10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(7):1419-1428. doi:10.7150/ijbs.34076 This issue Cite
Research Paper
TFAP2A Induced KRT16 as an Oncogene in Lung Adenocarcinoma via EMT
1. Jiangsu Cancer Hospital, Jiangsu Institute Of Cancer Research, Nanjing Medical University Affiliated Cancer Hospital; 42 Baiziting, Nanjing, Jiangsu, 210009, China (Corresponding Address)
2. School Of Medicine, Jiangsu University, Zhenjiang, Jiangsu, China
3. Key Laboratory Of Environmental Medicine Engineering, Ministry Of Education And School Of Public Health, Southeast University, Nanjing, Jiangsu, China
#Both authors contributed equally to this work
Received 2019-2-13; Accepted 2019-4-5; Published 2019-6-2
Abstract
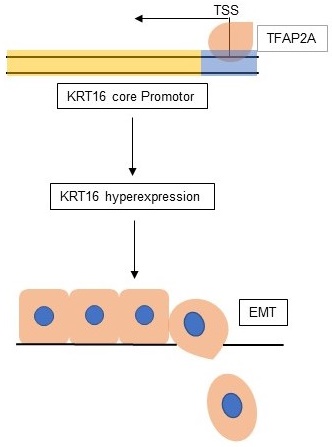
Objectives: keratin 16 (KRT16) is a type I cytokeratin that overexpressed in many kinds of cancers, but unlike other keratins, KRT16 was poorly studied, so the aim of current study was to determine the biological role of KRT16 in lung adenocarcinoma (LUAD).
Materials and Methods: by utilizing open access data, we determined KRT16 expression in LUAD. After that we evaluated the biological role of KRT16 in-vitro and in-vivo. We also explored the reason for KRT16 overexpression. Last, we explored the clinical significance of KRT16 in LUAD.
Results: we found KRT16 is overexpressed in LUAD and positively correlated with lymph node metastasis. Knockdown of KRT16 significantly influenced the LUAD cells' migration, invasion, proliferation and epithelial-mesenchymal transition (EMT). Moreover, TFAP2A could transcriptionally overexpress KRT16, which contributed to the TFAP2A tumorigenicity. Last, we determined that high level of KRT16 predicts poor prognosis of LUAD patients.
Conclusions: our data indicate that, TFAP2A induced KRT16 overexpression promotes tumorigenicity in LUAD via EMT, and KRT16 expression could serve as an independent prognosis marker.
Keywords: TFAP2A, KRT16, LUAD, EMT, Keratin
Introduction
Lung cancer is causing the highest morbidity and mortality among cancers in China and also constitutes a leading cause of cancer-related mortality worldwide [1, 2], the 5-year survival rate of lung cancer is lower than 15% in most countries [3], and the high mortality of lung cancer is mostly due to cancer metastasis [4]. Keratins are the intermediate filament (IF)-forming proteins, expressed in epithelial and endothelial cells, which constitute the cellular matrix, and responsible for the mechanical stability of cells [5]. Overexpressed Keratins are used as prognosis predictors in different types of cancers and play an important role in tumorigenesis [6-9]. KRT16 is located at chr.17q21.2, encodes for the type I cytoskeletal (CK)16 protein, and normally expressed in several epithelial tissues. Polioudaki, H., et al. reported that high KRT16 mRNA expression is observed in metastatic breast cancers and associated with poor RFS [10]. But unlike other keratins, KRT16 is not sufficiently researched. So, the purpose of the current study was to investigate the role of KRT16 in lung adenocarcinoma (LUAD) and explore the reason for KRT16 overexpression. We also identified an upregulated transcription factor (TF) TFAP2A, which contribute to the KRT16 overexpression. TFAP2A is an AP-2 family transcription factor, expressed early in embryogenesis and contributes to cell fate determination in the formation of neural crest and epidermis [11]. And aberrant expression of TFAP2A has been observed in various cancers [12, 13].
Materials and Methods
Data sources
The level 3 TCGA lung adenocarcinoma datasets containing 511 tumors and 58 adjacent normal lung tissues (57 paired tissues) were downloaded for clinical analysis, the “count” is the RSME normalized mRNA count[14], which represents the mRNA expression of genes. For volcano plot, F-test was used between tumor and normal in 20475 genes. And in receiver operating characteristic curve (ROC), the KRT16 gene expression unit is “log2(count+1)”. For GSEA, the latest official tool was downloaded from http://software.broadinstitute.org/gsea (Ver. 3.0).
Cell lines, cell culture, siRNA, and plasmids
PC9 cells were a generous gifted from Dr. Zhian Liu and H1650 cells were obtained from American Type Culture Collection (ATCC, USA). Cells were cultured in RPMI1640 media (Kaiji, Nanjing, China) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and cultured at 37°C in a humidified incubator containing 5% CO2. Transfection was followed by instruction of Lipofectamine 3000 (Invitrogen, USA). Nonsense RNAi (nsRNA) was used as a negative control (NC), qRT-PCR and western blot were used for evaluation the transfection efficiency. Two siRNAs were designed for KRT16 knockdown, siRNA1: GACCTGAGGAACAAGATCATT and siRNA2: AACAGCGAACTGGTACAGAGC, the siRNA for TFAP2A: GGGUAUUAACAUCCCAGUTT was adopted from a published article [15]. The following Nonsense siRNA was used as negative control: UUCUCCGAACGUGUCACGUTT. The human KRT16 targeting small hairpin RNA was designed based on siRNA2 sequence (which had a better efficiency) and inserted into pGFP-C-shLenti vector for lentivirus packaging, and the expression plasmid with full length of KRT16 and the luciferase reporter plasmids (firefly luciferase) containing the core promotor region of KRT16 were both customized and ordered from RiboBio (Guangzhou, China); and a Renilla luciferase expression plasmid was used for background control, 48 hours after co-transfection (plasmid with siRNAs), luciferase signal was activated by Dual-Luciferase® Reporter Assay System (Promega, Lot. No. E1910) and detected by a plate reader (Promega, GM3000); normalized luciferase values were determined by dividing firefly luciferase activity with the Renilla luciferase activity.
Cell proliferation, migration, invasion and wound healing assay
The Cell Counting Kit-8 (KGA317; KeyGen Biotech, Nanjing, China) was used to determine the cell proliferation rate. 2,000 cells/well (100 µl total volume) were seeded in 96-well plates and the absorbance was measured at 450 nm using a plate Reader (Promega, GM3000). The experiment was repeated in triplicate at various time points for 5 days. For migration and invasion assays, transfected cells (40,000 cells/well; 100 µl total volume) were seeded in the upper chamber of Transwell or Matrixgel assay inserts (8-mm pores; Millipore) containing 200 µl serum-free media. The lower chambers were filled with same media containing 10% FBS. After 24 hours in incubation, cells were fixed with methanol and stained with crystal violet. Cell migration or invasion was assessed by counting the number of stained cells in five random fields of each group. In wound healing assay, cells were seeded and transfected on six-well plates with si-KRT16 or si-NC, then 24 hours after transfection an artificial scratch wound on a confluent monolayer of cells was created with a 200μl pipette tip, after that serum-free medium was added, and cells were imaged at baseline and 24 hours. For colony formation assays, a total of 100 transfected cells were placed in a fresh 24-well plate and maintained in media containing 10% FBS, replacing medium every 3 or 4 days. After 10 days, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Visible colonies were manually counted. Experiments were repeated in triplicate.
Flow-cytometry analysis
Flow-cytometry analysis was performed for apoptosis rate measurement. Briefly, cells were washed and resuspended at a concentration of 1 × 106 cells/ml, then cells were stained with 7-AAD (Invitrogen, A1310) and Annexin V (eBioscience, BMS306FI-100). After 20min incubation at room temperature, cells were analyzed by a BD FACSCanto™ II flow cytometer. Each assay was performed in triplicate.
RNA preparation, reverse transcription, real-time quantitative PCR, and ChIP-qPCR
TRIzol was used for extracting total RNA from cultured cells, for qPCR 1µg total RNA was reverse transcribed to 20µl cDNA by ipsogen RT Kit (QIAGEN, Cat No: 679913). qPCR was performed with SYBR Select Master Mix (Applied Biosystems, Cat: 4472908). For ChIP-qPCR DNA preparation, the EpiTect ChIP OneDay Kit (QIAGEN, Cat No: 334471) was used based on official user guide, rabbit IgG (#PP64, Millipore) as the negative control, and H3K4me3 pull down as a positive control (the sequence of EIF4A2 was detected). The qPCR was performed using QuantStudioTM 3 Real-Time PCR System, the reaction protocol was 95 °C for 10 min, followed by 40 cycles of 92°C for 15 sec and 62.5°C for 1 min. Every sample was in triplicate and the expression was normalized to the reference gene expression (hGAPDH) using the 2-ΔΔCt method, and for ChIP enrichment fold change, samples were normalized to the IgG pulldown group and 2-(CtTarget - CtIgG) method was used. All the primers for qPCR are shown in supplementary file Table S1.
Protein preparation and western blot
The total protein was extracted using RIPA lysis buffer (Beyotime Biotechnology, P0013C) supplemented with protease inhibitors cocktail (Promega, G6521), protein concentration was measured by BCA kit (Beyotime Biotechnology, P0010S). After that as a standard western blot protocol, proteins were loaded and separated by 12% SDS-PAGE gel electrophoresis and incubated with primary and HRP-tagged secondary antibodies to detect the signal (antibody list in supplementary file Table S2).
Immunohistochemistry and in-vivo experiment
The tissue microarray (TMA) containing 88 cases of lung adenocarcinoma (LUAD) with matched adjacent normal tissue and 4 cases only have tumors were obtained from Shanghai Biochip Co., Ltd. (Cat. No. HLugA180Su03). Operations occurred between Jan 2008 and Jul 2013, all tissues were re-examined by an experienced pathologist after they were transferred from the local hospital and the TNM stage was determined in each patient. We also re-examined the tissues with the help from an experienced pathologist. This research was approved by the Human Research Ethics Committee of Nanjing Medical University. For IHC staining, briefly the slides were incubated with anti-human KRT16 antibody overnight at 4 °C, then sides were incubated using the secondary antibody for 2 hours in room temperature. After that the immunohistochemistry results were scored blindly by two researchers (including a pathologist). The staining intensity was scored according to 4 grades: 0 (No staining), 1 (weak staining), 2 (moderate staining), or 3 (intense staining). The percentage of positive cells multiply respective intensity scores was used as the final staining score (from 0 to 300)[16].
For nude mice experiment, eight 5-7 weeks old female nude mice were obtained from Nanjing Medical University affiliated animal facility. these mice were randomly separated into two groups NC and KRT16 knockdown, then 3.5×105 PC9 cells suspended in 200µl PBS were tail vein injected into mice, after 21 days, mice were sacrificed, and their lungs were harvest (one mouse in KRT16 knockdown group died at day 5 after injection, reason unknown), after H & E staining the tumor burden were counted under microscope. All animal studies were conducted following NIH animal use guidelines and protocols approved by Nanjing Medical University Animal Care Committee.
Statistical analysis
Data are presented as the mean ± standard deviation and analyzed using a Student's t-test or one-way ANOVA with post-hoc analysis. P<0.05 was considered to indicate a statistically significant difference. SPSS Statistics (version 20.0, IBM Corp), Excel (Office 365, Microsoft Office) and GraphPad Prism (version 7.04, GraphPad Software, Inc) software were used for statistical analyses and the production of graphs.
Results
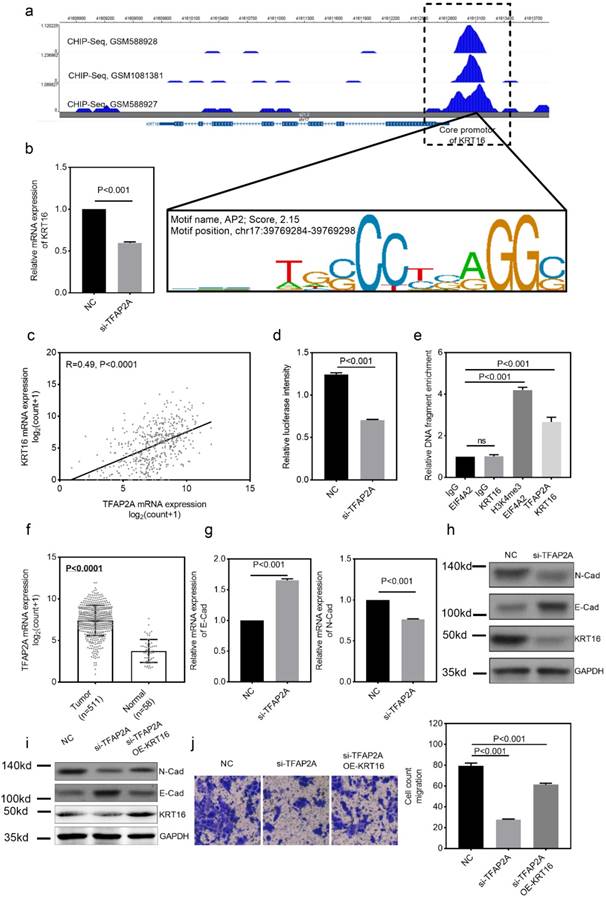
KRT16 is over-expressed in lung adenocarcinoma (LUAD) and correlates with lymph nodes metastasis
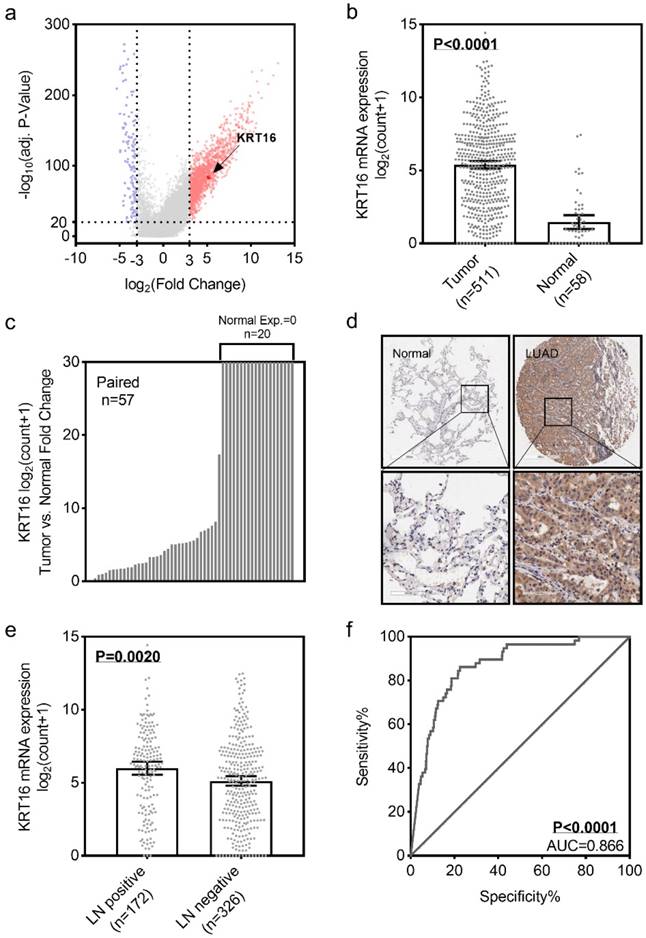
To identify the differential expression of KRT16 in LUAD, we first analyzed the TCGA_LUAD dataset containing 511 tumor samples and 58 normal samples (57 pairs) with clinical parameters. The bird view from volcano plot revealed that KRT16 is at a relatively significantly differential expression position from 20475 genes (Fig 1a & b).The red dots represent the significantly hyper-expressed genes in LUAD. On the opposite, these blue dots represent the significantly hypo-expressed genes in LUAD (Fig 1a). And receiver operating characteristic curve (ROC) indicates that KRT16 expression might serve as a marker for LUAD (Fig 1f). Among the 57 paired tissues (LUAD tumor tissues vs. their adjacent normal tissues), there are 20 normal tissues with null KRT16 expression but not their adjacent LUAD tissues (Fig 1c). As shown in Fig 1d, KRT16 protein level is higher in LUAD tissues (from the tissue microarray, TMA, quantification in Fig S1a). Moreover, the KRT16 expression is significantly higher in lymph node positive LUAD tissues, indicating that KRT16 might contribute to the tumor metastasis (Fig 1e).
KRT16 knockdown undermines the LUAD cell line migration, invasion and proliferation ability in-vitro
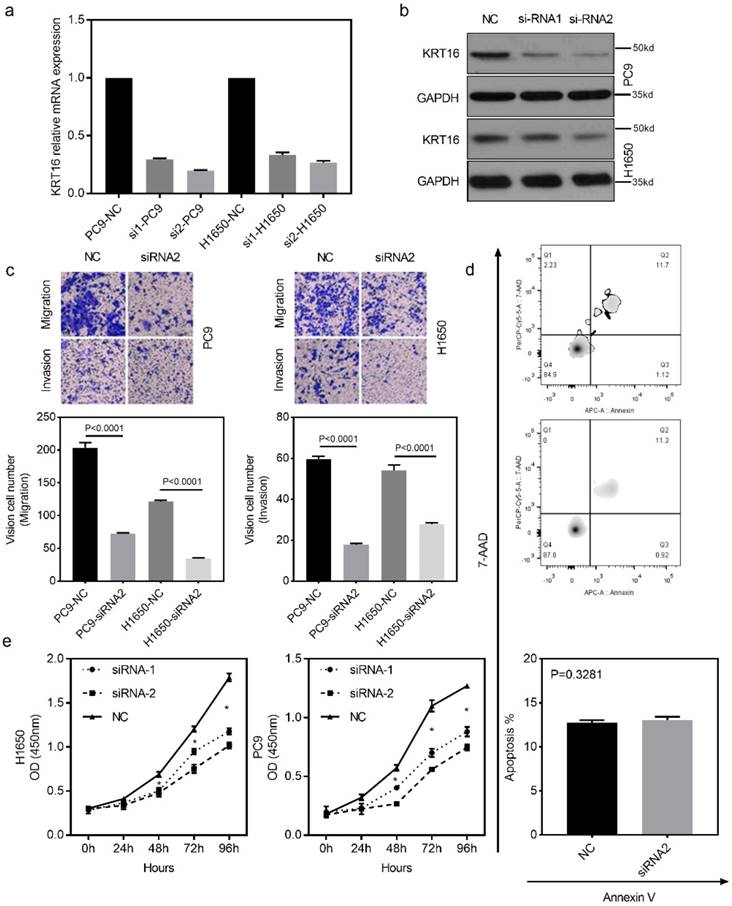
To investigate the biological function of KRT16 in vitro, two different siRNAs (siRNA-1 and siRNA-2) were designed for KRT16 knockdown. Both siRNA constructs were able to effectively knockdown KRT16 mRNA expression and protein level (Fig 2a & b). As shown in Fig 2c, the transwell and matrixgel assay revealed that KRT16 knockdown reduced migration and invasion ability in both PC9 and H1650 cells. The cell count kit 8 (CCK8) assay showed that the cell counts of these two cell lines were decreased by KRT16 knockdown (Fig 2e). In Fig 2d, no significant difference of apoptosis rate is observed between KRT16 knockdown and negative control (NC), thus KRT16 knockdown could undermine proliferation in-vitro. Last in colony formation assay, PC9 cells transfected with si-KRT16 have lower colony numbers (Fig S1b).
KRT16 is overexpressed in lung adenocarcinoma. a, volcano plot for differential expressed genes in lung adenocarcinoma (LUAD) TCGA dataset, left blue part were genes significantly downregulated in LUAD and right red part were genes significantly upregulated in LUAD, and KRT16 was at a relatively hyper-expression position. b & c, KRT16 was hyper expressed in LUAD compared with normal adjacent tissues, p<0.0001, and in the 57 paired samples, there were 20 pairs with null KRT16 expression in the normal tissues (but not their counterpart adjacent tumor samples). d, the KRT16 level was higher in tumor samples compared with adjacent normal samples. e, lymph node positive patients had higher KRT16 expression, p=0.002. f, receiver operating characteristic curve (ROC) shown KRT16 expression had a good specificity and sensitivity for diagnosis in LUAD, p<0.0001, area under curve (AUC)=0.866.
KRT16 knockdown decrease the migration, invasion and proliferation in-vitro. a & b, KRT16 was knocked down by two different siRNAs, both siRNAs could significantly knockdown KRT16 mRNA expression and protein level in PC9 and H1650 cell lines (the western blot quantification by Image J; PC14, NC 152, si1 119, si2 112; H1650, NC 148, si1 142, si2 125). c, transwell and matrixgel assay shown KRT16 knockdown could undermine the migration and invasion ability in PC9 and H1650. d, but KRT16 knockdown did not influence the cell death (apoptosis). e, KRT16 knockdown group had lower proliferation rate.
KRT16 is involved with epithelial mesenchymal transition (EMT) in LUAD
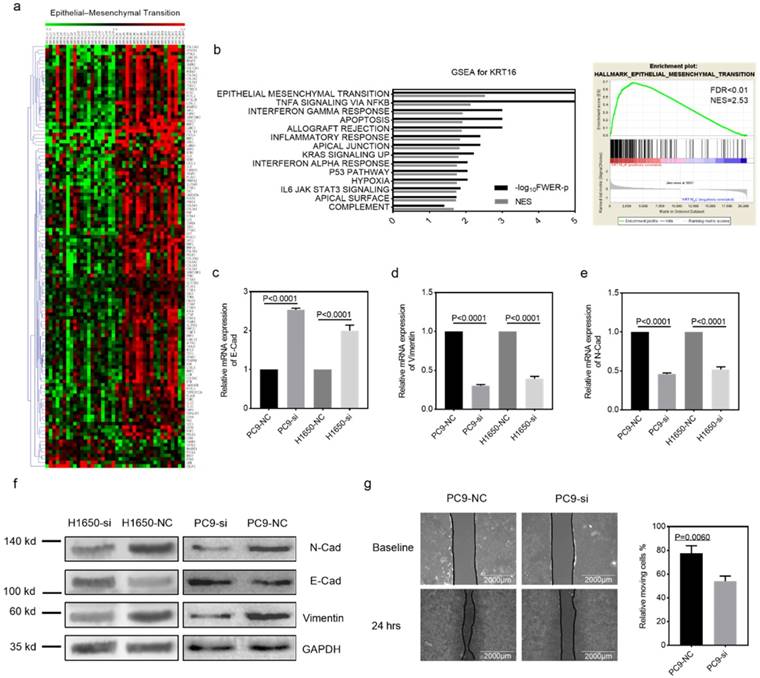
Gene set enrichment analysis. GSEA was introduced for potential mechanism exploring [17, 18], and the transcriptome data from 40 TCGA_LUAD tumor tissues were submitted for GSEA (based on KRT16 expression, top 20 versus bottom 20 from 511 TCGA_LUAD tumor samples). These results indicate that epithelial mesenchymal transition (EMT) might be the downstream mechanism of KRT16 phenotype in LUAD (Fig 3a & b). Given our findings that KRT16 expression is higher in LN positive tumors and KRT16 knockdown reduced cancer cell migration, invasion and colony formation, we sought to determine the expression of several EMT markers. Consistent with enrichment analyses, compared with NC, KRT16 knockdown have reduced N-Cad and Vimentin, and increased E-Cad (Fig 3c-f). Wound healing assay also yielded similar results that KRT16 knockdown impairs wound healing ability in PC9 cells (Fig 3g). These data indicated that KRT16 knockdown influences EMT in LUAD.
KRT16 is involved in epithelial mesenchymal transition (EMT). a & b, the TCGA_LUAD dataset (Top 20 KRT16 expression vs. Button 20 KRT16 expression) had been submitted for the Gene Set Enrichment Analysis (GSEA), these results indicating KRT16 was correlated with epithelial mesenchymal transition (EMT), Normalized Enrichment Score (NES)=2.53, FDR<0.01. c, d, e & f, EMT markers were measured by qPCR and western blot, KRT16 knockdown group had lower N-Cad and Vimentin but higher E-Cad compared with their negative control group. g, in PC9 cells, si-KRT16 impaired wound healing ability as compared with NC group (p=0.0060).
TFAP2A is the upstream transcription factor for KRT16 and KRT16 contributes to the oncogenic phenotype of TFAP2A
We also explored the potential mechanism for KRT16 hyper-expression in LUAD. One AP2 motif was identified on the core promotor region of the KRT16, so we sought to use the chromatin immunoprecipitation sequencing (ChIP-Seq) data from transcription factor (TF) that could bind to the AP2 motif. As shown in Fig 4a, three independent TFAP2A ChIP-Seq data (GSM588928, GSM1081381 and GSM588927 from MCF-7 cell line) all revealed a clear peak right on the core promotor of KRT16. Moreover, the expression of TFAP2A is positively correlated with KRT16 (Fig 4c). By utilizing duo-luciferase reporter system containing the core promotor region of KRT16, we found that TFAP2A knockdown could significantly decrease the relative luciferase intensity (Fig 4d) and TFAP2A knockdown could also decrease KRT16 expression (Fig 4b) in PC9 cell line. After that, we performed ChIP-qPCR in PC9 cells for validating. Our results revealed that KRT16 promoter DNA fragments were enriched by TFAP2A antibody pulldown (Fig 4e). And in Fig 4f, TFAP2A is also significantly hyper-expressed in LUAD. We came to the conclusion that TFAP2A should be an important TF for KRT16 and contribute to the KRT16 hyper expression in LUAD. Because TFAP2A was reported involved in EMT [12], we measured EMT markers in PC9 cells with TFAP2A knockdown, as shown in Fig 4b, g & h. TFAP2A knockdown significantly reduced KRT16 and N-Cad, and increased E-Cad in PC9 cell line, so TFAP2A might exert its oncogenic actively through KRT16 via EMT. Last, we performed a rescue assay in which a plasmid containing the whole length of KRT16 was introduced, KRT16 overexpression could partially rescue the phenotype in TFAP2A knockdown PC9 cells (Fig 4i & j).
TFAP2A is the upstream transcription factor for KRT16. a, CHIP-Seq peaks from three independent experiments indicate TFAP2A could bind on the core promotor region of KRT16 with an AP2 motif on chr17:29769284-39769298. b, TFAP2A knockdown decreased the KRT16 expression in PC9. c, TCGA_LUAD data indicates the expression of TFAP2A and KRT16 was positively corelated (Pearson R=0.49, P<0.0001). d, a luciferase reporter plasmid containing the core promotor sequence of KRT16 was utilized, after TFAP2A knockdown the relative luciferase intensity was decreased (a plasmid constantly expressing Renilla luciferase was used as a reference background signal) in PC9. e, ChIP-qPCR indicated TFAP2A pull down could enrich KRT16 promoter sequence in PC9 cells. f. TCGA_LUAD data, the TFAP2A expression is higher in tumor tissues compared with normal tissues. g & h, TFAP2A knockdown PC9 cells have less KRT16 and N-Cad level and more E-Cad level. i & j, TFAP2A siRNA and/or KRT16 overexpression vector was co-transfected into PC9 cells, the KRT16 overexpression could partially rescue the migration phenotype in TFAP2A knockdown PC9 cells.
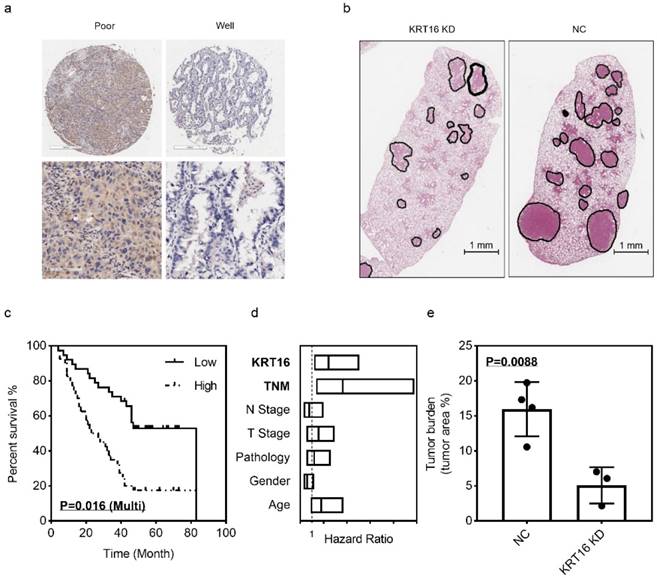
Prognostic value of KRT16 and KRT16 knockdown suppresses tumor formation in-vivo
The LUAD tissue microarray (TMA), which contained 92 tumor samples with follow up survival data, was stained with KRT16. After scoring, the multivariant cox regression survival analysis was performed, several clinical parameters including age, gender, pathology, T stage, N stage, TNM stage and KRT16 level were enrolled, as shown in Fig 5c & d, TNM stage and KRT16 expression could serve as independent survival markers, TNM stage high vs. low, p=0.01, HR and 95% CI: 3.642 (1.355 - 9.791); KRT16 high vs. low, p=0.016, HR and 95%CI: 2.448 (1.183 - 5.065). And as shown in Table 1, high level of KRT16 was corrected with lymph node positive and advanced TNM stage.
In the in-vivo experiment, PC9 cells were infected with KRT16 shRNA or NC shRNA, then injected into seven nude mice via tail vein (3 KRT16 shRNA mice vs. 4 NC shRNA mice). After 21 days of tumor growing, mice were sacrificed, and lungs were harvest. After H & E staining, the tumor burden was counted, as shown in Fig 5b & e. Comparing with control group, the KRT16 knockdown group has lower tumor burden, indicating that KRT16 knockdown could significantly undermine tumor metastasis and formation in-vivo.
Correlation between KRT16 level and clinicopathologic characteristics
| Groups | KRT16 | Pearson χ2 | P-value | ||
|---|---|---|---|---|---|
| High | Low | ||||
| Gender | Male | 28 | 23 | 1.1 | 0.294277 |
| Female | 18 | 23 | |||
| Age | ≤60 | 19 | 19 | 0 | 1 |
| >60 | 27 | 27 | |||
| Differentiation | Well | 29 | 32 | 0.44 | 0.508155 |
| Moderate-Poor | 17 | 14 | |||
| T stage | T1-T2 | 24 | 36 | 3.12 | 0.077335 |
| T3-T4 | 12 | 7 | |||
| Lymph node* | Negative | 6 | 28 | 7.83 | 0.005148 |
| Positive | 22 | 24 | |||
| TNM stage* | I-II | 8 | 33 | 18.97 | 0.000014 |
| III-IV | 19 | 7 | |||
P<0.05 was considered as significant, *Significant correlation.
KRT16 hyper expression predicts poor prognosis and KRT16 knockdown could undermine tumor formation in-vivo. 92 patients were enrolled in the multivariant cox regression survival analysis, a, some poor differentiated LUAD tissues had higher KRT16 level; c, in the K-M plot, KRT16 staining score was positively corelated with prognosis (p=0.016, multivariant cox regression); d, the forest plot of multivariant cox regression analysis shown TNM stage and KRT16 level could serve as independent indicators for survival (TNM stage high vs. low, p=0.01, HR and 95% CI: 3.642 (1.355 - 9.791); KRT16 high vs. low, p=0.016, HR and 95%CI: 2.448 (1.183 - 5.065). b & e seven nude mice were enrolled for tail vein injection (PC9 cells, KRT16 knockdown vs. negative control), compared with negative control group, the KRT16 knockdown group had significantly lower tumor burden (p=0.0088).
Discussion
In the present study, we demonstrate KRT16 is overexpressed in LUAD and especially overexpressed in lymph node positive patients. KRT16 expression has a good specificity and sensitivity for LUAD diagnosis. In the in-vitro experiment we found KRT16 knockdown could undermine the LUAD cells' migration, invasion, colony formation and proliferation ability, but not the apoptosis rate. By utilizing GSEA, we identified influencing EMT could be the downstream mechanism for KRT16 knockdown. As reported previously, keratin family are hyper-expressed in many kinds of cancers and play an improtant role in EMT. Thus, several EMT markers were measured between KRT16 knockdown and negative control, we found that KRT16 knockdown leads to higher E-Cad level but lower N-Cad and Vimentin level.
By exploring the upstream mechanism of KRT16 overexpression, we found the transcription factor TFAP2A, which is also hyper-expressed in LUAD and influencing EMT, is positively correlated with KRT16, and three independent published ChIP-Seq data all indicate TFAP2A binds to the core promotor region of KRT16. To validate whether TFAP2A could transcriptionally activate KRT16 expression, we designed the duo-luciferase system containing the sequence of KRT16 core promotor sequence. We found TFAP2A knockdown could significantly decrease the relative luciferase intensity; moreover, for better validation, ChIP-qPCR was performed, indicating that the KRT16 promoter sequence was enriched in the TFAP2A pull down DNA fragments. As a result, KRT16 hyperexpression in LUAD is induced by transcription factor TFAP2A. TFAP2A is an AP-2 family transcription factor which binds to the consensus sequence 5'-GCCNNNGGC-3' and is overexpressed in lung adenocarcinoma. TFAP2A modulates the expression of multiple downstream target genes involved in the development and progression of human cancers. TFAP2A was reported influencing epithelial-mesenchymal transition (EMT) in different types of cancers [12, 19, 20] and TFAP2A knockdown could undermine the migration and invasion ability in cancer cells. As it's reported in other types of cancers, TFAP2A influences tumor migration via EMT. We designed the migration rescue assay. We found TFAP2A knockdown could influence EMT and decrease the cancer cells' migration in LUAD. After we overexpressed KRT16 in the TFAP2A knockdown cells, we found the migration phenotype was partly rescued in LUAD cells, indicating TFAP2A could exert its oncogenic ability via KRT16.
Last, in-vivo nude mice tail vein oncogenesis experiment and survival analysis on human LUAD sample TMA (with follow up data) was performed. The in-vivo experiment indicates that KRT16 knockdown could significantly undermine tumor formation in nude mice lungs. In survival analysis, multivariate cox regression indicates LUAD patients with strongly expressed KRT16 have a poorer prognosis than those with absent or weaker KRT16 expression.
Our data indicate KRT16 hyperexpression in LUAD is induced by transcription factor TFAP2A. KRT16 knockdown in-vitro decreases the cancer cell migration, invasion and proliferation via EMT, and KRT16 knockdown could undermine the tumor formation ability in-vivo. Moreover, KRT16 may serve as an independent prognosis maker in LUAD.
Abbreviations
Keratin 16 (KRT16); Lung adenocarcinoma (LUAD); Epithelial-mesenchymal transition (EMT); Gene set enrichment analysis (GSEA); Haematoxylin and eosin (H & E); Immunohistochemistry (IHC); Quantitative real-time polymerase chain reaction (qRT-PCR); The Cancer Genome Atlas (TCGA); Chromatin immunoprecipitation (ChIP); Receiver operating characteristic curve (ROC); Normalized enrichment score (NES).
Supplementary Material
Supplementary figure and tables.
Acknowledgements
SB, LY designed the study. LY, QP, LG and ZW performed the experiments. WY, LD performed the statistical analysis. LY, SB, LD and ZY wrote the manuscript. All authors read and approved the final manuscript. This research was supported by Jiangsu province social development key research and development plan (Grant number: BE2017694) and National Natural Science Foundation of China (81673132 and 81472939).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen WQ, Zuo TT, Zheng RS, Zeng HM, Zhang SW, He J. [Lung cancer incidence and mortality in China in 2013]. Zhonghua Zhong Liu Za Zhi. 2017;39:795-800
2. Tan WL, Jain A, Takano A, Newell EW, Iyer NG, Lim WT. et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol. 2016;17:e347-e62
3. Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol. 2008;3:819-31
4. Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19:1450-64
5. Chou CF, Riopel CL, Rott LS, Omary MB. A significant soluble keratin fraction in 'simple' epithelial cells. Lack of an apparent phosphorylation and glycosylation role in keratin solubility. J Cell Sci. 1993;105( Pt 2):433-44
6. Saha SK, Kim K, Yang GM, Choi HY, Cho SG. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. Int J Mol Sci. 2018:19
7. Ricciardelli C, Lokman NA, Pyragius CE, Ween MP, Macpherson AM, Ruszkiewicz A. et al. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget. 2017;8:17819-32
8. Wu H, Wang K, Liu W, Hao Q. Recombinant adenovirus-mediated overexpression of PTEN and KRT10 improves cisplatin resistance of ovarian cancer in vitro and in vivo. Genet Mol Res. 2015;14:6591-7
9. Mockler D, Escobar-Hoyos LF, Akalin A, Romeiser J, Shroyer AL, Shroyer KR. Keratin 17 Is a Prognostic Biomarker in Endocervical Glandular Neoplasia. Am J Clin Pathol. 2017;148:264-73
10. Polioudaki H, Agelaki S, Chiotaki R, Politaki E, Mavroudis D, Matikas A. et al. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer. 2015;15:399
11. Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP. et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238-41
12. Dimitrova Y, Gruber AJ, Mittal N, Ghosh S, Dimitriades B, Mathow D. et al. TFAP2A is a component of the ZEB1/2 network that regulates TGFB1-induced epithelial to mesenchymal transition. Biol Direct. 2017;12:8
13. Shi D, Xie F, Zhang Y, Tian Y, Chen W, Fu L. et al. TFAP2A regulates nasopharyngeal carcinoma growth and survival by targeting HIF-1alpha signaling pathway. Cancer Prev Res (Phila). 2014;7:266-77
14. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323
15. Zhou X, Wang Z, Zhang Z, Cui Q, Wang Y, Wang G. Delivery of AP-2alpha siRNA into cultured bovine trophoblast cells by electroporation repressed key placenta-specific gene expression. Gene. 2012;499:169-75
16. Wang X, Sun Q, Chen C, Yin R, Huang X, Wang X. et al. ZYG11A serves as an oncogene in non-small cell lung cancer and influences CCNE1 expression. Oncotarget. 2016;7:8029-42
17. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-50
18. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267-73
19. Zhang D, Li H, Jiang X, Cao L, Wen Z, Yang X. et al. Role of AP-2alpha and MAPK7 in the regulation of autocrine TGF-beta/miR-200b signals to maintain epithelial-mesenchymal transition in cholangiocarcinoma. J Hematol Oncol. 2017;10:170
20. Huang W, Chen C, Liang Z, Qiu J, Li X, Hu X. et al. AP-2alpha inhibits hepatocellular carcinoma cell growth and migration. Int J Oncol. 2016;48:1125-34
Author contact
![]() Corresponding author: Email: shenbo987edu.cn, Tel: +86-25-8328-3598.
Corresponding author: Email: shenbo987edu.cn, Tel: +86-25-8328-3598.

 Global reach, higher impact
Global reach, higher impact