10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(8):1610-1617. doi:10.7150/ijbs.32518 This issue Cite
Research Paper
Specifically blocking the fatty acid synthesis to inhibit the malignant phenotype of bladder cancer
1. Department of Urology, Shenzhen Second People′s Hospital, the First Affiliated Hospital of Shenzhen University, Shenzhen 518000, Guangdong, China.
2. Shantou University Medical College, Shantou 515041, China
3. Department of Urology, Peking University First Hospital, Institute of Urology, Peking University, National Urological Cancer Center, Beijing 100034, China
4. Shenzhen Key Laboratory of Fertility Regulation, Center of Assisted Reproduction and Embryology, The University of HongKong, Shenzhen Hospital, Shenzhen, China
5. University of South China, Hengyang 421000, Hunan Province, China
*Equal contributors
Received 2018-12-23; Accepted 2019-1-29; Published 2019-6-4
Abstract
Fatty acid synthesis is regulated by transcription factors SREBPs and their escort protein SCAP. Malignant cells become dependent on de novo lipogenesis, which sustains rapid proliferation and resistance to cellular stress. Increasing evidence showed SCAP participated in various disease processes including malignant tumors, which regulate transcription factors SREBPs Tumorigenesis is associated with incur glucose consumption and lipogenesis. In our study, we discovered that SCAP was upregulated in BC tissues. SCAP knockdown by CRISPR-Cas9 inhibit the cell proliferation, invasion and migration. Additionally, the cell apoptosis was facilitated. What's more, downregulation of SCAP could weaken the cancer-promoting effects of estrogen on BC. Our study revealed that SCAP played a carcinogenic role in BC and lipogenesis might promote the initiation of BC by inducing SCAP. Thus, Targeting SCAP may provide a promising means of treating BC and a new perspective for the tumorigenesis of bladder cancer.
Keywords: fatty acid synthesis, tumorigenesis, cell proliferation, lipogenesis
Introduction
Bladder cancer (BC) is the most common malignant tumor in urinary system, ranking fourth in the incidence rate of male and the most expensive malignant tumor in the United States, and seventh in female. About 95% of BC is derived from bladder epithelium and most of them are malignant[1, 2]. At present, the commonly used therapeutic methods including chemotherapy and radiotherapy are not satisfactory[3]. Conventional radiation and chemotherapy often result in the emergence of drug-resistant tumor clones, which in turn aggravates the malignant phenotype of BC[4, 5]. Therefore, it is of great clinical significance and value to develop new therapeutic approaches for BC.
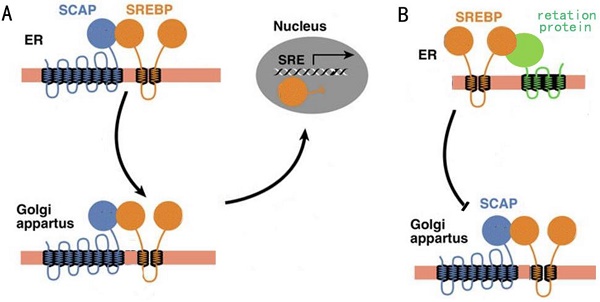
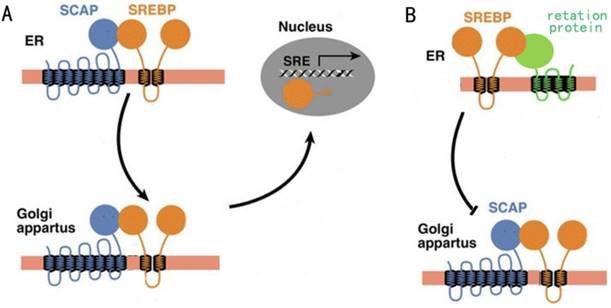
Regulation of fatty acid synthesis balance in mammalian cells is a complex network with multiple pathways and levels, and this regulation network involves many important genes, proteins and small molecular metabolites[6]. Lipid homeostasis represented by cholesterol and fatty acids follows a strict negative feedback regulation, which the key proteins in the lipid synthesis pathway can sense changes in lipid levels within the cells, and turn on or off the transcription of enzymes related to the lipid synthesis pathway[7]. In this regulation mechanism, the most critical regulator is sterol regulatory element-binding proteins (SREBPs) family[8]. SREBPs are a kind of transmembrane transcription regulator located in the endoplasmic reticulum (ER). In the case of low intracellular lipid level, the SREBP cleavage-activating protein (SCAP) located in the endoplasmic network can combine with the SREBPs and carry SREBPs from ER to golgi body by means of membrane transport (Fig. 1A). When intracellular lipid expression level is high, SCAP located on ER membrane can sense this signal, so SCAP binds to another retention protein on ER membrane, causing it to no longer bind to SREBPs (Fig. 1B). The result of this change was that SREBPs could not be activated into nucleus due to the lack of SCAP binding, lipid synthesis gene was shut down, and lipid level was decreased[9].
Previous studies have found that lipid synthesis is more active in many human tumor cells, which is closely related to tumor growth[10, 11]. It was found from model study of human glioma cells by Cheng et al., that glucose was promote by stabilizing that SCAP protein and activating srebp-1, thereby increasing lipid synthesis and promote tumor growth[12]. Flaveny et al. found that targeting lipid synthesis pathway key liver-x-receptor (LXR) with small molecule inhibitory agents in human prostate cancer, colon cancer and lung cancer, thereby reducing the expression of srebp-1 can significantly inhibit the growth of tumor[13]. In addition, a statin inhibitor or genetic intervention that interferes with the synthesis of key lipid cholesterol can also be effective in inhibiting tumors[14, 15]. Based on these findings, we speculated that inactivation of SCAP inhibits lipid synthesis and inhibits BC cell growth.
In our study, we not only revealed the general rules and mechanisms of lipid metabolism involved in the occurrence and development of BC, but also identified special regulation nodes of lipid metabolism in BC cells, laying an important foundation for the development of drugs targeting the lipid metabolism pathway of BC.
Result
SCAP is overexpressed in BC specimens and is associated with the clinical pathological features
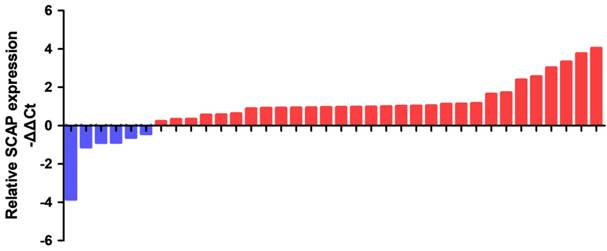
The relative expression levels of SCAP in 36 BC tissues and the adjacent tissues were detected by the quantitative real-time PCR assay (qRT-PCR assay). The specific patient information has been presented in the table 1. Our results showed that SCAP was highly expressed in the majority of 36 BC tumor specimens relative to the corresponding adjacent normal tissue (Fig. 1). What's more, we found that the increased expression of SCAP was positively related to tumor invasion, late TNM stage, the lymph node metastasis and high histological grade of the BC patients. However, there is no direct link between the SCAP expression and the age and the gender of the BC patients. In summary, we concluded that the overexpression of SCAP played a cancer-promoting role in BC.
Correlation between SCAP expression and clinicopathological features of BC patients. (*p<0.05, **p< 0.01, ***p< 0.001; *p<0.05 was considered significant.)
| Features | Total | SCAP expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Gender | ||||
| Female | 6 | 1 (16.7%) | 5 (83.3%) | 0.512 |
| Male | 30 | 9 (30.0%) | 21 (70.0%) | |
| Age | ||||
| ≤60 | 7 | 2 (28.6%) | 5 (71.4%) | 0.811 |
| >60 | 29 | 7 (24.1%) | 22 (75.9%) | |
| Histological grade | ||||
| Low | 13 | 8 (61.5%) | 5 (38.5%) | 0.019* |
| High | 23 | 5 (21.7%) | 18 (78.3%) | |
| Depth of invasion (T) | ||||
| Ta, T1 | 10 | 7 (70.0%) | 3 (30.0%) | 0.010** |
| T2, T3, T4 | 26 | 6 (23.1%) | 20 (76.9%) | |
| Lymph node metastasis (N) | ||||
| N0 | 33 | 5 (15.2%) | 28 (84.8%) | 0.033* |
| N1, N2, N3 | 3 | 2 (66.7%) | 1 (33.3%) | |
| TNM stage 0/I II/III/IV | 6 30 | 4 (66.7%) 4 (13.3%) | 2 (33.3%) 26(86.7%) | 0.005*** |
SCAP is upregulated in BC cells and targeting knockdown of SCAP with CRISPR-Cas9
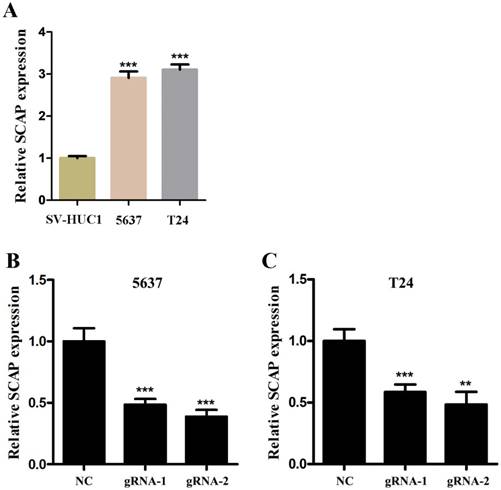
In order to study the relationship between the SCAP expression levels and the development of BC, BC cell line T24 and 5637 were taken as the main research objects. qRT-PCR assay was used to measure the expression level of SCAP in our study. Total RNA of T24 and 5637 were extracted and reverse transcribed. The expression level of SCAP in T24 and 5637 cells was detected with qPCR. The normal bladder epithelial cell line (SV-HUC1) was used as a control in our study. According to the experimental results, we found that the expression levels of SCAP in T24 and 5637 were significantly higher than those in the control group (Fig. 2A).
In our study, to study whether SCAP can be a potential therapeutic target for BC, we used CRISPR-Cas9 to silence the gene of SCAP in T24 and 5637 to explore the effect of SCAP on BC. According to the gene sequence of SCAP, we designed two gRNAs according to different action targets. The specific gRNA target sequence has been shown in the supplemental table. Next, we applied plasmids of CRISPR-Cas9 with gRNA1 and gRNA2 to T24 and 5637 cells, respectively. The effect of CRISPR-Cas9 was verified by qRT-PCR. We found that both gRNA1 and gRNA2 can induce Cas9 protein to silence the gene of SCAP within the cells, but gRNA2 is more obvious than gRNA1 (Fig. 2B, Fig. 2C).
A: The activity of SCAP when intracellular lipid levels are low. B: The activity of SCAP when intracellular lipid levels are high.
The relative expression levels of SCAP were detected by RT-qPCR in a total of 36 BC patients. SCAP was upregulated in the majority of BC tissues compared to the matched normal tissues (p<0.05).
Knockdown of SCAP can inhibit cell proliferation, invasion and migration, and induce cell apoptosis of BC cells
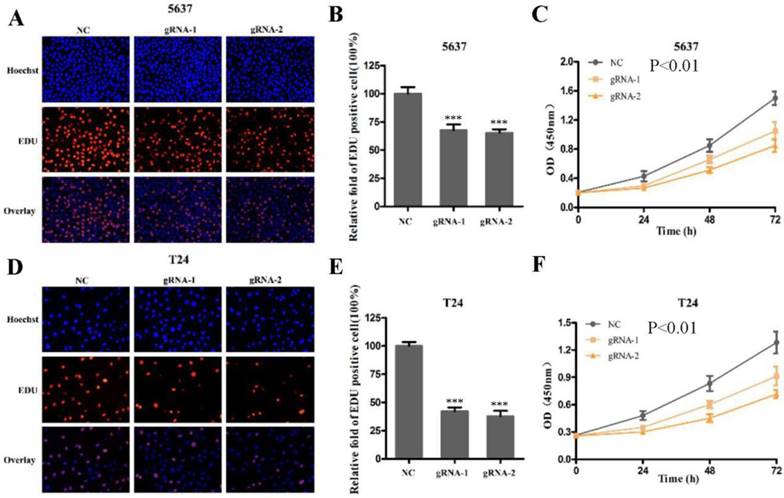
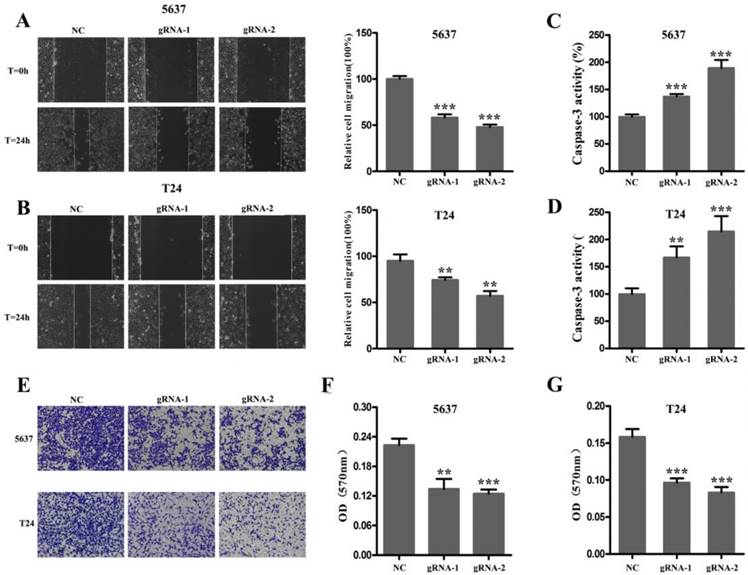
Next, we used CRISPR-Cas9 to study the effect of SCAP on BC. After transfecting Cas9-gRNA1, Cas9-gRNA2 or Cas9-gRNA-comtrol into the BC cell lines T24 and 5637 for 24h, we used the same method to study changes in the malignant phenotype of BC cells. In our study, EDU incorporation assay and CCK-8 assay were preformed to evaluate the changes of cell proliferation in BC cell line T24 and 5637. We observed that when SCAP was knocked down by CRISPR-Cas9, both the EDU incorporation assay and CCK-8 assay showed that the proliferation of the 5637 (Fig. 3A, Fig. 3B, Fig. 3C) and T24 (Fig. 3D, Fig. 3E, Fig. 3F) were suppressed. What's more, the cell migration were significantly suppressed by wound-healing assay in T24 (Fig. 4A) and 5637 (Fig. 4B). Increased cell apoptosis was showed in 5637 and T24 by the outcome of caspase-3 (Fig. 4C, Fig. 4D). In addition, a weaken cell invasion was shown by transwell assay in T24 (Fig.4E, Fig.4F) and 5637 (Fig.4E, Fig.4G). Therefore, knockdown of SCAP significantly inhibited the malignant phenotype of BC cells.
Knockdown of SCAP can inhibit tumorigenesis of BC cells in vivo
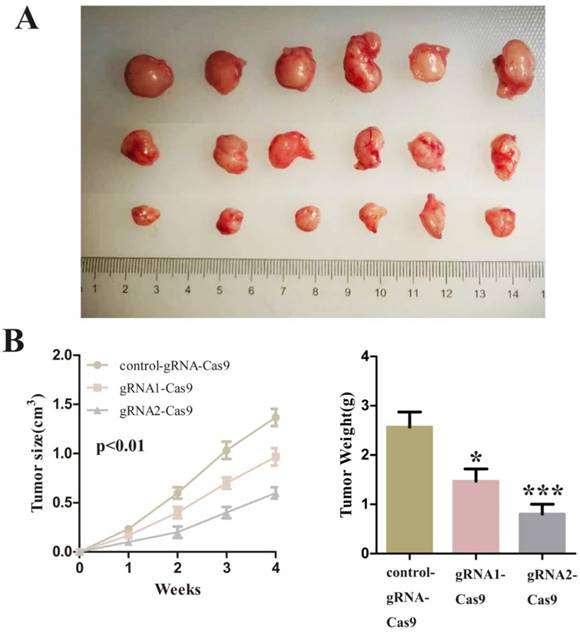
Finally, we decided to verify the impact on the tumor after SCAP was down-regulated in vivo. We injected T24 cells transfected with control-gRNA-Cas9, gRNA1-Cas9 and gRNA2-Cas9 subcutaneously into the nude mice of 4 weeks old. After these tumor cells grew stably under the skin of nude mice and formed a relatively obvious tumor cell mass. By observing the changes of tumor cell mass in each group of nude mice, we concluded that inhibition of SCAP in tumor cells can suppress tumor proliferation in vivo. The experimental phenomena demonstrated that the knockdown of SCAP could dramatically slow down the growth of tumors in vivo (Fig. 5A, Fig. 5B, Fig. 5C). All these results supported that inhibition of SCAP could suppress the growth of BC cells in vivo.
A SCAP was up-regulation in BC cells 5637 and T24 compared to human bladder epithelial immortalized cells SV-HUC1. B, C We knocked down SCAP in 5637 and T24 by using CRISPR-Cas9 in the guidance of two gRNAs, respectively. SCAP expression was significantly downregulated by CRISPR-Cas9 as shown in the figure. Data are shown as mean ± SD (***p< 0.001).
Knockdown of SCAP by CRISPR-Cas9 inhibited cell growth of BC cells. A, B, C Cell proliferation was detected by EDU assay and CCK8 in 5637 after transfection for 48h. D, E, F Cell proliferation was detected by EDU assay and CCK8 in T24 after transfection for 48h. Data are shown as mean ± SD (***p< 0.001).
After 48 hours of transfection, the ability of migration, invasion and apoptosis of 5637 and T24 was measured. A, B The migration of 5637 and T24 cells was significantly inhibited which was detected by wound-healing assay. C, D Down-regulation of SCAP notably promoted the apoptosis of 5637 and T24 which was proved by measuring the activity of caspase 3. E, F, G Down-regulation of SCAP decreased cell invasion detected by transwell assays. Data are shown as mean ± SD (**p< 0.01, ***p< 0.001).
Discussion
The CRISPR-related (CAS) system is a special system that exists in certain bacteria to remove foreign genetic material. As the most characteristic member of the Type II CRISPR system, nuclease Cas9 can be widely used in mammalian cells for double-stranded DNA cleavage and gene knockout under the guidance of single-directed RNA[16-18]. The CRISPR-Cas9 system acts directly on DNA molecules and has a stronger gene knockdown efficiency than other genetic tools such as siRNA[19]. Because of the powerful functions of CRISPR-Cas9 system, its emergence has also greatly promoted the development of the field of genetic editing[18].
Fat metabolism is one of the most important metabolic pathways in the body and plays an important role in maintaining the homeostasis of the cell environment[20]. Fat metabolism mainly consists of fatty acid metabolism and lipid droplet metabolism. Previous studies have shown that fat metabolism has a certain relationship with the occurrence and development of tumors, among which fatty acid metabolism is essential for the maintenance of the micro-environment of malignant tumors, and lipid droplet metabolism is involved in the occurrence and development of various malignant tumors[21]. More and more evidences have shown that lipid metabolism in tumor cells has certain specific changes, which can affect the synthesis and degradation of tumor cell membranes, and also regulate the synthesis and decomposition of lipids in tumor cells through signaling pathways, so as to maintain the balance of energy in the body[22]. Changes in fat metabolism can affect many cellular processes, including cell growth, proliferation, differentiation, and apoptosis[23].
This is the first report of the role of SACP in BC. Our study explored the expression pattern and clinical significance of SCAP in BC, and further explored the possible role of SCAP in BC cells. In addition, our study also confirmed that SCAP can be considered as a promising biomarker for the diagnosis of BC. The expression of SCAP in BC was significantly increased, and its expression level was positively correlated with histological grade and TNM stage of BC. Furthermore, we also studied the role of SCAP in BC cells, and found that silencing SCAP expression by targeting the SCAP gene CRISPR-Cas9 system can inhibit tumor cell proliferation, inhibit cell migration, and induce apoptosis. In order to more accurately demonstrate that inhibition of SCAP expression can inhibit tumor growth, we performed a subcutaneous tumor formation experiment in nude mice. The results of animal experiments also demonstrated that inhibition of SCAP expression inhibits tumor proliferation.
As a key gene in the tumor fatty acid metabolism pathway, the down-regulation of SCAP can effectively destroy the nutritional metabolism of BC cells. The discovery of SCAP has made it possible to develop new tumor drugs targeting BC cells. Targeting SCAP provides a good treatment strategy for clinical treatment of BC. It is worth noting that our research shows that SCAP plays a carcinogenic role in BC and plays a key role in the regulation of BC.
We first discovered that SCAP is more highly expressed in BC than normal tissue. Down-regulation of SCAP inhibits the malignant phenotype of BC. Our results indicate that SCAP is carcinogenic in BC. In conclusion, this study demonstrates that SCAP may be involved in the development of BC by affecting the progression of BC.
Methods
Patient samples
36 BC patients who got radical cystectomy were included in our research. BC tissues and the paired normal tissues were snap-frozen in liquid nitrogen at once after surgery. The informed consent was also got from all patients. Our study was approved by the institutional research ethics committee of Shenzhen Second People's Hospital.
A Tumors developed by subcutaneous injection of control-gRNA-Cas9, gRNA1-Cas9 or gRNA2-Cas9. The representative images of the three were showed using a ruler as a reference. B The sizes of tumors were measured every week and recorded. C The weights of tumor tissues were detected and analysed one month later.
Real-time quantitative PCR
We extracted the total RNA from the transfected cells or tissues by utilizing TRIzol reagent (Invitrogen, Grand Island, NY, USA), and the operation steps mainly according to the instruction of manufacturer. We converted the cDNA from total RNA by utilizing the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the instructions. The primer sequences were as follows: SCAP primers sense: 5'-TCTGTGGCTCAGTTCCTTTTGT-3' and reverse: 5'-ATGTAGGAAGTGCCAGAGCC-3'. GAPDH primers forward: 5'-CGCTCTCTGCTCCTCCTGTTC-3', reverse: 5'-ATCCGTTGACTCCGACCTTCAC-3'. We used a standard SYBR Green PCR kit (Takara, Dalian, China) for performing quantitative PCR (qPCR), and the final reaction volume was 20 ul according to the manufacturer's instructions, including 1 ul of First-Strand cDNA, 0.4 ul of sense primer, 0.4 ul of reverse primer, 10 ul of 2× All-in-OneTM qPCR Mix (Takara, Dalian, China), 0.4 ul of 50× ROX Reference Dye and 7.8 ul of double-distilled water. The reactions were performed in triplicate by using the ABI PRISM 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). GAPDH was chosen as the internal control. The average value in each triplicate was used to calculate the relative amount of HIF1A-AS2 using the comparative ΔCt method.
Cell lines and cell culture
Human BC cells 5637, T24 and human normal bladder epithelial cells (SVHUC-1) were bought from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The 5637 cells were cultured with RPMI-1640 (1640) (Invitrogen, Carlsbad, CA, USA). The T24 cell lines were cultured with DMEM (Invitrogen, Carlsbad, CA, USA). The SV-HUC-1 cells were cultured with F12K medium (Invitrogen, Carlsbad, CA, USA). 1640, DMEM and F12K media were all mixed with 10% fetal bovine serum (FBS). Then plates were grown in an incubator at 37℃ with an atmosphere of 5% CO2.
Cell proliferation assays
5-ethynyl-20-deoxyuridine (EdU) assay kit (Ribobio, Guangzhou, China) and Cell Counting Kit-8, CCK-8 (Beyotime Institute of Biotechnology, Shanghai, China) were used for cell proliferation evaluation. The operation procedure is performed mainly according to the instruction of manufacturer.
Cell migration assay
Cell migration was evaluated by wound-healing assay, which mainly according to the reported methods. Briefly, cells were seeded in 6-well plates and incubated in an incubator to get 100% confluence before assay. A sterile 200 ul pipette tip was utilized to generate a clear line in the wells. We used a digital camera system to take pictures from each well quickly. After one day, pictures were taken again. We counted the migration distance at the time of 0 h and 24 h.
Caspase 3 ELISA assay
The level of the cell apoptosis was detected by measuring the activity of caspase 3 by using the caspase 3 enzyme-linked immunosorbent assay (ELISA) assay kit (Cusabio, Wuhan, China), and the specific operation steps mainly according to the instructions of manufacturer. We measured the OD values by using a microplate reader (Bio-Rad).
Transwell assay
The cells were treated with Lipofectamine 2000-encapsulated plasmid control-gRNA-Cas9, gRNA1-Cas9 or gRNA2-Cas9 for 48h. T24 and 5637 cells added with 200μl serum-free medium were plated into the upper chambers containing matrigel (1:8, 50μl/well, BD Bioscience, San Jose, CA, USA). The lower chambers were supplemented with complete medium. After 48 hours of cultivation, cells under the surface of the lower chamber were washed by using 1 × PBS, fixed with methanol for 20 min, stained with 0.1% crystal violet for 25 min. Once dried, the invaded cells were observed though the inverted microscope and imaged. After that, each chamber with the invaded cells was soaked into 1 ml 33% acetic acid for 10 min to wash out the crystal violet. 100μl of 33% acetic acid were added into each well of 96-well plates, and then the absorbance was detected at 570 nm by a microplate reader (Bio-Rad).
In vivo tumor formation assays
The operation steps of animal studies were mainly according to institutional guidelines. The 4 weeks old male nude mice were bought from Shanghai Experimental Animal Center, Chinese Academy of Sciences, Shanghai, China. T24 cells (1×106 in 100 ul PBS) were subcutaneously injected into the right side of each mouse's abdomen. The mices were subcutaneously injected with 3ug of Lipofectamine 2000-encapsulated plasmid control-gRNA-Cas9, gRNA1-Cas9 or gRNA2-Cas9 every 3 days. We measured the tumor size every week. Tumor weight was detected one month later.
Statistical analysis
All experimental assays were repeated in triplicate. All data were shown as mean ± standard deviation (SD). All statistical analyses were counted by using SPSS 20.0 software (IBM, Chicago, IL, USA). The SCAP expression differences between BC tissues and the paired normal tissues were analyzed using paired samples t-test. CCK-8 assay data were analyzed by ANOVA. The independent samples t-test was used to analyze other data. P value of less than 0.05 was considered to be statistically significant.
Acknowledgements
This work was supported by the National Key Basic Research Program of China (973 Program) (2014CB745201), Shenzhen Knowledge Innovation Program (20160412141802805), National Natural Science Foundation of China (81772737), the Shenzhen Municipal Government of China (JCYJ20170413161749433, JSGG20160301161836370), the Sanming Project of Shenzhen Health and Family Planning Commission, SZSM201412018, SZSM201512037. The high level university's medical discipline construction 2016031638.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Berdik C. Unlocking bladder cancer. Nature. 2017;551:S34-s5
2. Grayson M. Bladder cancer. Nature. 2017;551:S33
3. Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W. et al. Bladder cancer. Lancet (London, England). 2016;388:2796-810
4. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nature medicine. 2015;21:827
5. Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nature medicine. 2015;21:560-2
6. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cellular and molecular life sciences: CMLS. 2016;73:377-92
7. Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition (Burbank, Los Angeles County, Calif). 2000;16:202-8
8. Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331-40
9. Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harbor perspectives in biology. 2011:3
10. Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS oncology. 2013;2:289-99
11. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature reviews Cancer. 2007;7:763-77
12. Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X. et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer cell. 2015;28:569-81
13. Flaveny CA, Griffett K, El-Gendy Bel D, Kazantzis M, Sengupta M, Amelio AL. et al. Broad Anti-tumor Activity of a Small Molecule that Selectively Targets the Warburg Effect and Lipogenesis. Cancer cell. 2015;28:42-56
14. Sukhanova A, Gorin A, Serebriiskii IG, Gabitova L, Zheng H, Restifo D. et al. Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer discovery. 2013;3:96-111
15. Jiang P, Mukthavaram R, Chao Y, Nomura N, Bharati IS, Fogal V. et al. In vitro and in vivo anticancer effects of mevalonate pathway modulation on human cancer cells. British journal of cancer. 2014;111:1562-71
16. Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS synthetic biology. 2013;2:604-13
17. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281-308
18. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE. et al. RNA-guided human genome engineering via Cas9. Science (New York, NY). 2013;339:823-6
19. Zhan H, Xie H, Zhou Q, Liu Y, Huang W. Synthesizing a Genetic Sensor Based on CRISPR-Cas9 for Specifically Killing p53-Deficient Cancer Cells. ACS synthetic biology. 2018;7:1798-807
20. Gibbons GF. Regulation of fatty acid and cholesterol synthesis: co-operation or competition? Progress in lipid research. 2003;42:479-97
21. Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nature reviews Cancer. 2016;16:732-49
22. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell metabolism. 2013;18:153-61
23. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews Cancer. 2013;13:227-32
Author contact
![]() Corresponding author: Wei Jiang, Weiren Huang, Zhiming Cai, E-mail:jiangwei01com (Wei Jiang); pony8980com (Weiren Huang); caizhiming2000com (Zhiming Cai)
Corresponding author: Wei Jiang, Weiren Huang, Zhiming Cai, E-mail:jiangwei01com (Wei Jiang); pony8980com (Weiren Huang); caizhiming2000com (Zhiming Cai)

 Global reach, higher impact
Global reach, higher impact