ISSN: 1449-2288
Int J Biol Sci 2019; 15(10):2065-2074. doi:10.7150/ijbs.35743 This issue Cite
Research Paper
Trends in Alzheimer's Disease Research Based upon Machine Learning Analysis of PubMed Abstracts
1. Key Laboratory of Symbolic Computation and Knowledge Engineering of the Ministry of Education, College of Computer Science and Technology, Jilin University, 130012, Changchun, China
2. Zhuhai Sub Laboratory, Key Laboratory of Symbolic Computation and Knowledge Engineering of the Ministry of Education, Zhuhai College of Jilin University, 519041, Zhuhai, China
3. Department of Electric Engineering and Computer Science, and Christopher S. Bond Life Sciences Center, University of Missouri, 65201, Columbia, USA
Abstract
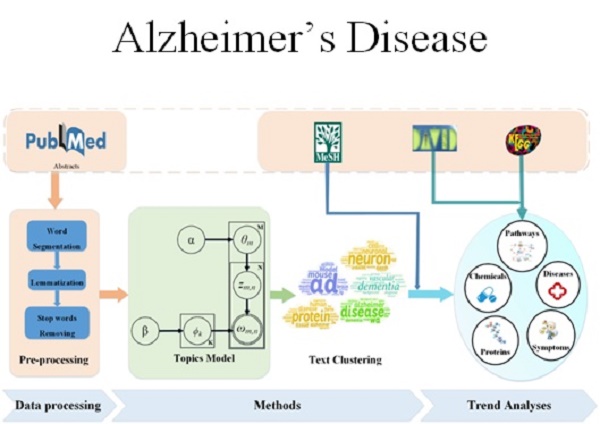
About 29.8 million people worldwide had been diagnosed with Alzheimer's disease (AD) in 2015, and the number is projected to triple by 2050. In 2018, AD was the fifth leading cause of death in Americans with 65 years of age or older, but the progress of AD drug research is very limited. It is helpful to identify the key factors and research trends of AD for guiding further more effective studies. We proposed a framework named as LDAP, which combined the latent Dirichlet allocation model and affinity propagation algorithm to extract research topics from 95,876 AD-related papers published from 2007 to 2016. Trends and hotspots analyses were performed on LDAP results. We found that the focus points of AD research for the past 10 years include 15 diseases, 15 amino acids, peptides, and proteins, 9 enzymes and coenzymes, 7 hormones, 7 carbohydrates, 5 lipids, 2 organophosphonates, 18 chemicals, 11 compounds, 13 symptoms, and 20 phenomena. Our LDAP framework allowed us to trace the evolution of research trends and the most popular areas of interest (hotspots) on disease, protein, symptom, and phenomena. Meanwhile, 556 AD related-genes were identified, which are enriched in 12 KEGG pathways including the AD pathway and nitrogen metabolism pathway. Our results are freely available at
Keywords: Alzheimer's disease, Latent Dirichlet Allocation, Affinity Propagation

 Global reach, higher impact
Global reach, higher impact