 (42%) | <0.001 |
| Low-risk group (<96 points) | 114 (34%) | 78 (±13) | 27 (24%) | |
| Medium-risk group (96-121 points) | 110 (33%) | 108 (±8) | 42 (38%) | |
| High-risk group (≥122 points) | 111 (33%) | 139 (±15) | 72 (65%) | |
| Discrimination assessment | ||||
| AUC (95% CI), final model in development | 0.76 (0.70, 0.81) | |||
| AUC (95% CI), final model in bootstrap validation (2000 replications) | 0.76 (0.70, 0.81) | |||
| AUC (95% CI), prognostic score alone | 0.72 (0.66, 0.77) | |||
| AUC (95% CI), prognostic score alone, bootstrap validation (2000 replications) | 0.70 (0.65, 0.76) | |||
| Calibration assessment** | ||||
| Hosmer-Lemeshow's goodness-of-fit test, final model | Chi-square = 6.17; p-value = 0.63 | |||
| Hosmer-Lemeshow's goodness-of-fit test, prognostic score alone | Chi-square = 14.4; p-value = 0.07 | |||
| Overall performance assessment*** | ||||
| Brier score, final model | 0.198 | |||
| Brier score, prognostic score alone | 0.212 | |||
Risk score ranged from 36-174. *Overall p-value. Pairwise p-values between low- vs. medium-risk, low- vs. high-risk and medium- vs. high-risk was 0.005, <0.001 and <0.001, respectively. **a non-significant Hosmer-Lemeshow goodness (p≥0.05) of goodness-of-fit test indicates good calibration.***Brier score: ranges 0-1, the smaller the score, the better performance.Abbreviation: SD, standard deviation; AUC, area under the Receiver Operating Characteristic curve.
Odds ratio (OR) for death by each risk group (n=335).
| Risk group | OR (95% CI) | P-value |
|---|---|---|
| Low-risk group (<96 points) | (reference) | |
| Medium-risk group (96-121 points) | 1.99 (1.11, 3.55) | 0.02 |
| High-risk group (≥122 points) | 5.95 (3.30, 10.72) | <0.001 |
Coefficient standard errors at 2000 replications were estimated by bootstrap.
In the final model and in the development of the prognostic scoring system, a sample of 335/400 (83.8%) patients having complete data for all five included variables were used. We found no significant differences in all demographic and clinical characteristics other than for gender (p = 0.04) and mortality (p = 0.03) between the 335 included patients in the final model and 65 excluded patients with incomplete data (Table S1).
Online calculator based on the prognostic score system
We have applied and created a free-of-charge online risk score calculator for HPV-RNA negative HNSCC patients. The calculator can be downloaded from the following link https://oaa.app.link/yWLO3m5QPP (a free OpenAsApp account registration prior to access the calculator is required) and can be used, on android or iOS mobile devices. Our model and online calculator predicts the survival probability (%) for an individual patient where a risk score (in points) and subsequently a risk group of low, medium or high are also provided.
Discussion
In this study, we present a newly developed and internally validated prognostic scoring system to predict all-cause mortality risk for HPV-negative HNSCC patients. Our final model included age, pT, pN, perineural invasion, and EAp53. Incorporated with clinicopathologic data and genomic features, our predictive prognostic score system can provide clinicians with additional information that would help allocate appropriate treatments and follow-up resources.
Our findings are consistent with the current literature, which indicates that pT and pN stage are contributive to mortality of HNSCC patients independent of HPV status [6, 7]. The pT and pN categories have been demonstrated to be independent factors for OS or PFS in both HPV-positive and HPV-negative OPSCC patients [7, 17]. We also found pN status as the most important prognostic factor for a substantial proportion of patients with pT1 - pT2 HNSCC in our recent 17-year retrospective single center cohort study [18]. These observations underscore the possible impact of traditional TNM classification on HNSCC outcome.
In line with other authors' findings, we found that older age at diagnosis was associated with overall mortality and is a significant risk predictor in our model [6, 7]. The incidence of HNSCC in the elderly patients is increasing not only for HPV-negative tumor but also for HPV-positive oropharyngeal cancer as seen in a recent report of the National Cancer Database of the United States [19]. For older patients, the reduced tolerability of multimodality therapy and general physical health issues and co-morbidities are main factors that would lead to inefficient treatments [20]. These general challenges are well-known and intensive multimodality treatment with a manageable safety profile is still unavailable.
Perineural invasion (PNI) occurs in up to 40% of HNSCC patients and the association with loco- regional recurrence, distant metastases and poor survival have also been documented [21, 22]. In our study, occurring in 37% of patients, PNI had significantly higher odds for mortality. To date, the biological mechanisms for PNI remains elusive and therapeutic escalation should be considered for improvement of patient's outcome [23]. Recently, an in vitro PNI model of HNSCC has been developed where a rapid evaluation of multiple treatment conditions has been performed to improve therapeutic selection [24]. In addition, quantitative characteristics of PNI as well as nerve parameters can also be used to tailor adjuvant treatment [25]. Future studies incorporating in vitro and in vivo models to investigate the underlying mechanism are needed.
One important consideration is the impact of genomic alterations on HNSCC outcome. In our model, EAp53 score appears to be a strong predictor of mortality. TP53 gene is mutated in 60% - 70% HNSCC and consequently leads to HNSCC oncogenesis mainly due to the loss of wild-type p53 (wtp53) function [3]. Moreover, TP53 variants are heterogeneous and gain of function p53 mutants are further identified to be independent of wtp53 function and being related to tumorigenesis and progression especially in HPV-negative HNSCC [26]. In head and neck cancer, an initial study demonstrated that high-risk patients stratified by EAp53 were associated with decreased survival and increased distant metastases [15]. Osman et al. demonstrated that a subgroup of HNSCC patients with higher EAp53 score was associated with decreased sensitivity to cisplatin treatment [27]. Additionally, EAp53 can also be used to predict the patient's response to treatment with cisplatin in an orthotopic mouse model of tongue cancer [27]. In line with these findings, EAp53 has successfully been associated with survival in advanced larynx cancer in a prospective study and its associations with lymphatic metastasis in larynx cancer and pN+ extra-nodal extension+ status in OSCC have also been reported [28, 29]. Tumors with high-risk mutations of the TP53 gene also represent a more aggressive biological phenotype identified with being more resistant to chemo- or radiotherapy [27, 30]. Notably, recent guidelines recommend that the screening strategies for TP53 assessment in human cancer are necessary in clinical practice given its clinical significance [31]. Importantly, reactivation of the p53 function enhances the cytotoxicity and tumor apoptosis of cisplatin treatment in preclinical studies and novel targeted therapies such as p53-MDM2 inhibitors have been described [32-34]. These data represent an important step for understanding EAp53 as potentially targetable molecular alterations and also for therapeutic selection and molecular monitoring of patients.
Smoking is a known risk factor for the development of HNSCC [3]. Although, smoking status did not significantly contribute to mortality in our model, one recent study has characterized the genetic smoking signature of HNSCC and identified a higher mutational load and TP53 mutation, not other driver mutations in smokers compared to non- smokers [35]. Moreover, the proportion of smokers was higher in TP53-mutated vs TP53 wild-type HNSCC patients [35]. However, in HPV-positive tumors, TP53 mutation did not differ greatly by smoking status [36]. Other investigators also assessed the immune feature of HNSCC and reported lower levels of immune infiltration in TP53-mutated tumors [35]. Additionally, lower response rates to immune checkpoint inhibitor therapy among smokers with HNSCC may be due to their genetic and immune signatures [37], which raise the possibility that tobacco-induced genetic and epigenetic changes, rather than as a discrete variable (positive vs. negative tobacco exposure), could confound treatment response and survival. HPV-positive OPSCC are more commonly infiltrated by HPV-specific T-cells and making it more sensitive to standard therapy [38]. Additional studies are warranted to further describe the smoking related molecular landscape in both HPV-positive and negative tumors.
For multimodality management of HNSCC, cisplatin is recommended as the radiosensitizer in adjuvant setting as well as in definitive setting and recently also for patients with advanced HPV-positive OPSCC [39]. However, it is likely that HNSCC patients who have a higher load of TP53 mutations may render the effectiveness of the cisplatin treatment [27]. Thus, for high-risk patients, there is a more pressing need of improving the effectiveness of treatment. For example, drug sensitivity testing utilizing organoids derived from HNSCC patients could help to allocate effective treatment [40]. We recently found Disulfiram (Antabuse®) traditionally used to treat alcohol dependence acts as potent radio-chemo sensitizer of HNSCC in preclinical studies (unpublished data). While more effective management strategies should be developed and customized to treat e patients, the treatment protocols currently in use and under clinical investigation can potentially be tailored for proper indication for individual patients with different mortality risk. For example, one study has assessed the optimal dosing schedule of concurrent cisplatin in locally advanced HNSCC and demonstrates that cisplatin at 100 mg/ m2 every 3 weeks performed better in loco-regional control, but with more toxicity, than cisplatin at 30 mg/m2 weekly as adjuvant chemoradiotherapy [41]. We found docetaxel in combination with cisplatin and 5‐fluorouracil effectively prolonged survival in patients with locally advanced and/or recurrent metastatic HNSCC with tolerable side effects [42]. Taken together, a favorable safety profile with versatile anticancer effects is urgently needed to obtain curative therapeutic efficacy in aggressive cases.
Our study is limited by the small sample size and the proportion of individuals lacking complete data. Given the excluded patients having a higher mortality than included patients, our data cannot rule out completely the possibility of selection bias. However, the similarity in most of the available demographic and clinical variables between the two cohorts suggests that the selection bias, if any, would be minimal. Second, we acknowledge that our study includes a more homogenous population which is mainly white. A future external validation study focus on a more heterogeneous population would be necessary to test the bias. Third, the treatment effect was not adjusted for in our analyses due to unknown information. Moreover, delays in postoperative radiotherapy or chemoradiotherapy have been identified in association with decreased survival for patients who underwent curative intent surgery [43]. Overall treatment time greater than approximately 14 weeks was found to consistently increase mortality risk [43]. Therefore, the benefit of treatment protocols must be evaluated in future prospective studies. Despite these potential limitations, we have improved upon previous models by adding genomic profiling and HPV-status. Our predictive model may provide further rationale for intervention as early as possible in the course of disease and initiating appropriate therapy at the time of diagnosis and refined follow-up protocols.
In summary, our predictive model has the potential to predict the mortality risk and stratify patients with HPV-negative HNSCC into 3 categories, low, medium, and high risk. We proposed that for low-risk patients, a management per routine protocols is sufficient. Intermediate risk patients may benefit from a closer follow-up such as follow-up every 3-month, imaging (CT/MRI) once a year or if they present with any change in pain of function within 6 weeks (less than three months). Patients with high-risk would need more aggressive treatments if available a closer follow-up adapted to the individual ability to cover costs, and personal wishes would be advised.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81702091), the Medical and the Health Science Project of Zhejiang Province (2019KY327), the Opening Project of Zhejiang Provincial Top Key Discipline of Clinical Medicine (No. LKFJ008) and Guangji Talents Foundation Award (E) of Zhejiang Cancer Hospital.
Author Contributions
Q.X., M.J.N. and A.E.A. conceived and designed the study. Q.X., D.Y. and N.D. accessed and analyzed the data. N.D. and E.A.G. developed the methodology. Q.X., N.D., B.S., A.M.K., M.J.N., A.E.A., E.A.G. wrote and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gatta G, Botta L, Sanchez MJ, Anderson LA, Pierannunzio D, Licitra L. et al. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer. 2015;51:2130-43
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7-30
3. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nature reviews Cancer. 2018;18:269-82
4. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136:E359-86
5. Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F. et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:2171-7
6. Emerick KS, Leavitt ER, Michaelson JS, Diephuis B, Clark JR, Deschler DG. Initial clinical findings of a mathematical model to predict survival of head and neck cancer. Otolaryngol Head Neck Surg. 2013;149:572-8
7. Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal DI, Weber RS, Lambert L. et al. Development and Validation of Nomograms Predictive of Overall and Progression-Free Survival in Patients With Oropharyngeal Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2017;35:4057-65
8. Ribeiro IP, Caramelo F, Esteves L, Menoita J, Marques F, Barroso L. et al. Genomic predictive model for recurrence and metastasis development in head and neck squamous cell carcinoma patients. Scientific reports. 2017;7:13897
9. Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC. et al. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA: a cancer journal for clinicians. 2017;67:122-37
10. Chang SW, Abdul-Kareem S, Merican AF, Zain RB. Oral cancer prognosis based on clinicopathologic and genomic markers using a hybrid of feature selection and machine learning methods. BMC Bioinformatics. 2013;14:170
11. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1
12. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
13. Chen WS, Bindra RS, Mo A, Hayman T, Husain Z, Contessa JN. et al. CDKN2A Copy Number Loss Is an Independent Prognostic Factor in HPV-Negative Head and Neck Squamous Cell Carcinoma. Front Oncol. 2018;8:95
14. Nulton TJ, Olex AL, Dozmorov M, Morgan IM, Windle B. Analysis of The Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget. 2017;8:17684-99
15. Neskey DM, Osman AA, Ow TJ, Katsonis P, McDonald T, Hicks SC. et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer research. 2015;75:1527-36
16. Nguyen DT, Graviss EA. Development and validation of a risk score to predict mortality during TB treatment in patients with TB-diabetes comorbidity. BMC Infect Dis. 2019;19:10
17. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF. et al. Human papillomavirus and survival of patients with oropharyngeal cancer. The New England journal of medicine. 2010;363:24-35
18. Qian X, Sinikovic B, Schreiber F, Ochsenreither S, Klinghammer K, Wollenberg B. et al. pN status predicts outcomes in surgically treated pT1-pT2 patients of various disease stages with squamous cell carcinoma of the head and neck: a 17-year retrospective single center cohort study. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies. 2018;275:2787-95
19. Rettig EM, Zaidi M, Faraji F, Eisele DW, El Asmar M, Fung N. et al. Oropharyngeal cancer is no longer a disease of younger patients and the prognostic advantage of Human Papillomavirus is attenuated among older patients: Analysis of the National Cancer Database. Oral oncology. 2018;83:147-53
20. van Deudekom FJ, Schimberg AS, Kallenberg MH, Slingerland M, van der Velden LA, Mooijaart SP. Functional and cognitive impairment, social environment, frailty and adverse health outcomes in older patients with head and neck cancer, a systematic review. Oral oncology. 2017;64:27-36
21. Arora A, Husain N, Bansal A, Neyaz A, Jaiswal R, Jain K. et al. Development of a New Outcome Prediction Model in Early-stage Squamous Cell Carcinoma of the Oral Cavity Based on Histopathologic Parameters With Multivariate Analysis: The Aditi-Nuzhat Lymph-node Prediction Score (ANLPS) System. Am J Surg Pathol. 2017;41:950-60
22. Laske RD, Scholz I, Ikenberg K, Meerwein C, Vital DG, Studer G. et al. Perineural Invasion in Squamous Cell Carcinoma of the Oral Cavity: Histology, Tumor Stage, and Outcome. Laryngoscope Investig Otolaryngol. 2016;1:13-8
23. Nair D, Mair M, Singhvi H, Mishra A, Nair S, Agrawal J. et al. Perineural invasion: Independent prognostic factor in oral cancer that warrants adjuvant treatment. Head & neck. 2018;40:1780-7
24. Huyett P, Gilbert M, Liu L, Ferris RL, Kim S. A Model for Perineural Invasion in Head and Neck Squamous Cell Carcinoma. J Vis Exp. 2017. 2017;5:119
25. Schmitd LB, Beesley LJ, Russo N, Bellile EL, Inglehart RC, Liu M. et al. Redefining Perineural Invasion: Integration of Biology With Clinical Outcome. Neoplasia. 2018;20:657-67
26. Zhou G, Liu Z, Myers JN. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. Journal of cellular biochemistry. 2016;117:2682-92
27. Osman AA, Neskey DM, Katsonis P, Patel AA, Ward AM, Hsu TK. et al. Evolutionary Action Score of TP53 Coding Variants Is Predictive of Platinum Response in Head and Neck Cancer Patients. Cancer research. 2015;75:1205-15
28. Sandulache VC, Michikawa C, Kataria P, Gleber-Netto FO, Bell D, Trivedi S. et al. High-Risk TP53 Mutations Are Associated with Extranodal Extension in Oral Cavity Squamous Cell Carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2018;24:1727-33
29. Scheel A, Bellile E, McHugh JB, Walline HM, Prince ME, Urba S. et al. Classification of TP53 mutations and HPV predict survival in advanced larynx cancer. Laryngoscope. 2016;126:E292-9
30. Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM. et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:290-300
31. Leroy B, Ballinger ML, Baran-Marszak F, Bond GL, Braithwaite A, Concin N. et al. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer research. 2017;77:1250-60
32. Roh JL, Ko JH, Moon SJ, Ryu CH, Choi JY, Koch WM. The p53-reactivating small-molecule RITA enhances cisplatin-induced cytotoxicity and apoptosis in head and neck cancer. Cancer letters. 2012;325:35-41
33. Jeay S, Ferretti S, Holzer P, Fuchs J, Chapeau EA, Wartmann M. et al. Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the p53-MDM2 Inhibitor HDM201. Cancer research. 2018;78:6257-67
34. Sandulache VC, Skinner HD, Ow TJ, Zhang A, Xia X, Luchak JM. et al. Individualizing antimetabolic treatment strategies for head and neck squamous cell carcinoma based on TP53 mutational status. Cancer. 2012;118:711-21
35. Desrichard A, Kuo F, Chowell D, Lee KW, Riaz N, Wong RJ. et al. Tobacco Smoking-Associated Alterations in the Immune Microenvironment of Squamous Cell Carcinomas. Journal of the National Cancer Institute. 2018;110:1386-1392
36. Mirghani H, Lacroix L, Rossoni C, Sun R, Auperin A, Casiraghi O. et al. Does smoking alter the mutation profile of human papillomavirus-driven head and neck cancers? Eur J Cancer. 2018;94:61-9
37. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L. et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. The New England journal of medicine. 2016;375:1856-67
38. Welters MJP, Ma W, Santegoets S, Goedemans R, Ehsan I, Jordanova ES. et al. Intratumoral HPV16-Specific T Cells Constitute a Type I-Oriented Tumor Microenvironment to Improve Survival in HPV16-Driven Oropharyngeal Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2018;24:634-47
39. Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ. et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet. 2019;393:40-50
40. Tanaka N, Osman AA, Takahashi Y, Lindemann A, Patel AA, Zhao M. et al. Head and neck cancer organoids established by modification of the CTOS method can be used to predict in vivo drug sensitivity. Oral oncology. 2018;87:49-57
41. Noronha V, Joshi A, Patil VM, Agarwal J, Ghosh-Laskar S, Budrukkar A. et al. Once-a-Week Versus Once-Every-3-Weeks Cisplatin Chemoradiation for Locally Advanced Head and Neck Cancer: A Phase III Randomized Noninferiority Trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2018;36:1064-72
42. Albers AE, Grabow R, Qian X, Jumah MD, Hofmann VM, Krannich A. et al. Efficacy and toxicity of docetaxel combination chemotherapy for advanced squamous cell cancer of the head and neck. Mol Clin Oncol. 2017;7:151-7
43. Tam M, Wu SP, Gerber NK, Lee A, Schreiber D, Givi B. et al. The impact of adjuvant chemoradiotherapy timing on survival of head and neck cancers. Laryngoscope. 2018;128:2326-32
Author contact
![]() Corresponding author: Xu Qian, MD, PhD, Division of Molecular Diagnostics, Department of Laboratory Medicine, Zhejiang Cancer Hospital, No.1 East Banshan Road, Hangzhou 310022, P.R. China. Email: Qian_michelle2014com
Corresponding author: Xu Qian, MD, PhD, Division of Molecular Diagnostics, Department of Laboratory Medicine, Zhejiang Cancer Hospital, No.1 East Banshan Road, Hangzhou 310022, P.R. China. Email: Qian_michelle2014com

 Global reach, higher impact
Global reach, higher impact