10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(9):1816-1834. doi:10.7150/ijbs.35520 This issue Cite
Research Paper
Impact of chronically alternating light-dark cycles on circadian clock mediated expression of cancer (glioma)-related genes in the brain
1. The Key Laboratory of Aquatic Biodiversity and Conservation of Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China.
2. Key Laboratory of Molecular Biophysics of the Ministry of Education, Wuhan, Hubei 430074, China
3. University of Chinese Academy of Sciences, Beijing, China.
4. The Department of Cerebrovascular Diseases, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, China.
5. Henan Medical Key Laboratory of Translational Cerebrovascular Diseases, Zhengzhou, China.
Received 2019-4-6; Accepted 2019-5-15; Published 2019-7-20
Abstract
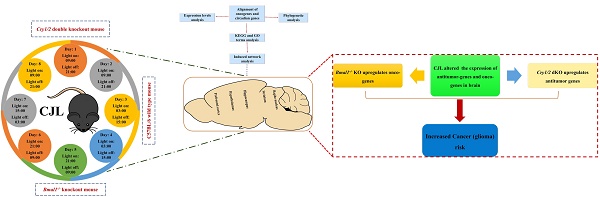
Disruption of the circadian rhythm is a risk factor for cancer, while glioma is a leading contributor to mortality worldwide. Substantial efforts are being undertaken to decrypt underlying molecular pathways. Our understanding of the mechanisms through which disrupted circadian rhythm induces glioma development and progression is incomplete. We, therefore, examined changes in the expression of glioma-related genes in the mouse brain after chronic jetlag (CJL) exposure. A total of 22 candidate tumor suppressor (n= 14) and oncogenes (n= 8) were identified and analyzed for their interaction with clock genes. Both the control and CJL groups were investigated for the expression of candidate genes in the nucleus accumbens, hippocampus, prefrontal cortex, hypothalamus, and striatum of wild type, Bmal1-/- and Cry1/2 double knockout male mice. We found significant variations in the expression of candidate tumor suppressor and oncogenes in the brain tissues after CJL treatment in the wild type, Bmal1-/- and Cry1/2 double knockout mice. In response to CJL treatment, some of the genes were regulated in the wild type, Bmal1-/- and Cry1/2 similarly. However, the expression of some of the genes indicated their association with the functional clock. Overall, our result suggests a link between CJL and gliomas risk at least partially dependent on the circadian clock. However, further studies are needed to investigate the molecular mechanism associated with CJL and gliomas.
Keywords: Circadian Rhythm, Clock Genes, Glioma, Oncogenes, Tumor Suppressor Genes.
Introduction
Disruption in biological rhythms is linked with various serious psychiatric and brain disorders including mood disorders, depression, anxiety, insomnia, suicidal ideation, Parkinson disease (PD), Alzheimer disease (AD), Prader-Willi syndrome (PWS), Smith-Magenis syndrome (SMS), autism spectrum disorders (ASDs) and attention-deficit hyperactivity disorder (ADHD) (1-3). Similarly, disrupted biological rhythms have been associated with heart diseases, obesity, diabetes, and gastrointestinal dysfunctions (4-6). Shiftwork increases the risk of breast cancer (7) and other malignancies which are caused by a bright light at night through mitigation of pineal hormone melatonin (8). Shiftwork mediated disruption in circadian rhythm can cause cancer by altering regulation of the myelocytomatosis viral oncogene human recombinant (C-myc), alpha protein (Gadd 45a), murine double minute oncogene (Mdm-2) and p53 encoding genes (8,9). Per1 and Per2 are associated with tumor growth (10), whereas altered Per2 expression dysregulates tumor suppressor genes (cMyc, cyclin D1, cyclin A, Mdm-2, and Gadd45a) and impairs apoptosis by p53 gene (11-13). Altered PER1, PER2, and PER3 promote colonic adenoma, colonic cancer, and breast cancer (14,15). The disrupted circadian system induces tumorigenesis in breast and prostate tissues (8), inhibits p53 and enhances the expression of MYC (12,16). Shiftwork suppresses melatonin and related hormones to cause malignancies, cardiovascular, and metabolic disorders (8).
Cancer, the second leading cause of death around the globe that killed 9.6 million people in 2018, has also been linked with the environmentally disrupted circadian clock. Although the preclinical data support this link; however, the precise molecular mechanisms underlying the relationship between cancer initiation/progression and clock disruption is yet to be understood (7,17,18). The International Agency for Research on Cancer (IARC) listed shift work as a carcinogen (group 2A) for disrupting the biological rhythms (19-21). Several clinical and epidemiological studies revealed a connection between disrupted circadian rhythms and cancer in prostate, breast, and reproductive organs (12,18,20,22). Similarly, several molecular evidence in laboratory studies summarized by Masri and Sassone-Corsi (2018) connect disruption of the circadian molecular machinery with hepatocellular carcinoma, lung cancer, lymphoma, and other tumor types (18). The core clock genes seem essential in tumorigenesis. The SCN, a master circadian clock, is an endogenous timekeeping system which further controls many peripheral clocks in the peripheral tissues of the body (23,24). Cancer affects the molecular and physiological regulation of brain regions, including nucleus accumbens (NAc), hippocampus, prefrontal cortex (PFC), hypothalamus, and striatum, which leads to the development of psychiatric disorders. Thus, alteration of cancer-related genes in these regions may accelerate the risk of brain cancers (25-29). Although circadian rhythm disruption accelerates tumor progression (8,18) however, there is minimal information available about the circadian rhythm disruption and cancer development in the various tissues of the brain. Therefore, the objectives of our study were to investigate the expression of some vital antitumor and oncogenes in the important brain regions of C57BL/6, Bmal1-/-, and Cry1/2 mice after exposure to chronic jetlag like conditions.
Material and methods
Animals
Male (C57/BL6) mice were obtained from Model Animal Research Center of Nanjing University. Bmal1-/- mice is a generous gift from Dr. Lili Chen. Cry1/2 double knockout mice were generous gifts from Dr. Erquan Zhang. All animals were bred at the SPF animal facility at College of Life Science & Technology, Huazhong University of Science & Technology. These mice were housed in standard cages. The Ambient temperature was 25 ± 1 °C, with food and water available ad libitum. Mice remained group housed throughout the experiment. All animal experiments were approved by the Institutional Animal Use and Care Committee at Tongji Medical College, Huazhong University of Science & Technology.
Light dark cycle conditions
Mice were maintained in light-tight housing cabinets. After acclimatization for one week under 12h light: 12h dark (12L:12D), mice were assigned randomly to LD. The CJL group was exposed to 6h phase advance every two days, whereas the lights on and off times were unchanged for the control group throughout the experimental period. The mice were exposed to jetlag conditions for 30 days. Normal food and water were provided throughout the experimental period.
Wheel running activity
Mice were randomized into 2 groups; CJL group n(WT=15, Bmal-/-=12, Cry1/2=10) and control group n(WT=15, Bmal-/-=8, Cry1/2=8). The average ages of the mice were 6-8 weeks. All mice were singularly caged and provided with an in-cage running wheel. All mice were given free access to running wheels over a 1-week acclimation period to determine running characteristics of each mouse and to ensure that our randomization was effective in terms of running time and distance. We determined during the acclimatization period that approximately only one percent of wheel activity occurred during the daytime. CJL mice were then exposed to altered light-dark cycles with 6 h phase advance for one month while the control mice were exposed to normal 12L:12D conditions.
Brain dissection
Tissues were dissected and snap frozen. The brains of mice and the brain regions were dissected following the previously published protocol by Spijker S. (2011) and mouse brain atlas. In brief, after cervical dislocation, the head was removed by using surgical scissors to cut from the posterior side of the ears. A midline incision was made in the skin through scissors. The skin was flipped over the eyes to free the skull. A small incision was made on the top of the skull with care to avoid damaging the brain. Then a cut was made through the most anterior part of the skull or frontal bone. Hence the brain was removed. Hippocampus was removed by placing the brain with the ventral side facing the metal plate. Closed small curved forceps were placed between the cerebral halves, and the brain was held with largely curved forceps. The forceps were gently opened to open the cortical halves. This step was repeated until the complete opening of the regions. After opening around 60% along the midline, the left cortex was opened from the hippocampus by repeatedly opening the forceps in closed position 30-40° counterclockwise. Small forceps were used to separate the hippocampus from the fornix. Hence, two halves of the hippocampus were separated. Medial prefrontal cortex and striatum extracted through coronal sections approximately 1.0 mm using a sharp and clean blade. Anterior commissure became visible after cutting the olfactory bulb. The first section contains motor cortex, while the subsequent section contains the anterior corpus callosum with a darker area in the prefrontal cortex (PFC). Next ventral-dorsal striata were separated. Hypothalamus was located via optic chiasm which was away from the anterior portion of the hypothalamus, followed by dissection of the mammillary nuclei from the posterior of the hypothalamus. For raphe nuclei, the two main sections (approximately 1.5-2.0 mm) were cut off from the brain and then a comparatively thinner section approximately, 1.0 mm was cut off. The midbrain was exposed by cutting off the upper gray matter regions, and raphe nucleus was exposed and scooped out. Nucleus accumbens (NAc) was dissected by following the previously published protocol with necessary modifications (30). In brief, the brain was trimmed to expose an angled parasagittal surface and cut sharp blade to yield approximately 1 mm thick brain slice that contained most of the ipsilateral fornix and the central portion of the NAc. The NAc was dissected out with care to avoid cutting out the other brain parts. All the brain regions were immediately transferred to liquid nitrogen soon after the dissection and stored in -80°C before further use. The brain slices were measured using vibratome. The fresh brain was used to extract higher quality RNA quality. For all dissection experiments, mice were transported to the experimental room and acclimatized for one hour. The brain dissection was performed at ZT(2-3). To avoid ambiguity and to provide equal opportunity, all the mice were dissected in three independent experiments by repeating the order of dissection according to mice type. Each experimental group consisted of 7 mice, while 2-3 mice/ group were dissected each time experiment. n(WT/control=7, WT/CJL=7; Bmal1-/-/control=7, Bmal1-/-/CJL=7; Cry1/2 double knockout-control=7, Cry1/2 double knockout-CJL=7).
RNA extraction
Total RNA was extracted using TRIzol™ reagent (ThermoFisher SCIENTIFIC, China) following the standard protocol provided by the manufacturer. 1 mL of TRIzol™ reagent per 50-100 mg of tissue was added, and samples were homogenized using a hand-held homogenizer. Samples having high-fat contents were centrifuged for 5 min at 12,000 × g and 4°C. The supernatant was transferred to a new tube. Samples were incubated for 5 min to permit complete dissociation of the nucleoprotein complex. 0.2 mL of chloroform (trichloromethane, CHCl3) was then added per 1 mL of TRIzol™ Reagent. After incubation for 2-3 min, the samples were centrifuged for 15 min at 12,000 × g and 4°C. The mixture was separated into a lower red phenol-chloroform, interphase, and a colorless upper aqueous phase. The aqueous phase containing the RNA was transferred into a new tube. An equal volume of ethanol was then added to the aqueous phase and incubated for 10 min. The samples were subsequently centrifuged for 10 min at 12,000 × g in 4°C. The supernatant was discarded with a micropipette. The pellet was washed with 1 mL of 75% ethanol per 1 mL of TRIzol™ Reagent used by vortexing briefly and then centrifuge for 5 min at 7500 × g in 4°C. After discarding the supernatant with a micropipette, the RNA pellet was air dried for 15-20 min. The pellet was resuspended in 20-50 μL of RNase-free water according to the weight of tissue used for RNA extraction. The tubes were heated at 55-60°C for 10 minutes. Then RQ1 buffer (RNAse free DNAse buffer) and RQ1 RNase free DNAse enzyme (SolarBio Science and Technology, Beijing, China) was added into the tubes and heated at 37°C for 30 min. Next, stop enzyme was added and heated at 65°C for 10 min to stop the reaction. The RNA concentration was determined using spectrophotometer by the following formula
Primers design
Primers were designed using PRIMER BLAST, PRIMER-3, and PRIMER-5 (Table 1). The average melting temperature was selected as 60°C, and PCR product size was 70-200 bp. Standard primer size selected was 20bp, whereas average GC content (%) was 50%. All the primers were purchased from AuGCT DNA-SYN Biotechnology, China.
List of primers
| GENE | FORWARD PRIMER SEQUENCE | REVERSE PRIMER SEQUENCE |
|---|---|---|
| Crem | 5'ACTCTAGCTCAGGTAGCAACA 3' | 5'AGGTGGTGTCCCTTCTTCCT 3' |
| Tspan32 | 5'ACTTCCTAGTCTTGCTGCTGG 3' | 5'GCCCAATAGCGCAGTGTTTC 3' |
| Bin1 | 5'TTGCCAAGGCAGAAGAGGAG 3' | 5'CTCCTGCAGATCCACGTTCA 3' |
| Robo1 | 5'GCTGGCGACATGGGATCATA 3' | 5'AATGTGGCGGCTCTTGAACT 3' |
| Dact1 | 5'CGGCCTAGCTCAGGGTTTTA 3' | 5'CCGCCTTTACATTCCAACCA 3' |
| Rap1gap | 5'GCTTCACCTTCGGTGCCTAT 3' | 5'CCCATCACTCCTCCACACAC 3' |
| Bcr | 5'CAGAATTCGCAGCAGTCCTTT 3' | 5'TGTTCCAAACGAGGAATCTGCT 3' |
| Prkaa2 | 5'GGCAAAGTGAAGACTACCAGG 3' | 5'TGTGACAGTAATCCACGGCA 3' |
| Fzr1 | 5'CGTGACCGCATGATCCTACA 3' | 5'ACACGAGCAGCTTGTTGTCA 3' |
| Gpx3 | 5'TTCCTGAAGAACTCCTGCCC 3' | 5'GTTCCAGCGGATGTCATGGA 3' |
| Gsk3b | 5'GCATTTATCATTAACCTAGCACCCT 3' | 5'GCTGCCATCTTTATCTCTGCTA 3' |
| Nf1 | 5'AGTGAAAGTAGTTACCGTGGTC 3' | 5'CCTCATAGTCACGCTTCGGT 3' |
| Dlg1 | 5'CTCTGGTCATCAGTGGGCTC 3' | 5'CTTTGGTTGCCCAGCAAGAC 3' |
| Klf5 | 5'AGCTGGTCCAGACAAGATGTG 3' | 5'ACTGGTCTACCACTGAGGCA 3' |
| Akt1 | 5'TGAGAAGAAGCTGAGCCCAC 3' | 5'TAGGAGAACTTGATCAGGCGG 3' |
| Araf | 5'AACCGCCGACAGTTCTACC 3' | 5'CCCTGGCCTTTCATCTACGA 3' |
| Akt2 | 5'CTGACTCCGAGAAGGCGTC 3' | 5'CAGTATCGTCTGTCACCGGC 3' |
| Ccnd1 | 5'TCAAGTGTGACCCGGACTG 3' | 5'GATGTCCACATCTCGCACG 3' |
| Kras | 5'TGAAGATGTGCCTATGGTCCTG 3' | 5'GCATCGTCAACACCCTGTCT 3' |
| Pdgfb | 5'GCTCCGTCTACGCGTCC 3' | 5'GAATGGGATCCCCCTCGG 3' |
| Pik3r1 | 5'CACCATTACAAAGAAAGCCGGA 3' | 5'GGGCAGTGCTGGTGGAT 3' |
| Prdm16 | 5'GAAGTCACAGGAGGACACGG 3' | 5'TCATTGCATATGCCTCCGGG 3' |
Quantitative PCR (qPCR)
cDNA was synthesized using high capacity RNA to cDNA kit (Transgen, Biotech, China). qPCR was performed using Trans Master Mix (Transgen, Biotech, China) and an applied biosystems 96 well thermal cycler (ThermoFisher SCIENTIFIC). Primer sequences are listed in Table S1. Master mix was prepared according to the manufacturer's protocol with certain modifications according to the requirements. For each reaction, 10 μL qPCR mix (2X), 0.4 µL forward primer (1nmol), 0.4 µL of reverse primer (1nmol) and 0.4 μL reference dye were used per reaction. 1 µL or 1.4 µL of cDNA or RNA (200 or 100 ng/µL) was added to each reaction. The reactions were then run using the following condition: initiation temperature at 94 °C for 2-10 min followed by 40 cycles with denaturation temperature set at 94 °C for 15 seconds and annealing temperature at 60 °C for 1 min. After completion of the reaction, fold changes of expression of genes of interest were normalized to GAPDH endogenous reference gene and then normalized to control samples, and calculated using the ΔΔCt method (31).
Data resources and cancer-associated genes selection
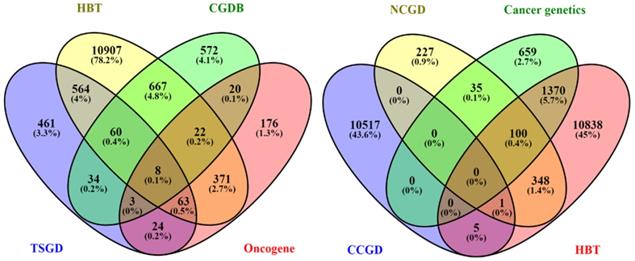
We used the Candidate Cancer Gene Database (CCGD) to download available cancer-associated genes (http://ccgd-starrlab.oit.umn.edu/about.php) (32). A total of 10523 genes were found in the database. For confirmation, these genes were aligned to a total of 2164 genes downloaded from cancer genetics database (http://cancer-genetics.org/genes_download.txt). The aligned 1213 genes were further aligned with 698 protein-coding oncogenes downloaded from oncogene database (http://www.ongene.bioinfo-minzhao.org/) considering the genes reported in human (33). For the sake of removing ambiguity, we further aligned the selected 272 genes with a total of 713 cancer genes downloaded from the Network of Cancer Gene Database (NCG6.0) (https://www.facs.org/quality-programs/cancer/ncdb) (34). At this stage, we finalized 185 genes which were mutually present in these datasets. Genes lists were downloaded from Human Brain Transcriptome (35,36) through online browsing (http://hbatlas.org/pages/hbtd) and aligned to obtain to the region-specific genes. Finally, 133 genes were identified among all the datasets. For antitumor genes, we first downloaded Tumor Suppressor Gene Database (TSGD) via an available online link (https://bioinfo.uth.edu/TSGene/). A total of 1217 genes were downloaded (37). These genes were aligned with genes downloaded from HBT to select genes expression in the brain. To find out the genes under the regulation of the circadian system, we further aligned these genes with the genes sets of Circadian Gene Database (CGDB) (38) (http://cgdb.biocuckoo.org/links.php) and Circadian Expression profile database (http://circadb.hogeneschlab.org/human) (39). Finally, a total of 68 antitumor genes were identified mutually in related databases. In the case of HBT, the genes in pituitary and nerve tibial were combined and aligned with a regional module of HBT, and successful candidates were further processed.
Sequence extraction and phylogenetic analysis
To find out sequences for the selected genes, we used the Genebank/FASTA sequence (NCBI) database (https://www.ncbi.nlm.nih.gov/nuccore). We used the browse function NCBI (Gene) to search for the sequences of a total of 201 genes through their gene IDs. For the sake of simplicity, we selected only one variant of each gene for phylogenetic analysis. All the pre-selected genes were subjected to MEGA 6.0.6 software for phylogenetic analysis (40), and all those sequences that clad at the same level were taken as relevant genes. The phylogenetic tree was constructed using sequence alignment and phylogeny options. Duplicates and poorly aligned sequences were removed before building a tree.
Cancer-associated genes annotations and bioinformatics analysis
The selected genes were subjected to functional annotation analysis using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/summary.jsp) for finding the associated KEGG pathways (41). Top ten pathways were selected, and topmost genes were compared with the similar clads of the phylogenetic tree. The gene symbols were converted to ensemble IDs using BioDBnet (https://biodbnet-abcc.ncifcrf.gov/db/dbOrthoRes.php) before their KEGG analysis. To find the interactions of molecular pathways, the genes were subjected to CPDB database (http://cpdb.molgen.mpg.de/). Induced network modules and enrichment analyses were carried out to determine the specific interactions and genetic level, post-genetic level, and biochemical level (42,43). Based on the KEGG analysis and phylogenetic tree representation, 33 genes were selected.
Tissue level expression and pattern analysis
The selected 22 representative cancer-related genes were further analyzed for their specific expression in brain and other tissues. Mean expression values of genes in the human brain were downloaded from HBT, whereas the tissue-specific expression values were downloaded using the NCBI gene browser using mus musculus as the target species (https://www.ncbi.nlm.nih.gov/gene). The data were subjected to the “expression-based pretty heat map” tool of imageGP, an online tool for converting numerical data into heatmaps using log2 through Pearson distance matrix method with default remaining conditions (http://www.ehbio.com/ImageGP/index.php/Home/-Index/PHeatmap.html). Overall, genes identified through different databases were subjected to Venn diagrams for their proper representation using Venny 2.1 (http://bioinfogp.cnb.csic.es/-tools/venny). Furthermore, SCNseq online database was used to determine the ZT timepoint for maximum and minimum expression of selected genes in SCN (http://www.wgpembroke.com/shiny/SCNseq/).
Statistical analysis
All results presented were analyzed using Excel (Microsoft Software, 365) and GraphPad Prism 7 (GraphPad Software, La Jolla, California, USA). Student's T-test, one-way and two-way ANOVA have performed accordingly. P value <0.05 was considered significant.
Results
Identification and selection of candidate genes
Oncogenes and tumor suppressor genes were selected in the peer-reviewed literature using relevant databases, including CCGD, cancer genetics database, OGDB, NCG6.0, HBT, TSGD, CGDB, and CircaDB (Figure 1). The schematic overview of the experimental process has been shown in Figure 1.
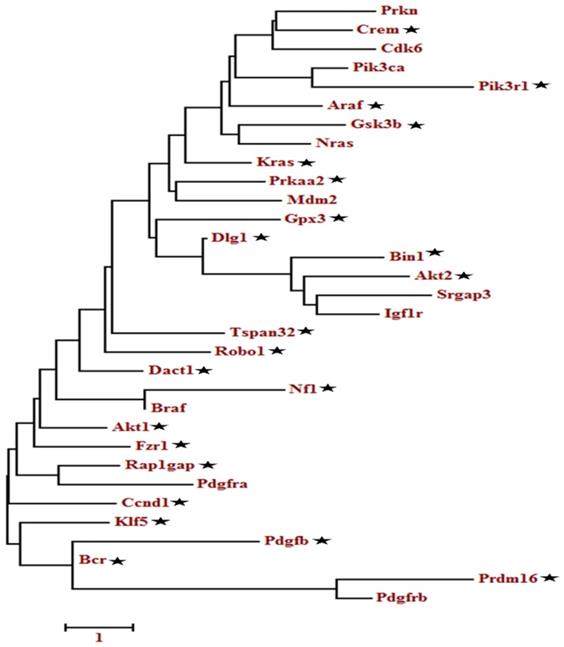
A total of 133 oncogenes and 68 tumor suppressor genes were found in all the databases (Table S4). KEGG pathway analysis showed that among the 201 unigenes (both oncogenes and tumor suppressor genes), 33 genes were implicated in the glioma pathway (Figure S1). The pathway-based analysis further revealed that four tumor suppressor genes (Per2, Prkaa2, Npas2, Arntl) were associated with circadian rhythm pathway. The phylogenetic analysis of the selected 201 genes (tumor suppressor and oncogenes) as depicted in Table S4, showed a relationship between oncogenes and tumor suppressor genes (Figure S2-S4). Among the closely related genes, 32 genes found appeared repeatedly in topmost cancer pathways (especially glioma) during KEGG analysis. These identified genes were further analyzed using phylogenetic analysis to determine the candidate genes (Figure 2). Finally, 22 candidate genes were selected based on the criteria that at least one representative gene was selected per clade. The ZT timepoints for maximum and minimum expression levels were determined using the online database SCNseq (Table 2).
Distribution of genes downloaded using different databases. Venn diagram shows the number of downloaded genes from all selected databases (HBT, CGDB, TSGD, Oncogene, NCGD, Cancer genetics, CCGD, and HBT) and the distribution of genes across the databases. The matched genes among all databases were selected for further process.
Molecular phylogenetic analysis of the selected tumor suppressor and oncogenes genes. The evolutionary history was inferred by using the Maximum likelihood method based on the Tamura-Nei model. The tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log-likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved, 32 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6.
The evolutionary history was inferred by using the Maximum likelihood method based on the Tamura-Nei model. The tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log-likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved, 32 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6.
Final selected genes associated with glioma
| Gene symbol | Ensemble ID | Mean expression in brain | highest expression time-point | lowest expression time-point |
|---|---|---|---|---|
| Tumor suppressor genes | ||||
| Crem | ENSMUSG00000063889 | 4.410036144 | ZT2 | ZT14 |
| Tspan32 | ENSMUSG00000000244 | 5.689827594 | ZT6 | ZT14 |
| Bin1 | ENSMUSG00000024381 | 4.722469534 | ZT6 and 10 | ZT22 |
| Robo1 | ENSMUSG00000022883 | 5.971814277 | ZT6 and 14 | ZT2, 10 and 18 |
| Dact1 | ENSMUSG00000044548 | 4.794540791 | ZT10 | ZT2 and 14 |
| Rap1gap | ENSMUSG00000041351 | 7.611098895 | ZT14 | ZT2 and ZT22 |
| Bcr | ENSMUSG00000009681 | 9.403224037 | ZT4 | ZT6 and 10 |
| Prkaa2 | ENSMUSG00000028518 | 8.473877937 | ZT6 | ZT18 |
| Fzr1 | ENSMUSG00000020235 | 7.298708704 | ZT10 | ZT22 |
| Gpx3 | ENSMUSG00000018339 | 2.796361742 | ZT10 | ZT22 |
| Gsk3b | ENSMUSG00000022812 | 2.892956828 | ZT2 | ZT18 |
| Nf1 | ENSMUSG00000020716 | 5.168002969 | ZT6 | ZT22 |
| Dlg1 | ENSMUSG00000022770 | 4.647283501 | ZT2 | ZT10 |
| Klf5 | ENSMUSG00000005148 | 4.792437773 | ZT10 | ZT2 and 22 |
| Oncogenes | ||||
| Akt1 | ENSMUSG00000001729 | 2.866842546 | ZT10 | ZT22,2 |
| Akt2 | ENSMUSG00000004056 | 5.15949289 | ZT14 | ZT22,2 |
| Araf | ENSMUSG00000001127 | 9.892118727 | ZT2,22 | ZT14 |
| Ccnd1 | ENSMUSG00000070348 | 4.013195242 | ZT6 | ZT14 |
| Kras | ENSMUSG00000030265 | 3.594204131 | ZT2,6,10,22 | ZT14,18 |
| Pdgfb | ENSMUSG00000000489 | 3.809463804 | ZT14 | ZT22,2 |
| Pik3r1 | ENSMUSG00000041417 | 6.597822674 | ZT2-14,22 | ZT14 |
| Prdm16 | ENSMUSG00000039410 | 9.218920632 | ZT6,18 | ZT10,22 |
Candidate cancer-related genes are predicted to interact with clock genes
To systematically study the complex biological function and associated pathways of the genes, we mapped the assembled 22 (candidate) unigenes against the KEGG analysis tool using DAVID database and found 15, 15 and 12genes associated with glioma, prostate cancer, and chronic myeloid leukemia, respectively (Table S2). These genes were associated with 83 KEGG pathways (Table S2). We further investigated the interaction between clock genes and selected candidate genes, at the gene level, protein level, and biochemical level using the CPDB online database. We found that the clock genes (Arntl, Per1, Per2) interact with the candidate tumor suppressor and oncogenes via Gsk3b, Crem, and Mdm2 (Figure 3). Mdm2 and Crem are associated with glioma, leukemia, prostate cancer, chronic myeloid leukemia, and several other cancer pathways, whereas, Gsk3b is an antitumor gene associated with prostate cancer (Table S3). A putative schematic network was generated to summarize and characterize the selected functional genes in cancer pathways (Figure 3 A-C, Figure 4).
The induced network was analyzed using the CPDB online database by considering the gene interaction, protein interaction, and biochemical interaction without intermediate links. The interactions between cancer-related genes and clock genes were determined. (A)- MDM2 is associated with PER2 in glioma (B)- CREM is associated with PER1 in myeloid leukemia and (C)-GSK3 is associated with ARNTL in prostate cancer. The cancer genes interacted with clock genes have been indicated with a blue line and green arrow.
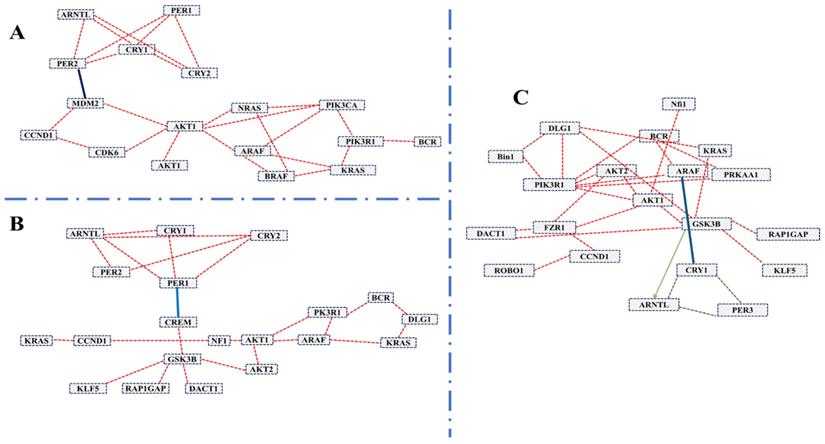
Schematic network based on CPDB analysis. This figure shows the putative schematic diagram for the interaction of clock genes with cancer-associated genes. Mdm2 and Gsk3b were detected to provide the connection between cancer-related genes and clock genes.
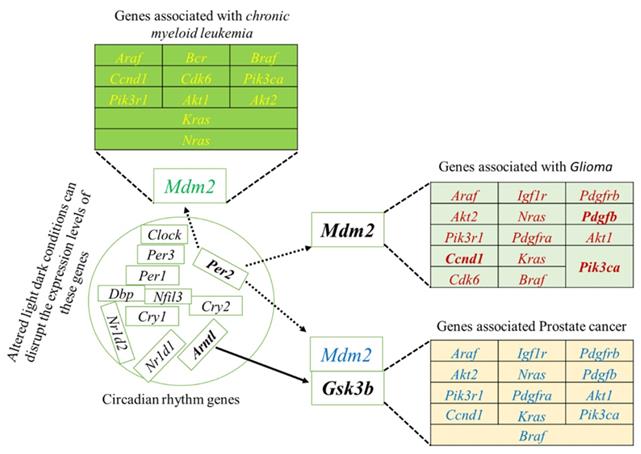
The selected candidate genes express in various organs
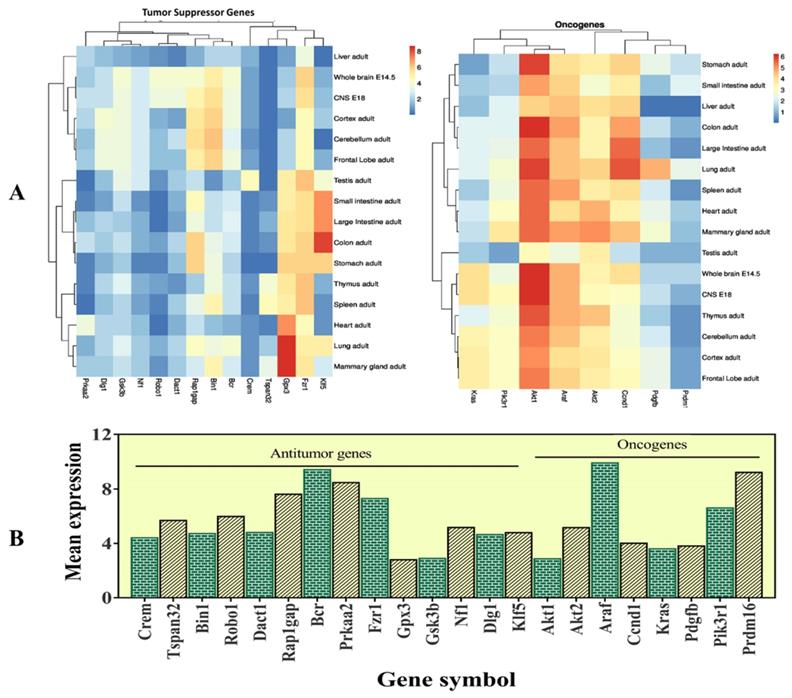
The selected 22 candidate genes (Table 2) were searched using the NCBI browser for their expression pattern in different organs and brain areas. The HBT downloaded data was searched for mean expression levels of genes in the human brain. The expression patterns showed that 22 selected genes related to cancer and tumor suppression were expressed in the central nervous system (CNS), whole brain, cortex, cerebellum, liver, intestine, and frontal lobe. Most of them were found with low expression levels in the entire brain. However, Crem and Tspan32 showed the lowest and Fzr1, and Bin1 showed the highest expression levels. Overall, the highest expression levels were observed for Gpx3 (mammary glands and lungs), Klf5 (colon), and Gsk3b (brain) (Figure 5). Among the selected genes, 15 (Araf, Braf, Kras, Ccnd1, Cdk6, Igf1r, Nras, Pik3ca, Pik3r1, Pdgfra, Pdgfrb, Pdgfb, Akt1, Akt2, and Mdm2) were related to glioma and leukemia (Figure 4).
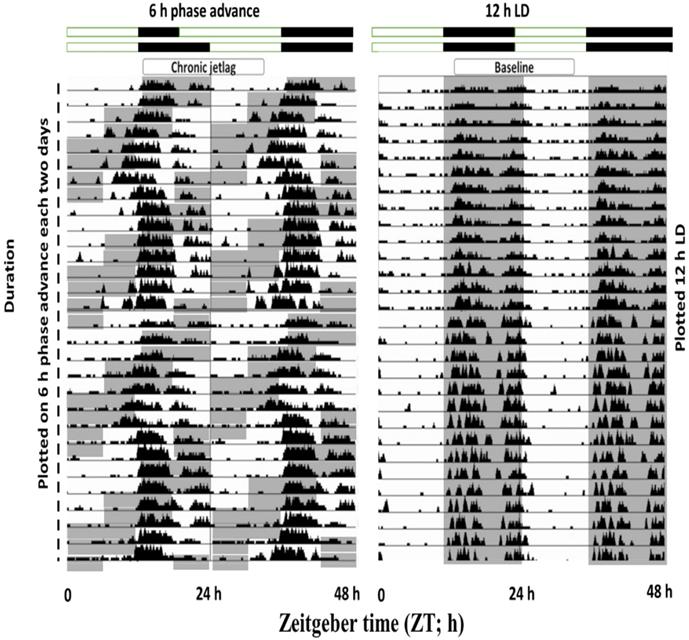
Exposure to CJL conditions shifted the phase of wheel-running activity
To understand the effects of shiftwork on mice physiological behavior, we first employed an established CJL model by exposing mice to a 6 h phase shift every 2-3 days (figure 6). Wheel running activities were recorded which confirmed that activity was shifted by 6 h phase advance of the LD cycle. Those mice showed abnormal wheel run activity were excluded from the experimental process.
CJL altered the brain region-specific expression levels of candidate genes in mice with intact circadian clock
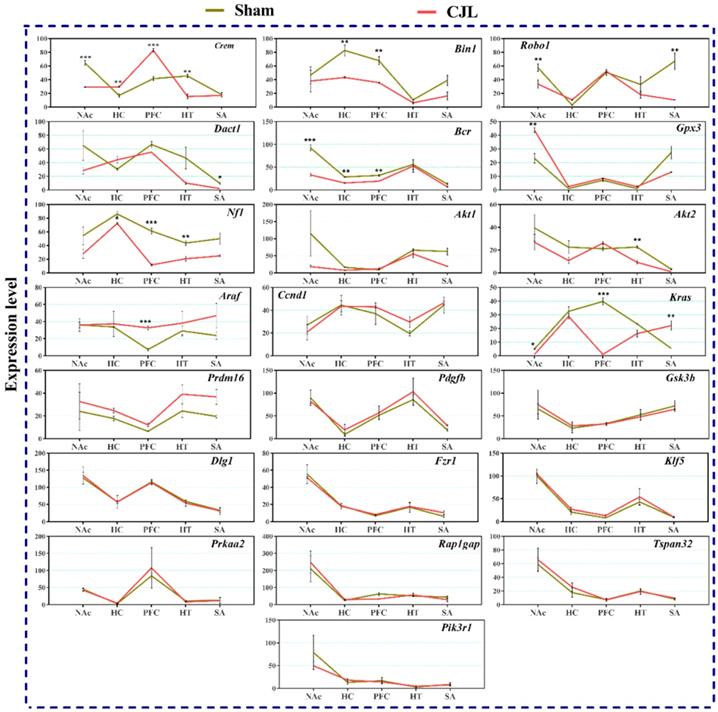
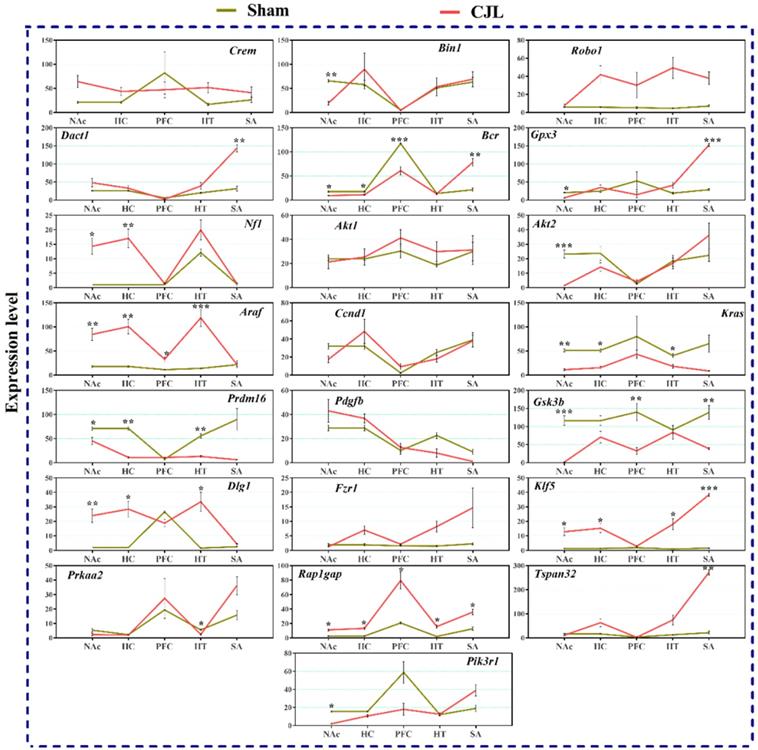
We investigated the mRNA levels in C57BL/6 male mice, exposed to CJL for a period. We found that Crem was downregulated in NAc and hypothalamus and upregulated in PFC and hippocampus compared to control. In the CJL group, Bin1 was downregulated in the hippocampus and PFC. Both the Robo1 and Dact1 were downregulated in the striatum. However, Robo1 was upregulated in the hippocampus. Our results revealed that the Bcr was downregulated in NAc, hypothalamus, and PFC. Similarly, Nf1 was downregulated in the hippocampus, PFC and hypothalamus. In contrast, Gpx3 was upregulated in NAc. Akt2 was downregulated in hypothalamus whereas Akt1 remained the same throughout the brain regions after CJL in comparison with control. Furthermore, Kras was upregulated in striatum and NAc. The levels of Araf, Prdm16, Ccnd1, Pdgfb, Pik3r1, Gsk3b, Dlg1, Fzr1, Klf5, Prkaa2, Rap1gap, and Tspan32 remained unaltered in all the brain regions in response to CJL treatment compared with control (Figure 7).
This figure represents the expression levels of the selected genes. A- heatmap of the selected genes to determine their RPKM values in different organs of the body and different brain regions. B- mean expression levels in the brain.
CJL altered the brain region-specific expression levels of candidate genes in cry1/2 knockout mice
Cry1/2 double knockout mice exposed to CJL showed significant variations in the expression of selected genes in the different brain areas, as shown in figure 8. In summary, Dact1 and Tspan32 were upregulated in the striatum. In NAc, several genes (Pik3r1, Akt2, Gpx3, Kras, Gsk3b, and Bin1) were downregulated. The genes (Araf1, Dlg1, Klf5, and Rap1gap) showed upregulations while Bcr, Kras, and Gsk3b were downregulated in three or more brain regions after CJL. However, the genes (Crem, Robo1, Akt1, Ccnd1, Pdgfb, and Fzr1) showed no variations in any of the brain region (Figure 7).
CJL altered the brain region-specific expression levels of candidate genes in Bmal1-/- knockout mice
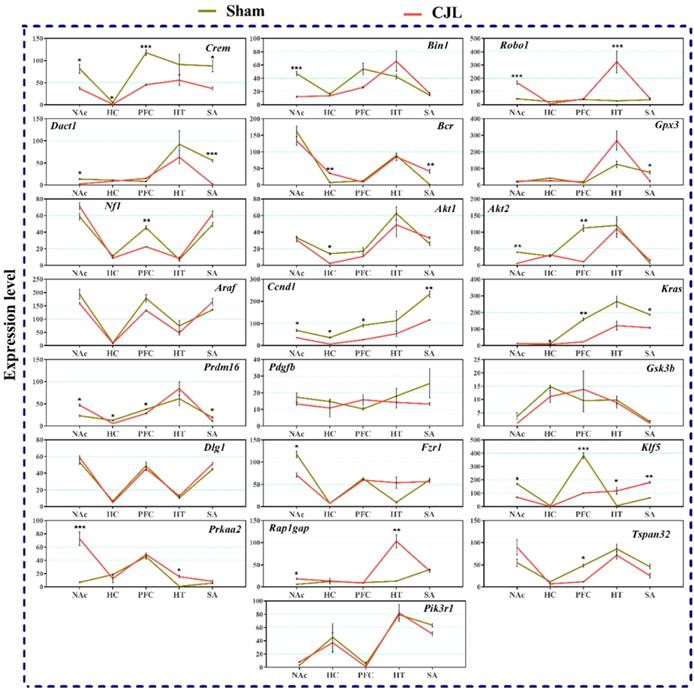
Figure 9 indicate variations in mRNA levels in the brain regions of Bmal1-/- knockout mice exposed to CJL. In short, five of the genes (Ccnd1, Kras, Klf5, Crem, and Prdm16) showed downregulations in three or more brain regions. Overall, several genes (Bin1, Dact1, Gpx3, Nf1, Akt1, Akt2, Fzr1, and Tspan32) showed downregulation, and several genes (Robo 1, Bcr, Klf5, Prkaa2, and Rap1gap) showed upregulation in one or more brain region after CJL (Figure 9).
Clock genes (Bmal1, Cry1/2) regulate the expression of cancer-related genes in the brain
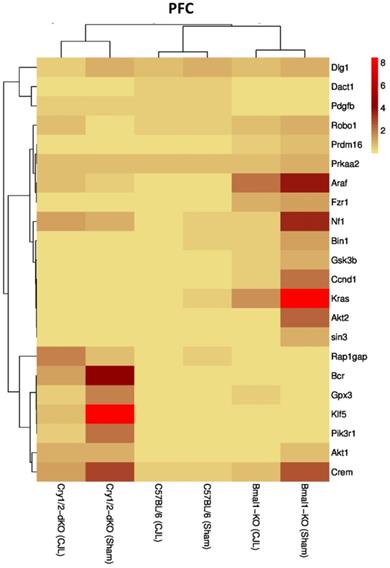
We further investigated the association of candidate cancer-related genes clock (Bmal1 and Cry1/2) genes. We compared expression levels of candidate cancer genes from Bmal-/- and Cry1/2 knock mice with C57BL/6 mice. This analysis revealed that some of the tumor suppressor and oncogenes were differentially regulated, such as Kras was upregulated in PFC of Bmal-/- at baseline line level. Similarly, Klf5 was upregulated in Cry1/2 knockout mice. Most of the genes showed similar regulation when compared at a baseline level and post CJL level (Table S1). Overall, the genes were profoundly affected by CJL in Bmal-/- when compared sham group (Figure 10-14).
Discussion
Complex body function regulation and optimization need a time frame clock (44). Cancer, the second leading cause of death around the globe, is linked with the environmentally disrupted circadian clock, and this link is supported by pre-clinical data (7,17,18). In the current study, we observed significant variations in the expression of several glioma-related genes in the different brain regions of wild type and clock gene mutant mice on CJL exposure. Increasing evidence by epidemiological studies strongly suggests a relationship between abnormal circadian rhythms through shiftwork and the initiation and progression of cancer (7,17,18), whereas the experimental studies support an association between clock genes and increased risk of cancer in animal models (12,45,46). However, the precise connections between chronic jetlag and expression pattern of genes associated with glioma risks in brain regions, are yet to be understood to minimize the risks and develop treatment strategies. To our knowledge, this first study was designed to investigate alterations in the expression of glioma-related genes in response to CJL in the hypothalamus, hippocampus, striatum, nucleus NAc, and prefrontal cortex using wild type, Bmal1-/- knockout, and Cry1/2 double knockout mice. Our results demonstrate that CJL exposure in mouse brain significantly alters the expression of genes associated with cancer prognosis and tumor suppression. In previous studies, the abnormal regulation of Bmal and other core clock genes have shown an increase in the risk of different types of cancers (45,47-49).
Double-plotted actograms from baseline and CJL mice groups. Baseline mice maintained under 12-hour light/dark conditions (Right side). CJL group was exposed to 6 h phase advance each 2-3 days. Gray-shaded areas of actograms delineate lights off (dark).
Similarly, the expression of cancer-linked genes also altered in response to environmental alteration (44,50). Our findings support the concept that CJL in the brain induces an alteration in the glioma-linked gene expression, which could cause an increase in cell proliferation (50,51). The genes we studied in our models have broad cellular functions and are strongly associated with glioma risks, specifically, glioma (Table S3). The up and downregulations of these genes may not indeed indicate cancer progression or development. However, the links of these genes to CJL in the brain provides evidence of a potential mechanism through which CJL might ultimately contribute to glioma.
Glioma and other brain tumors are usually considered a disease of deliberate death, where the average survival is maximum few years after diagnosis (52-55). Due to the direct interaction of altered light-dark cycles with the circadian clock, shiftwork or jetlag like conditions may increase the risks in developing gliomas. Several studies have demonstrated the increased risks of different cancers associated with abnormal circadian rhythms (18,20,56,57). For instance, Bmal1 gene regulates the molecular system to prevent cancer progression in several tissues (47,49). Our data demonstrate aberrant expression of cancer-related genes in the brain regions of wild type and clock gene mutant mice in response to CJL. Therefore, CJL may either cause localized cell proliferation during the initial exposures or aggravate pre-existing neoplastic lesions.
CJL altered mRNA levels of genes associated with cancer in wild type mice. This figure shows mRNA levels in brain regions (NAc, hippocampus, PFC, hypothalamus, and striatum) extracts from sham and CJL treated animals (n = 7 per group), as assayed by three independent qPCR assays. Expression levels in CJL treated mice were normalized to selected expression levels in baseline mice. Results are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA.
Abnormal circadian rhythms can downregulate immune function, which in turn can impair the immune response against tumor cells (58,59). Similarly, clock genes disruption can facilitate the dissemination of tumor cells in peripheral body parts (60). However, only the downregulated immune function cannot account for cancer progression. It may also include the signals triggered by brain regions (hypothalamus and hippocampus) that may impact the synthesis of mitogenic factors and neurotransmitter release to promote tumorigenesis by acting directly on receptors present in cancer cells. They also suppress hormonal pulsatility, which can assist cell proliferation (50,53,60,61). In the case of our study, the decreased mRNA levels of tumor suppressor genes provide evidence that CJL is inducing the progression of glioma in the brain.
The expression analysis results indicated alterations in cancer-related genes in wild type mice exposed to CJL. In NAc Crem, Robo1, Bcr, and Kras were downregulated whereas, Gpx3 was upregulated. In hippocampus Bin1, Bcr, and Nf1 were downregulated, and Crem was upregulated. In PFC Bin1, Bcr, Nf1, and Kras were downregulated whereas Crem and Araf were upregulated. In hypothalamus Crem, Bcr, Nf1, and Akt2 were downregulated. In striatum Robo1, and Dact1 were downregulated while Kras was upregulated. The expression analysis results indicated alterations in cancer-related genes in Bmal-/- mice exposed to CJL.
CJL altered mRNA levels of genes associated with cancer in Cry1/2 mice. This figure shows mRNA levels in brain regions (NAc, hippocampus, PFC, hypothalamus, and striatum) extracts from sham and CJL treated animals (n = 7 per group), as assayed by three independent qPCR assays. Expression levels in CJL treated mice were normalized to selected expression levels in baseline mice. Results are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA.
In NAc Crem, Bin1, Akt2, Dact1, Ccnd1, Fzr1, and Klf5 were downregulated, whereas Robo1, Prdm16, and Prkaa2 were upregulated. In hippocampus Crem, Akt1, Ccnd1, Kras, and Prdm16 were downregulated whereas Bcr was upregulated. In PFC Crem, Nf1, Akt2, Ccnd1, Kras, Prdm16, Klf5, and Tspan32 were downregulated. In the hypothalamus, Robo1, Prkaa2, Klf5, and Rap1gap were upregulated. In the striatum, Crem, Dact1, Gpx3, Ccnd1, Kras, and Prdm16 were downregulated whereas Bcr and Klf5 were upregulated. The expression analysis results indicated alterations in cancer-related genes in Cry1/2 double knockout mice exposed to CJL. In NAc, Bin1, Gpx3, Bcr, Akt2, Kras, Prdm16, Gsk3b, and Pik3r1 were downregulated whereas Nf1, Araf, Dlg1, Klf5, and Rap1gap were upregulated. In the hippocampus, Bcr, Kras and Prdm16 were downregulated while NF1, Araf, Dlg1, Klf5, and Rap1gap were upregulated. In PFC Bcr, and Gsk3b were downregulated whereas Araf and Rap1gap were upregulated. In the hypothalamus, Kras, Prdm16 and Prkaa2 were downregulated, whereas Araf, Dlg1, Klf5, and Rap1gap were upregulated. In the striatum, Gsk3b was downregulated whereas Dact1, Bcr, Gpx3, Klf5, Rap1gap, and Tspan32 were upregulated. Crem (NAc), Bcr (hippocampus), Nf1 and Kras (PFC), and Dact1 (striatum) were similarly regulated in wild type and Bmal1-/- mice. Bcr and Kras (NAc), and Bcr (hippocampus and PFC) were similarly regulated in wild type and Cry1/2 double knockout mice. Bin1 and Akt2 (NAc), Kras and Prdm16 (hippocampus), Klf5 and rap1gap (hypothalamus), and Bcr and Klf5 (striatum) were similarly regulated in Bmal1-/- and Cry1/2 double knockout mice.
CJL altered mRNA levels of genes associated with cancer in Bmal-/- mice. This figure shows mRNA levels in brain regions (NAc, hippocampus, PFC, hypothalamus, and striatum) extracts from sham and CJL treated animals (n = 7 per group), as assayed by three independent qPCR assays. Expression levels in CJL treated mice were normalized to selected expression levels in baseline mice. Results are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA.
Comparative expression analysis of cancer-related genes in PFC. This figure represents the expression levels of candidate cancer genes in clock gene mutant mice in comparison with mice with the intact clock. The results represented here show fold change values of qPCR analysis. The values were normalized to the selected values of one group per brain region.
Comparative expression analysis of cancer-related genes in NAc. This figure represents the expression levels of candidate cancer genes in clock gene mutant mice in comparison with mice with the intact clock. The results represented here show fold change values of qPCR analysis. The values were normalized to the selected values of one group per brain region.
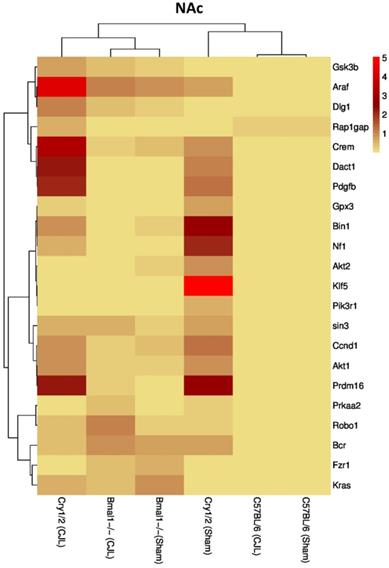
The expression of Crem, Dact1, Ccnd1, Fzr1, Robo1, Prdm16, Prkaa2, Gpx3, Bcr, Kras, Gsk3b, Pik3r1, Nf1, Araf, Dlg1, Klf5, and Rap1gap was inconsistent in NAc, Crem, Akt1, Ccnd1, Bcr, Nf1, Araf, Dlg1, Klf5 and Rap1gap in hippocampus, Crem, Nf1, Akt2, Ccnd1, Kras, Prdm16, Klf5, Tspan32 Bcr, Gsk3b, Araf and Rap1gap in PFC, Robo1, Kras, Prdm16 and Prkaa2, Araf, Dlg1, in hypothalamus, and Gsk3b Dact1, Gpx3, Rap1gap and Tspan32 of clock gene mutant mice. As the expression level of glioma-related genes was found differentially expressed in the comparison between WT, Bmal1-/- and Cry1/2-KO knockout mice, we suggest that clock genes may have a direct or indirect association with expression patterns of these genes (Table 1). For instance, the upregulation of Crem in Bmal1-/- mice in PFC suggests that its expression is associated with Bmal1 directly or indirectly as this gene was downregulated in PFC of wild type mice.
Furthermore, Cry1/2-KO the gene was found unaltered suggesting that the Crem's expression is dependent on Cry1/2 gene/s. Crem has been previously implicated as a key player in tumor regulation/suppression (62,63), hence its altered expression also indicates that CJL can increase cancer risk if Bmal1 is nonfunctional while the non-functionality of Cry1/2 has an opposite effect. Similarly, lacking functional Bmal1may increase the CJL mediated glioma risk via downregulating the previously reported tumor suppressor genes Bin1(53), Tspan32 (64,65), Dact1 (66,67), Fzr1 (68), Gpsx3 (55,69), and Gsk3b (70), whereas lacking functional Cry1/2, may play a role in preventing CJL mediated cancer via downregulating the oncogenes Akt1 (71,72), Kras (73,74), Pik3r1 (75)and Prdm (76,77), which have been reported as oncogenes that promote cancer metastasis and proliferation.
In contrast, the presence of functional Cry1/2 and Bmal1may play a role in preventing CJL mediate cancer development by interacting with cancer-associated genes that were reported playing role in cancer including Tspan32 (64,65), Rap1gap (78), Prkaa2 (79,80), Fzr1(68), Gsk3b (70), Dlg1(61), Klf5 (81,82), Akt1 (72,79,83), Araf (84), Ccnd1(85), Pik3r1(75) and Prdm16 (76,77).
Our results indicated that the tumor suppressor genes were downregulated, whereas the oncogenes were upregulated in WT mice, unlike the clock genes mutant mice. Although, some genes were regulated in a similar pattern such as Kras, Rap1gap, Araf, and Dlg1 were upregulated only. In contrast, Bin1, Fzr1, Akt1, Akt2, and Pik3r1 were downregulated only. Despite the lack of knowledge, several studies have identified multiple molecular and physiological pathways that affect the circadian clock system and thus leads to cancer progression (18,49,86).
The genes found altered in this report may be tissue-specific, as evident from the different genes found upregulated in one region while unaltered and downregulated in another area on CJL treatment. For instance, Bcr was downregulated in PFC and hippocampus while upregulated in the striatum. We detected that Bmal1 has a direct link with tumor invasion in the brain through interaction with Mdm2 and other cancer-related genes (Figure 3). Mdm2 is a proto-oncogene that plays an essential role in human sarcomas, which is overexpressed in a wide variety of cancers. Its protein forms a complex with the p53 protein. Hence, its oncogenic potential is p53-dependent (87).
Considering the importance of the Bmal1 gene in cancer prevention and its association with the clock system, we investigated the expression of tumor suppressor genes to determine if Bmal-/- differentially express these genes. In wild type mice (on CJL treatment), Crem downregulated in NAc and hypothalamus, Bin1 in PFC and hippocampus, Robo1 in NAc and striatum, Dact1 in the striatum, Bcr in NAc, hippocampus, and PFC, and Nf1 in the hippocampus, PFC and hypothalamus. Similarly, in Bmal-/- knockout mice, Bin1 was downregulated in NAc, Bcr was downregulated in NAc, hippocampus, Prkaa2 in the hypothalamus, and Gpx3 in NAc. In contrast, Dact1, Bcr, and Gpx3 were upregulated in the striatum, Nf1 in NAc and hippocampus, Dlg1 in NAc, hippocampus, and hypothalamus, Klf5 in NAc, hippocampus, hypothalamus, and striatum, Rap1gap in NAc, hippocampus, PFC, hypothalamus, and striatum and Tspan32 in the striatum.
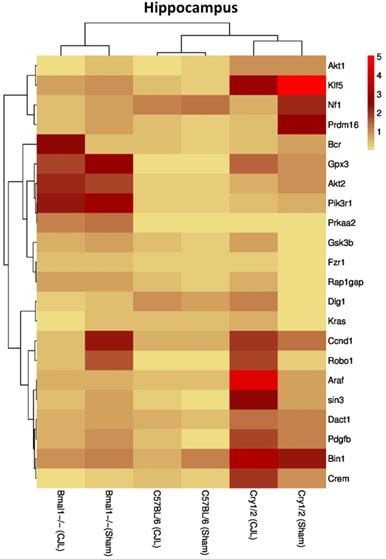
Comparative expression analysis of cancer-related genes in the hippocampus. This figure represents the expression levels of candidate cancer genes in clock gene mutant mice in comparison with mice with the intact clock. The results represented here show fold change values of qPCR analysis. The values were normalized to the selected values of one group per brain region.
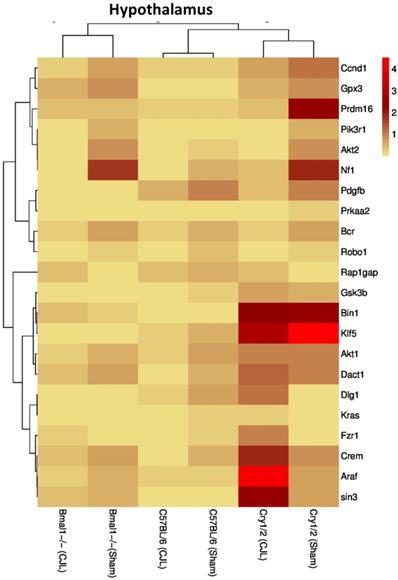
Comparative expression analysis of cancer-related genes in the hypothalamus. This figure represents the expression levels of candidate cancer genes in clock gene mutant mice in comparison with mice with the intact clock. The results represented here show fold change values of qPCR analysis. The values were normalized to the selected values of one group per brain region.
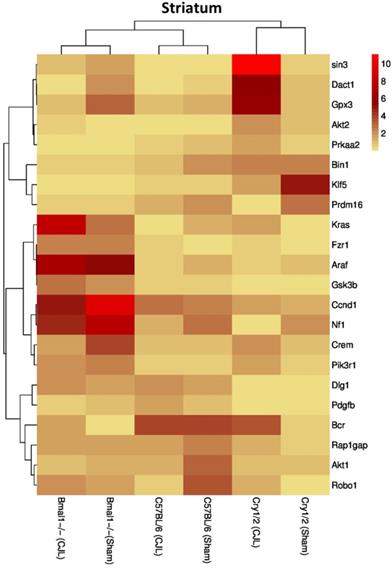
Comparative expression analysis of cancer-related genes in the striatum. This figure represents the expression levels of candidate cancer genes in clock gene mutant mice in comparison with mice with the intact clock. The results represented here show fold change values of qPCR analysis. The values were normalized to the selected values of one group per brain region.
In our study, Prkaa2 and Ccnd1, and Nf1 showed upregulation in Bmal1-/- mice and downregulation in Cry1/2-KO mice and vice versa. These genes are linked with cancer (79,85,88). This regulation suggests a relationship between Cry1/2 and Bmal1 and Prkaa2, Ccnd1and Nf1 in promoting CJL mediated cancer. CJL can significantly alter the expression of multiple genes affecting several molecular functions leading to increased cell proliferation, mitogenic signaling, altered bioenergetics, apoptosis, DNA repair, and extracellular matrix. Furthermore, cancer is a multistep and highly complex process. CJL alone cannot provide all the specific characteristics for cellular proliferation and transformation into a malignant phenotype. However, it may dysregulate certain pro- and anti-proliferative cellular processes. We observed CJL induce upregulation of oncogenes and downregulation of TSGs. Our findings are in support of the previous findings that shiftwork increases cancer risks (19,22). In an earlier study, Bmal1 has been found associated with tumor suppression (47); hence its non-functionality may upregulate oncogenes. However, these findings contradict with our gene expression data. The observed higher expression of TSGs in case of Bmal1-/- mouse could be a compensatory response by other clock genes or anticancer enzymes. The brain is sensitive to signals that affect tumorigenic growth, and therefore, sustains a cell death/proliferation equilibrium (52,85). The upregulated expression of oncogenes and downregulation of TSGs is noteworthy.
Further analysis of tumor suppressor and oncogenes in C57BL/6 mice compared with Bmal1-/- and Cry1/2 mice showed significant differences among mRNA levels of several genes specific to brain regions. These differences indicated that clock genes are directly linked with glioma risk. The present study has some limitations. First, the tissues were dissected following standard protocols. However, being small size and hard to separate closely adjacent tissues, it might be possible that this surrounding may have had residual cells with the processed regions although brain regions we used were well isolated from other tissues. To reduce contamination, we dissected using microdissection. Secondly, we examined only male mice (Bmal1-/- and Cry1/2-KO) while female mice can also be considered in future experiments.
Conclusion
This study illustrates the effects of the disrupted circadian clock via CJL on the expression of cancer-related genes in 5 different brain regions in wild type and clock mutant male mice. As indicated by the functions and related molecular pathways, the candidate genes were found mainly associated with glioma. CJL may be an early stimulus that induces glioma and other cancers by influencing circadian clock that may, in turn, affects a wide variety of cellular functions. These results suggest the association of clock genes (Bmal1, Cry1, and Cry2) with the modification in the expression of glioma-related genes. The intact circadian clock resists the alteration in expression of some glioma promoting genes, but not others, provide support for future investigations related to glioma diagnosis, prognosis, and treatment in association with light-dark cycles regulation and a circadian clock function.
Supplementary Material
Supplementary figures and tables.
Abbreviations
CJL: Chronic jetlag; Bmal1: Brain and muscle ARNT like protein 1; Cry1/2: Cryptochrome ½; TSGs: Tumor suppressor genes; NAc: Nucleus Accumbens; PFC: Prefrontal cortex.
Acknowledgements
The authors are thankful to Prof. Dr. Luoying Zhang (PhD in circadian rhythm-related studies from United States) for providing the resources and guidance related to circadian disruption and clock gene mutation and resources for conducting the experimental work at key laboratory of molecular biophysics and laboratory of circadian rhythm and sleep genetics, college of life sciences, Huazhong University of science and technology. The first author is thankful to the CAS-TWAS Ph.D. fellowship program for the financial resources and support for the research studies.
Author's contributions
All authors have read, finalized, and approved the manuscript. SK: Conceptualization, experimental work, formal analysis, data curation, writing - original draft, and methodology. YL: data curation, experimental work. RS: Conceptualization, experimental work, and data curation. GN: manuscript revision and editing. MX: conceptualizations, data confirmation, Supervision. HH: Supervision.
Funding
The project was supported by National Science Foundation of China (grant number 31870917, 81471174, 81520108011, and 81870942), National Key Research and Development Program of China (grant no: 2018YFC1312200), and Innovation Scientists and Technicians Troop Constructions Projects of Henan Province of China for MX. The funding contributed to study design, data analysis, and paper processing.
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Use and Care Committee at Tongji Medical College, Huazhong University of Science & Technology.
Availability of data and material
The datasets generated during the current study can be obtained from the relevant databases (the web links have been provided in the methods section). The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Logan RW, McClung CA. Rhythms of life: circadian disruption and brain disorders across the lifespan. Nat Rev Neurosci. 2019;20:49-65
2. Khan S, Nabi G, Yao L, Siddique R, Sajjad W, Kumar S. et al. Health risks associated with genetic alterations in internal clock system by external factors. Int J Biol Sci. 2018;14:791-8
3. Khan S, Duan P, Yao L, Hou H. Shift-work mediated disruptions of Circadian rhythms and sleep homeostasis cause serious health problems. Int J Genomics. 2018;2018:1-27
4. Takeda N, Maemura K. Circadian clock and cardiovascular disease. J Cardiol. 2011;57:249-256
5. Jagannath A, Taylor L, Wakaf Z, Vasudevan SR, Foster RG. The genetics of circadian rhythms, sleep and health. Hum Mol Genet. 2017;26:128-138
6. Maury E. Off the Clock : From circadian disruption to metabolic disease. Int J Mol Sci. 2019;20:1597-1621
7. Filipski E, Innominato PF, Wu M, Li XM, Iacobelli S, Xian LJ. et al. Effects of light and food schedules on liver and tumor molecular clocks in mice. J Natl Cancer Inst. 2005;97:507-17
8. Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev. 2013;17:273-84
9. Khan S, Ullah MW, Siddique R, Nabi G, Manan S, Yousaf M. et al. Role of Recombinant DNA Technology to Improve Life. Int J Genomics. 2016:2016
10. Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4
11. Park SC, Park I, Kim H, Lee JM. N-Terminal Domain Mediated Regulation of ROR α 1 Inhibits Invasive Growth in Prostate Cancer. Int J Mol Sci. 2019;20:1-14
12. Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJM. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423-31
13. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375-82
14. Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE. et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786-93
15. Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17:539-45
16. Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5:e10995
17. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
18. Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat Med. 2018;24:1795-803
19. Fritschi L, Glass DC, Heyworth JS, Aronson K, Girschik J, Boyle T. et al. Hypotheses for mechanisms linking shiftwork and cancer. Med Hypotheses. 2011;77:430-6
20. Samulin Erdem J, Notø HØ, Skare Ø, Lie JAS, Petersen-Øverleir M, Reszka E. et al. Mechanisms of breast cancer risk in shift workers: Association of telomere shortening with the duration and intensity of night work. Cancer Med. 2017;6:1988-1997
21. Stevens RG, Hansen J, Costa G, Haus E, Kauppinen T, Aronson KJ. et al. Considerations of circadian impact for defining 'shift work'in cancer studies: IARC Working Group Report. Occup Environ Med. 2011;68:154-62
22. Flynn-Evans EE, Mucci L, Stevens RG, Lockley SW. Shiftwork and prostate-specific antigen in the national health and nutrition examination survey. J Natl Cancer Inst. 2013;105:1292-7
23. Mohawk JA, Green CB, Takahashi JS. Central and Peripheral Circadian Clocks in Mammals. Annu Rev Neurosci. 2012;35:445-62
24. Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. PNAS. 2013;110:9950-5
25. Totterdell S, Smith AD. Convergence of hippocampal and dopaminergic input onto identified neurons in the nucleus accumbens of the rat. J Chem Neuroanat. 1989;2:285-98
26. Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J. et al. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156:1072-83
27. Kesler SR, Kent JS, O'Hara R. Prefrontal cortex and executive function impairments in primary breast cancer. Arch Neurol. 2011;68:1447-53
28. Jehn CF, Kühnhardt D, Bartholomae A, Pfeiffer S, Schmid P, Possinger K. et al. Association of IL-6, hypothalamus-pituitary-adrenal axis function, and depression in patients with cancer. Integr Cancer Ther. 2010;9:270-5
29. Bergouignan L, Lefranc JP, Chupin M, Morel N, Spano JP, Fossati P. Breast cancer affects both the hippocampus volume and the episodic autobiographical memory retrieval. PLoS One. 2011;6:1-9
30. Matthews RT, Coker O, Winder DG. A novel mouse brain slice preparation of the hippocampo-accumbens pathway. J Neurosci Methods. 2004;137:49-60
31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402-8
32. Abbott KL, Nyre ET, Abrahante J, Ho YY, Vogel RI, Starr TK. The candidate cancer gene database: A database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43:844-8
33. Liu Y, Sun J, Zhao M. ONGene: A literature-based database for human oncogenes. J Genet Genomics. 2017;44:119-21
34. Palmieri T, Nulsen J, Yakovleva A, Dressler L, Venkata SK, Ciccarelli FD. et al. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019;20:1-12
35. Weinberger DR, Guennel T, Sousa AMM, Cheng F, Sedmak G, Huttner A. et al. Functional and Evolutionary Insights into Human Brain Development through Global Transcriptome Analysis. Nature. 2011;478:483-9
36. Kawasawa YI, Coppola G, State MW, Krsnik Ž, Bogdanović D, Mane SM. et al. Functional and Evolutionary Insights into Human Brain Development through Global Transcriptome Analysis. Neuron. 2009;62:494-509
37. Zhao M, Sun J, Zhao Z. TSGene: A web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:970-6
38. Li S, Shui K, Zhang Y, Lv Y, Deng W, Ullah S. et al. CGDB: A database of circadian genes in eukaryotes. Nucleic Acids Res. 2017;45:397-403
39. Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;4:1009-13
40. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725-9
41. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57
42. Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. Consensus Path DB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:712-7
43. Kamburov A, Wierling C, Lehrach H, Herwig R. Consensus Path DB - A database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:623-8
44. VoPham T, Weaver MD, Vetter C, Hart JE, Tamimi RM, Laden F. et al. Circadian Misalignment and Hepatocellular Carcinoma Incidence in the United States. Cancer Epidemiol Biomarkers Prev. 2018;27:719-27
45. Sato F, Bhawal UK, Yoshimura T, Muragaki Y. DEC1 and DEC2 crosstalk between circadian rhythm and tumor progression. J Cancer. 2016;7:153-159
46. Yeh CM, Shay J, Zeng TC, Chou JL, Huang T, Lai HC. et al. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol. 2014:2101-2107
47. Zhao S, Wang X, Jiang X, Lu Y, Lu Z, Yang L. et al. The circadian clock gene Bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett. 2015;371:314-25
48. Zeng Z, Luo H, Yang J, Wu J, Chen D. et al. Overexpression of the Circadian Clock Gene Bmal1 Increases Sensitivity to Oxaliplatin in Colorectal Cancer. Clin Cancer Res. 2013;20:1042-1052
49. Chen Y, Zhou X, Chen L, Zhao J, Cheng B, Tang Q. et al. Circadian Clock Gene Bmal1 Inhibits Tumorigenesis and Increases Paclitaxel Sensitivity in Tongue Squamous Cell Carcinoma. Cancer Res. 2016;77:532-44
50. Flores IE, Sierra-Fonseca JA, Davalos O, Saenz LA, Castellanos MM, Zavala JK. et al. Stress alters the expression of cancer-related genes in the prostate. BMC Cancer. 2017;17:1-10
51. Gelman IH. Emerging roles for SSeCKS/Gravin/AKAP12 in the control of cell proliferation, cancer malignancy, and barriergenesis. Genes Cancer. 2010;1:1147-56
52. Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L. et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the microenvironment. Cancer Cell. 2017;32:42-56
53. Vashishtha V, Jinghan N, Yadav AK. Antagonistic role of GSK3 isoforms in glioma survival. J Cancer. 2018;9:1846-1855
54. Bondale N, Castro MG, Wibowo M, Assi H, Klein J, Baker GJ. et al. Lentiviral-Induced High-Grade Gliomas in Rats: The Effects of PDGFB, HRAS-G12V, AKT, and IDH1-R132H. Neurotherapeutics. 2014;11:623-35
55. Yao DM, Zhou JD, Zhang YY, Yang L, Wen XM, Yang J. et al. GPX3 promoter is methylated in chronic myeloid leukemia. Int J Clin Exp Pathol. 2015;8:6450-6457
56. Yang H, Bernd K. Are There Thresholds in Glioblastoma Cell Death Responses Triggered by Temozolomide. Int J Mol Sci. 2019;20:1562-1582
57. Van Dycke KCG, Rodenburg W, Van Oostrom CTM, Van Kerkhof LWM, Pennings JLA, Roenneberg T. et al. Chronically Alternating Light Cycles Increase Breast Cancer Risk in Mice. Curr Biol. 2015;25:1932-1937
58. Abreu M, Basti A, Genov N, Mazzoccoli G, Relógio A. The reciprocal interplay between TNFα and the circadian clock impacts on cell proliferation and migration in Hodgkin lymphoma cells. Sci Rep. 2018;8:1-15
59. Guissoni Campos LM, Buchaim RL, da Silva NC, Spilla CSG, Hataka A, Pinato L. Suprachiasmatic Nucleus and Subordinate Brain Oscillators: Clock Gene Desynchronization by Neuroinflammation. Neuroimmunomodulation. 2017;24:231-241
60. Iwamoto A, Kawai M, Furuse M, Yasuo S. Effects of chronic jet lag on the central and peripheral circadian clocks in CBA/N mice. Chronobiol Int. 2014;31:189-198
61. Sugihara T, Nakagawa S, Sasajima Y, Ichinose T, Hiraike H, Kondo F. et al. Loss of the cell polarity determinant human Discs-large is a novel molecular marker of nodal involvement and poor prognosis in endometrial cancer. Br J Cancer. 2016;114:1012-1018
62. Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J. et al. Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types. PLoS Genet. 2013;9:1-19
63. Mandl C, Ciccolini F, Hölzl-Wenig G, Tucker KL, Liss B, Parlato R. Regulation of proliferation and histone acetylation in embryonic neural precursors by CREB/CREM signaling. Neurogenesis. 2014;1:1-13
64. Naudin C, Smith B, Bond DR, Dun MD, Scott RJ, Ashman LK. et al. Characterization of the early molecular changes in the glomeruli of Cd151 -/- mice highlights induction of mindin and MMP-10. Sci Rep. 2017;7:1-12
65. Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014;19:862-871
66. Hou L, Li X, Liu B, Li R, Wang H, Linghu H. et al. DACT1 Overexpression in type I ovarian cancer inhibits malignant expansion and cis-platinum resistance by modulating canonical Wnt signalling and autophagy. Sci Rep. 2017;7:1-12
67. Cheng H, Lu M, Mao LJ, Wang JQ, Li W, Wen RM. et al. Relationships among MTHFR a1298c gene polymorphisms and methylation status of Dact1 gene in transitional cell carcinomas. Asian Pacific J Cancer Prev. 2012;13:5069-74
68. Cuppen E, Xu H, Prinsen MBW, Muñoz J, van Mourik T, Boxem M. et al. Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells. Nat Commun. 2015;6:5906-5917
69. Guo W, Zhang S, Cao S, Li J, Zhai W, Zhang G. et al. Methylation of promoter and expression silencing of GPX3 gene in hepatocellular carcinoma tissue. Clin Res Hepatol Gastroenterol. 2014;39:198-204
70. Qiu H jiang, Lu X he, Yang S sha, Weng C yin, Zhang E keng, Chen F chao. MiR-769 promoted cell proliferation in human melanoma by suppressing GSK3B expression. Biomed Pharmacother. 2016;82:117-23
71. Goto A, Dobashi Y, Tsubochi H, Maeda D, Ooi A. MicroRNAs associated with increased AKT gene number in human lung carcinoma. Hum Pathol. 2016;56:1-10
72. Zhu J, Wang M, He J, Zhu M, Wang JC, Jin L. et al. Polymorphisms in the AKT1 and AKT2 genes and oesophageal squamous cell carcinoma risk in an Eastern Chinese population. J Cell Mol Med. 2016;20:666-677
73. Tsuchida N, Murugan AK, Grieco M. Kirsten Ras* oncogene: Significance of its discovery in human cancer research ras for viral origin, RAS for human origin and Ras for both viral and human origins. Oncotarget. 2016;7:46717-46733
74. Lavrado J, Brito H, Borralho PM, Ohnmacht SA, Kim NS, Leitão C. et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci Rep. 2015;5:1-10
75. Jiang T, Xiang L, Zhang S, Zhang T, Ai X, Huang Z. et al. Overexpression of PIK3R1 promotes hepatocellular carcinoma progression. Biol Res. 2018;51:1-10
76. Tan SX, Hu RC, Liu JJ, Tan YL, Liu WE. Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells. Int J Clin Exp Pathol. 2014;7:2305-2311
77. Feng T, Feng N, Lv W, Wang Y, Li W, Yu X. et al. Low expression of LINC00982 and PRDM16 is associated with altered gene expression, damaged pathways and poor survival in lung adenocarcinoma. Oncol Rep. 2018;40:2698-2709
78. Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X. et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339-4349
79. Mucaki EJ, Zhao JZL, Lizotte DJ, Rogan PK. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduct Target Ther. 2019;4:1-12
80. Puig-Oliveras A, Castelló A, Folch JM, Ballester M, Fernández AI, Revilla M. Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci Rep. 2016;6:1-12
81. Zhao C, Li Y, Qiu W, He F, Zhang W, Zhao D. et al. C5a induces A549 cell proliferation of non-small cell lung cancer via GDF15 gene activation mediated by GCN5-dependent KLF5 acetylation. Oncogene. 2018;37:4821-4837
82. Sun Z, Han Q, Zhou N, Wang S, Lu S, Bai C. et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7:884-894
83. Chen X, Wang K, Xu Z, Li S, Liu Q, Fu D. et al. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70
84. Lee JW, Soung YH, Kim SY, Park WS, Nam SW, Min WS. et al. Mutational analysis of the ARAF gene in human cancers. Apmis. 2005;113:54-7
85. Chen DG, Zhu B, Lv SQ, Zhu H, Tang J, Huang C. et al. Inhibition of EGR1 inhibits glioma proliferation by targeting CCND1 promoter. J Exp Clin Cancer Res. 2017;36:1-12
86. Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X. et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. 2011;127:531-40
87. Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci. 2002;95:15608-15612
88. Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:1-19
Author contact
![]() Corresponding author: xumengzhouedu.cn (MX); houhwac.cn (HH)
Corresponding author: xumengzhouedu.cn (MX); houhwac.cn (HH)

 Global reach, higher impact
Global reach, higher impact