Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(12):2664-2675. doi:10.7150/ijbs.34743 This issue Cite
Research Paper
BmBlimp-1 gene encoding a C2H2 zinc finger protein is required for wing development in the silkworm Bombyx mori
1. State Key Laboratory of Silkworm Genome Biology; Key Laboratory of Sericultural Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs; College of Biotechnology, Southwest University, Chongqing 400715, China.
2. College of Plant Protection, Southwest University, Chongqing 400716, China.
Received 2019-3-9; Accepted 2019-7-28; Published 2019-10-12
Abstract
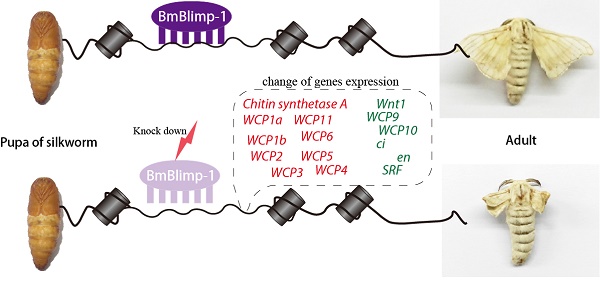
Cys2-His2 zinc finger (C2H2-ZF) proteins represent the most common class of transcription factors. These factors have great potential for the management of developmental progression by regulating the specific spatiotemporal expression of genes. In this study, we cloned one C2H2-ZF protein gene of Bombyx mori, BGIBMGA000319, that is orthologous to B-lymphocyte-induced maturation protein-1 (Blimp-1); we thus named it as Bombyx mori Blimp-1 (BmBlimp-1). In the silkworm, the BmBlimp-1 gene is specifically upregulated during day 2 of the pupal to adult stage and is highly expressed in wing discs on day 3 of the pupa. Knockdown of its expression level in the pupal stage results in a crumpled-winged silkworm moth. Using the predicted DNA-binding sequences of BmBlimp-1 to search the silkworm genome to screen target genes of BmBlimp-1, 7049 genes were identified to have at least one binding site of BmBlimp-1 on their 1 kb upstream and downstream genome regions. Comparisons of those genes with a reported pupal wing disc transcriptome data resulted in 4065 overlapping genes being retrieved. GO enrichment analysis of the overlapping genes showed that most of the genes were enriched in the binding term. Combining functional annotation and real-time quantitative PCR, 15 genes were identified as the candidate target genes of BmBlimp-1, including several wing cuticular protein genes, chitin synthase A, and wing disc development genes, such as Wnt1, cubitus interruptus (ci) and engrailed (en). Moreover, the amino acid sequence of the zinc finger motif of Blimp-1 gene was highly conserved among the 15 insect species. We propose that BmBlimp-1 is an important regulatory factor in silkworm wing development.
Keywords: Bombyx mori, Blimp-1, wing development, transcription regulation
Introduction
Insects are the most diverse taxonomic group in the world, with more than one million described species. These account for more than half of all known living beings [1,2]. Meanwhile, insects are the only invertebrates that have the ability to fly. The flight ability is one of the most important reasons for their diversity. Their wings are not only the core structures for flight, but also play important roles in orientation, protection, communication, and courtship [3]. The molecular mechanism of insect wing development has long been an area of special interest.
The larvae of holometabolous insects need to undergo dramatic morphological changes in order to develop into adults. Moreover, their wings, as the flying organs of adults, develop from wing imaginal discs present inside the body of the larvae [4]. From Drosophila melanogaster, as a standard research organism in insect wing research, a good foundation of knowledge concerning wing imaginal disc development has been collected. Previous research found that two major hormones, 20-hydroxyecdysone (20E) and juvenile hormone, induce and regulate multiple biological processes, including growth, molting and reproduction. JH maintains the larval status, and 20E induces periodic ecdysis and metamorphosis [5]. Hormones can regulate the development of wings by initiating or shutting off the expression of relevant genes [6]. Previous research has shown that a series of genes are involved in wing early development of Drosophila. These genes include transcription factors such as engrailed (en), Serum Response Factor (SRF), cubitus interruptus (ci), Ultrabithorax (Ubx), distal-less (Dll), oculomotor-blind (omb), apterous (ap), scalloped (sd) and vestigial (vg). Moreover, wing development involves secreted proteins such as Wingless (Wg), Decapentaplegic (Dpp), Hedgehog (Hh), and Fringe (Fng), receptors such as Notch (N) and Epidermal growth factor receptor (Egfr), and the ligands Delta (Dl) and Serrate (Ser) [7-16]. However, many of the factors involved in the molecular mechanisms of wing development remain unknown.
Drosophila belongs to Order Diptera, in which the hind wings have degenerated into halteres. However, the Lepidoptera possess more typical insect wings. The morphology of Lepidoptera wings is an emergent representative of other holometabolous insect wings [17]. In Lepidoptera, represented by butterflies, the wings have more abundant color schemes than those of flies. Some wing development-related genes also control the wing color pattern in butterflies, for example, the HOX family gene Ubx [18]. Therefore, the development of Lepidopteran wings is an area of current focus.
The silkworm Bombyx mori is an economically important insect that has been domesticated for 5000 years. At present, the silkworm is used as a model species of Lepidoptera for basic research [19,20]. In the silkworm, several novel wing development-related genes have been discovered. Twelve wing disc cuticle proteins (WCPs) have been identified (BmWCP1a, BmWCP1b, BmWCP2, BmWCP3, BmWCP4, BmWCP5, BmWCP6, BmWCP7, BmWCP8, BmWCP9, BmWCP10, and BmWCP11) [21]. Cuticular protein and chitin are the main components of the insect exoskeleton, which plays an important role in providing stability of the cuticle layer during the wing development process [22]. Moreover, dysfunction of adhesion between upper and lower layers of wings causes abnormal wing development. Laminins are a major component of the basal lamina, loss of function of which can produce the blistered wings phenotype in the silkworm [23].
In this study, we found that the BGIBMGA000319 gene encodes a Cys2-His2 (C2H2) type zinc finger protein orthologous to that of the B-lymphocyte-induced maturation protein-1 (Blimp-1) gene. The Blimp-1 gene has been identified as a master locus that regulates plasma cell development and immunoglobulin-secreting B-lymphocyte differentiation, and the gene is involved in the differentiation of the myeloid lineage [24-26]. Moreover, the expression of the Blimp-1 gene in primordial germ cells is essential for mouse embryonic development. Loss of function of the gene can lead to embryonic death in mice [27,28]. The Blimp-1 gene participates in determining cell fates in the embryo and plays important roles in multiple hematopoietic lineages. In this article, we report that Bombyx mori Blimp-1(BmBlimp-1) is specifically upregulated during metamorphosis stages, and that it is highly expressed in the pupal wings on day 3 of the pupal stage. RNA interference (RNAi) resulted in abnormal wings. We thus conclude that the gene participates in the development of the wings.
Results
Structural features and phylogenetic analysis of BmBlimp-1
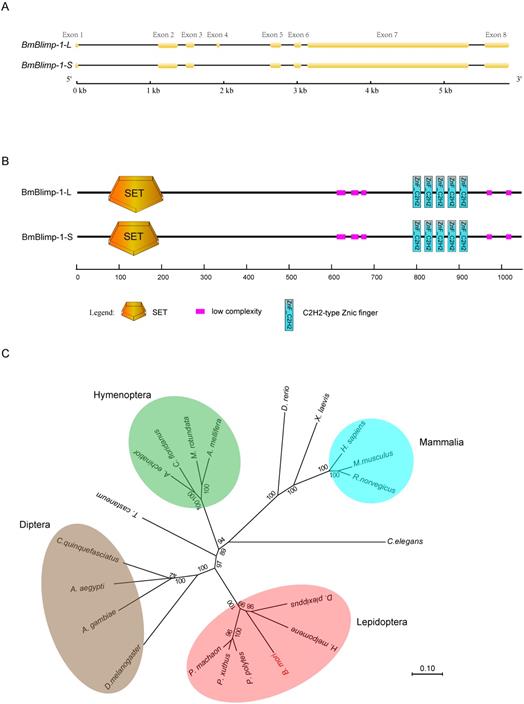
In the silkworm genome database (silkDB, http://www.silkdb.org) [29], a predicted gene BGIBMGA000319 that is specifically upregulated from the pupal to adult stages was identified by microarray analysis. We speculated that this gene might be involved in pupal development. To explore the function of the BGIBMGA000319 gene, we first cloned the ORF (open read frame) of this gene from the wild type strain Dazao. Sequence alignment found that the BGIBMGA000319 gene had alternative splicing; the long transcript was 3213 bp in length and contained eight exons. The short transcript was 3171 bp, which lacked the fourth exon (Fig. 1A). Translation of amino acid (AA) sequences based on the nucleotide sequence indicated that the two transcripts encoded 1070 and 1056 amino acid residues, respectively. Conservative domain analysis of these two predicted proteins was performed using SMART (Simple Modular Architecture Research Tool) software [30]. The two proteins shared the same domain that contained one SET (Su(var)3-9, Enhancer-of-zeste and Trithorax) domain and five C2H2-type zinc fingers. The only difference was that the short transcript contained 14 fewer amino acid residues of the SET domain than the long transcript (Fig. 1B). The SET domain can interact with dual-specificity phosphatases (dsPTPases) and contribute to epigenetic effects. The SET domain-containing proteins are involved in histone methylation, which plays an important role in organism growth and development and regulation of gene expression [31]. In addition, the C2H2-type zinc finger is considered as a common DNA-binding domain, and genes carrying this domain perform major transcription regulation.
To analyze the homologous genes of BGIBMGA000319 using its amino acid sequence, we performed a BLAST against the GenBank non-redundant protein sequences (nr) database and found a PR domain zinc finger protein 1(PRDM1) of human that had a sequence homologous to that of the BGIBMGA000319 gene. The total BLAST score was 511, the E-value was 2e-72, and the percent identity was 66.48%. The PRDM1 protein was also called Blimp-1(B lymphocyte-induced maturation protein 1), and it can regulate the differentiation of mature B cells into plasma cells and can inhibit the expression of genes related to the proliferation of B cells in humans [32]. The orthologous gene of Blimp-1 was found in other Lepidopteran species, such as Papilio machaon (Pm), Papilio xuthus (Px), Danaus plexippus (Dp), Heliconius melpomene (Hm), and Papilio polytes (Pp) (Table S1). A phylogenetic tree was constructed based on the nucleotide sequences for Blimp-1 from 21 species (Fig. 1C). The result showed that the Blimp-1 gene has only one copy in these species' genomes, and that Lepidoptera, Diptera and Hymenoptera were clustered into a separate group.
Structure and phylogenetic trees of the BmBlimp-1 gene. (A) The BmBlimp-1 gene generates two transcripts, a long transcript named BmBlimp-1-L, and a shorter transcript named BmBlimp-1-S. The yellow squares indicate exons, and the black lines indicate introns. (B) Conservative domain analysis revealed that the two transcripts contained the same number domains; the SET domain of the BmBlimp-1-S was 14 amino acids less than that of the BmBlimp-1-L. (C) A neighbor-joining phylogenetic tree of the complete nucleotide sequence of BmBlimp-1 genes among 21 species was constructed using the MUSCLE program and was visualized using the MEGA7 software.
Expression pattern analysis of BmBlimp-1
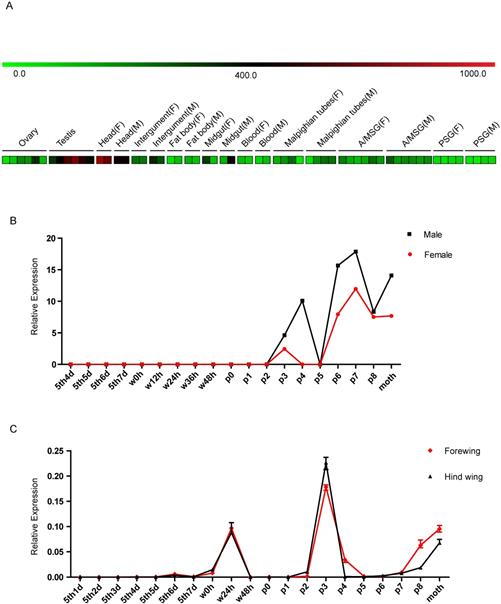
Based on sequence and homology analyses of the BGIBMGA000319 gene, we named it Bombyx mori Blimp-1 (BmBlimp-1). The expression pattern of BmBlimp-1 was retrieved from the microarray data of the silkworm [33] (Table S2). In multiple tissue chip data of silkworm larvae on day 3 of the fifth instar, the BmBlimp-1 gene is expressed in low levels in the testis and head, and no expression was detected in eight other tissues (Fig. 2A). In the metamorphosis stage microarray data for the whole body during the later larval to adult stages, the BmBlimp-1 gene was notably upregulated in pupae and adult stages; expression began on day 2 of the pupa, and the expression peak appeared on the day 7 of the pupa (Fig. 2B). Interestingly, we found an EST (expressed sequence tag) of the BmBlimp-1 gene, accession number is FS926355, in the EST database of the silkworm in Kaikobase [34]; the gene was cloned from the wings of pupal stage moths. This indicates that the BmBlimp-1 gene was expressed in the wings during the pupal stage. Therefore, expression analysis of the BmBlimp-1 gene was performed by real-time PCR from the larva to the adult stages in wing discs of the silkworm. The results showed that the expression pattern of forewing and hindwing was consistent, and this gene displayed peak expression on day 3 of the pupa and upregulation at wandering 24 hours, P7, P8 and day 1 of the adult (Fig. 2C). This suggests that the BmBlimp-1 gene may be related to the development of pupal wings in the silkworm.
Expression profiling of the BmBlimp-1 gene in multiple tissues and developmental stages of the silkworm. (A) Expression data were downloaded from the BmMDB database. The heat map was constructed using the Mev program. The multiple larval tissues were examined from the third day of the fifth instar, and each tissue had at least two duplications. A value ≥ 400 indicates that the gene was considered as expressed in the tissue. A/MSG: anterior/median silk gland; PSG: posterior silk gland; F: female; M: male. (B) The BmBlimp-1 gene microarray data during development stages from day 4 of larvae to adult. The developmental time points include day 4 of the fifth instar (5th4d), 5th5d, 5th6d,5th7d, the beginning of the wandering stage for spinning (W0), 12 hours after wandering (W12h), W24h, W36h, W48h (spinning finished), beginning of pupation (P0), one day after pupation (P1), P2, P3, P4, P5, P6, P7, P8 and day 1 of the adult moth. The expression level of the BmBlimp-1 gene was calculated by comparing gene expression with that in the 5th3d control. (C) The expression pattern of the BmBlimp-1 gene in wing discs from day 1 of the fifth instar larvae to adult.
Knockdown of BmBlimp-1
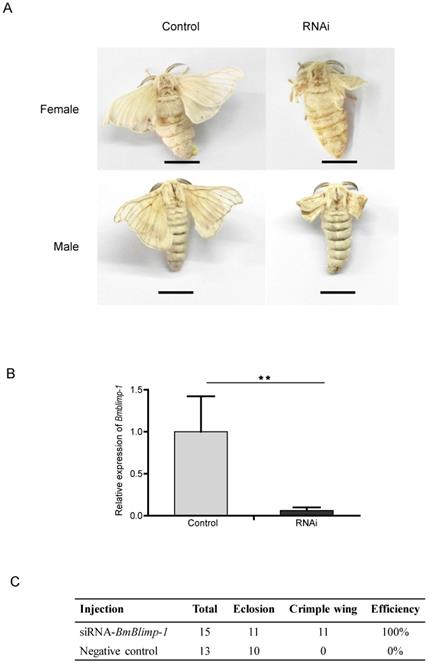
Remarkably, the BmBlimp-1 gene displayed peak expression in wing discs on day 3 of the pupae, and then was quickly downregulated to almost no expression. This suggests that the predicted transcript factor may perform important regulatory roles in pupae for 3 days. To study the function of the BmBlimp-1 gene, we synthesized siRNA to knock down the expression level of this gene in the pupae. The individuals of wild type strain Dazao, who pupate for about 36 hours, were selected. Using glass needles, we injected 5 μg siRNA of BmBlimp-1 into silkworm blood, and the controls were injected with the same quantity of scrambled negative control siRNA. The wings of the experimental group individuals were crumpled and unable to expand, while the control group had normal wings (Fig. 3A). The qRT-PCR showed that the expression level of the Bmblimp-1 gene was significantly downregulated in the experimental group (Fig. 3B). This indicates that the normal expression of the BmBlimp-1 gene can maintain the normal development of silkworm wings.
RNAi of BmBlimp-1 gene in Silkworm. (A) Phenotypes of silkworm adults after knockdown of the BmBlimp-1 gene. The bar indicates 1 centimeter (cm). (B) qRT-PCR of the BmBlimp-1 gene between control and experimental groups. Statistical analyses were performed using Student's t-test (n = 3). The asterisk indicates statistical significance. (C) Statistics of gene knockdown.
Downstream target genes analysis of BmBlimp-1
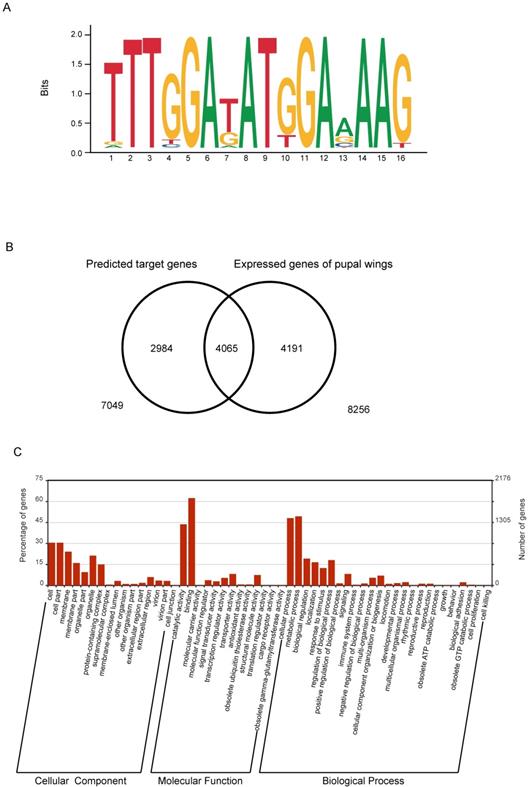
The RNA interference experiment showed that knockdown of the expression level of the BmBlimp-1 gene could cause wing dysplasia in the silkworm. However, BmBlimp-1 is a C2H2-type zinc finger gene that may function by regulating the transcription of other functional genes. Research has shown that the DNA-binding sequence of the C2H2-ZF motif depends on the amino acid residue sequence of its α-helical component. Considering the first amino acid residue of the α-helical region in the C2H2-ZF domain as position 1, the four positions -1, 2, 3 and 6 are the key amino acids for recognition and binding of DNA via interaction with the hydrogen donors and acceptors exposed in the major groove. In addition, in a group of tandem zinc fingers, the recognition sequences of individual zinc fingers may be modified due to adjacent ZFs. [35,36]. Based on empirical calculations of pairwise amino acid-nucleotide interaction energies, the web server http://zf.princeton.edu provides a method for predicting position weight matrices (PWMs) representing DNA-binding specificities for C2H2-ZF proteins [37]. Therefore, we predicted the DNA-binding sequence of the BmBlimp-1 gene on this web site. The predicted DNA-binding sequence was represented as a sequence logo (Fig. 4A). Using FIMO (Find Individual Motif Occurrences) software of the MEME suite [38], we searched the silkworm genome using the predicted DNA-binding sequence, screening the range of one kilobase upstream and downstream of the predicted genes. We identified 7,049 genes that had at least one putative binding site within 1 kb up/downstream and considered each as a possible downstream target gene of BmBlimp-1. To investigate the possible downstream genes of BmBlimp-1 in the pupal wing disc, we compared a reported transcript data of silkworm pupal wings [39]. In all, 4065 overlapped genes were found (Fig. 4B), and these genes were used as possible target genes regulated by the BmBlimp-1 gene during pupal development. The 4065 genes were annotated with Gene Ontology (GO), and 2902 had a GO term. The GO enrichment map was constructed (Fig. 4C); the genes were enriched in binding, metabolic process, cellular process and catalytic activity terms. Furthermore, the 4065 genes were subjected to a BLAST search of the non-redundant sequence database (nr) to functional annotation (Table S3). Multiple reported genes related to wing development were found, including wing cuticular proteins, Wingless, and engrailed, and those genes were considered as candidate target genes of BmBlimp-1 for wing development (Table 1).
Predicted target genes of BmBlimp-1. (A) DNA-binding sequences of BmBlimp-1. (B) Venn diagram of putative target genes and pupal wing transcriptome data; there are 4055 overlapping genes. (C) GO enrichment analysis of overlapping genes generated by the WEGO program.
Candidate target genes of BmBlimp-1 related to wing development
| Gene | SilkDB ID | Sequence location |
|---|---|---|
| WCP1b | BGIBMGA000429 | nscaf1681: 3265316-3270930 (+) |
| WCP2 | BGIBMGA000265 | nscaf1681: 3249518-3250498 (-) |
| WCP3 | BGIBMGA000269 | nscaf1681:3231459-3232623 (-) |
| WCP9 | BGIBMGA000325 | nscaf1681: 697463-702380 (-) |
| WCP10 | BGIBMGA001444 | nscaf2136: 2851152-2860721(-) |
| WCP11 | BGIBMGA000324 | nscaf1681: 710443-712032 (-) |
| chitin synthase A | BGIBMGA002963 | nscaf2589: 6004788-6016095 (-) |
| decapentaplegic (dpp) | BGIBMGA010384 | nscaf2993: 6692859-6696343 (-) |
| engrailed (en) | BGIBMGA009644 | nscaf2964: 4195216-4196156 (-) |
| wingless (wg) | BGIBMGA006146 | nscaf2847: 4290086-4300264 (+) |
| apterous (ap) | BGIBMGA002127 | nscaf2210: 3499610-3502050 (+) |
| Serrate (Ser) | BGIBMGA000783 | nscaf1705: 3574-11539 (-) |
| asense (ase) | BGIBMGA001002 | nscaf1898: 5940257-5941471 (-) |
| cubitus interruptus (ci) | BGIBMGA004545 | nscaf2800: 2013357-2027060 (-) |
| Serum Response Factor (SRF) | BGIBMGA001384 | nscaf2110: 8727-9565 (-) |
| BGIBMGA001382 | nscaf2108: 36519-37205 (+) |
Analysis of the expression levels of wing disc development-related genes
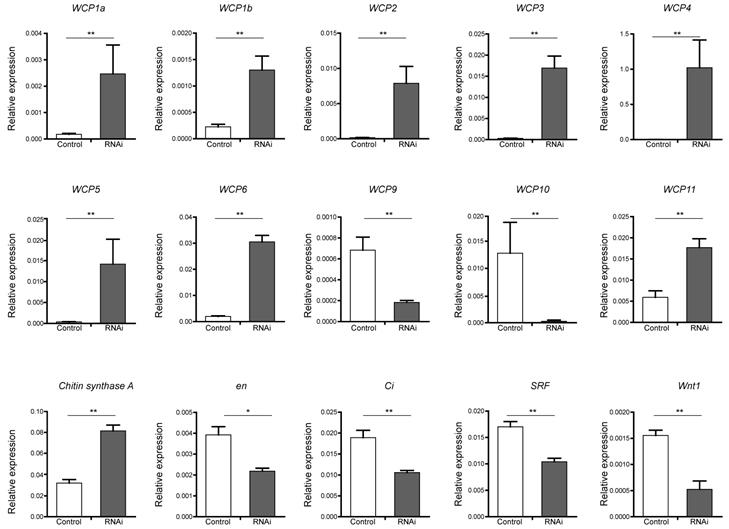
To verify candidate target genes of the BmBlimp-1 protein, we employed real-time fluorescence quantitative PCR to measure the expression levels in RNAi individuals and a control group. Chitin synthase A gene and 13 WCP genes were investigated. The results showed that eight wing cuticular protein genes (WCP1a, WCP1b, WCP2, WCP3, WCP4, WCP5, WCP6 and WCP11) and the chitin synthase A gene were significantly upregulated in the RNAi group (Fig. 5). Meanwhile, WCP9 and WCP10 were significantly downregulated. The expression levels of WCP7 and WCP8 were very low (not shown in the figure). In addition, eight wing disc development-related genes, decapentaplegic (dpp), engrailed (en), apterous (ap), Serrate (Ser), asense (ase), cubitus interruptus (ci), Serum Response Factor (SRF) and Wnt1, were investigated. Among the factors related to the development of wing discs, we found that Wnt1, en, ci and SRF were significantly downregulated (Fig. 5), and that ap, ase and Dpp were not significantly different (data not shown).
Expression level of candidate target genes of BmBlimp-1 by qRT-PCR. The genes, WCP1a, WCP1b, WCP2, WCP3, WCP4, WPC5, WCP6, WCP9, WCP10, WCP11, chitin synthase A, cubitus interruptus (ci), engrailed (en), Serum Response Factor (SRF) and Wnt1 were significantly differentially expressed in the RNAi treatment group compared with the control group. All data are mean ± S.D (n = 3). Statistical analyses were performed using Student's t-test (n = 3). The asterisk indicates statistical significance.
Conservation Analysis of Zinc Finger Motifs of BmBlimp-1. (A) Amino acids of zinc finger motif analysis among 21 species. (B) Wnt1 gene structure and DNA-binding site position of Blimp-1 among seven species. (C) DNA sequences of the binding sites of Blimp-1 among the seven species, and the sequences logo generated by these DNA-binding sequences.
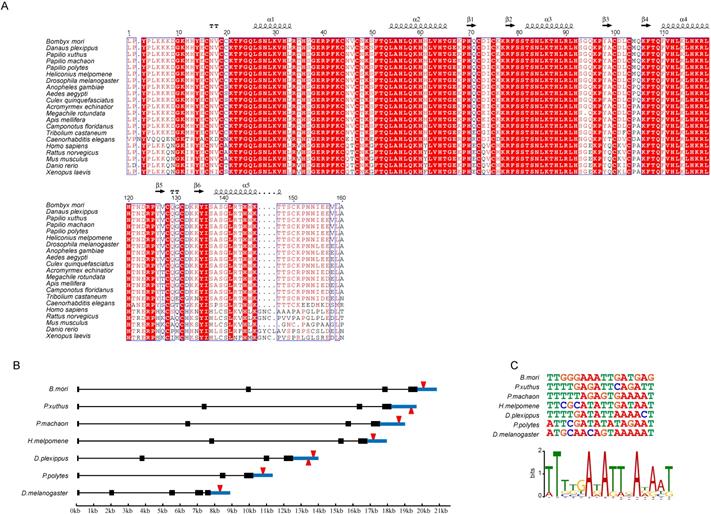
Zinc Fingers Conservation and Binding sites analysis of Blimp-1
Genes with important functions tend to be conserved among species. To investigate the degree of conservation of DNA-recognition sequences of the Blimp-1 gene among species, we compared the AA sequences of zinc finger domains of the Blimp-1 gene among 21 species. The AA sequences of the ZF domains of the Blimp-1 gene were aligned (Fig. 6A), illustrating that the sequences of this protein were highly conservative among 15 insects and Caenorhabditis elegans. The AA sequences of α-helical regions were completely consistent among 15 insect species, and in C. elegans only one alanine was changed to proline in the fifth α-helix. In addition, the entire region of the five zinc finger domains shared the same AA residues among the silkworm, Danaus plexippus (Dp), Papilio xuthus (Px), Papilio machaon (Pm) and Papilio polytes (Pp), and there is only one amino acid difference between the silkworm and Heliconius melpomene (Hm), an asparagine changed to serine. Furthermore, among four vertebrates (Homo sapenis, Mus musculus, Rattus norvegicus and Danio rerio), the AA sequences of the α-helical regions are highly conserved. The tandem zinc fingers of Blimp-1 genes are strongly conserved among species, suggesting the persistence of the highly conserved recognition sequences, and the proteins may regulate some of the same target genes. Therefore, we selected the Wnt1 gene from the candidates mentioned above; this gene encodes an important secretory protein. The structure of the Wnt1 gene was analyzed among seven species (Bm, Px, Pm, Hm, Dp, Pp and Dm) (Fig. 6B). The Wnt1 genes in these species have similar structures, and this gene has the longest intron in the silkworm among the species examined. Meanwhile, the DNA-binding sequences of Blimp-1 were predicted for the seven species. A binding sites analysis of Blimp-1 to the respective Wnt1 genes was performed. We compared the binding sites of the Blimp-1 proteins to the Wnt1 genes in each species. The results showed that a conserved DNA-binding site of Blimp-1 was present within approximately 1 kb downstream of the Wnt1 gene in the seven species (Fig. 6C). This suggests that the Blimp-1 protein may bind to and regulate the expression of the Wnt1 gene in a similar manner among the seven species.
Discussion
In this study, we found a C2H2 zinc finger protein gene BGIBMGA000319 that is highly expressed during the silkworm pupal stage. As this gene has sequence homology with the B-lymphocyte-induced maturation protein-1 (Blimp-1) gene in vertebrates, we named it BmBlimp-1. RNAi experiments found that knockdown of the expression level of this gene at the peak expression stage causes abnormal wing development in the silkworm. Using DNA-binding motif sequence prediction, silkworm whole-genome analysis, and comparative transcriptome data of wing discs of silkworm pupae, we screened for genes related to wing development and detected genes with significant changes in expression levels in the knockdown group. The candidate target genes of BmBlimp-1 were identified, and these genes included Wnt1, engrailed (en), asense (ase), cubitus interruptus (ci), Serum Response Factor (SRF), wing cuticular protein genes (WCP1a, WCP1b, WCP2, WCP3, WCP4, WCP5, WCP6, WCP9, WCP10 and WCP11) and chitin synthase A. Furthermore, we found that the zinc finger motif of the Blimp-1 gene is highly conserved among multiple insect species, which suggests that this gene may perform homologous and important functions among species.
The Blimp-1 gene participates in determining cell fate and plays important roles in multiple hematopoietic lineages in vertebrates. In Drosophila, the Blimp-1 gene is induced directly by 20-hydroxyecdysone (20E), and its gene product exists during the high-ecdysteroid phase. This gene can bind to and regulate the transcription of ftz-f1, a nuclear receptor transcription factor that plays an important role in molting and metamorphosis. Knockdown of Blimp-1 can lead to pre-pupal lethality, while prolonging its expression results in delay of pupation [40]. In embryogenesis, Blimp-1 is critically required for the development and maturation of the tracheal system [41]. In the silkworm, BmBlimp-1 is also upregulated during the pupal stage. This suggests that a similar mechanism is shared between the silkworm and the fruit fly, in which BmBlimp-1 may be directly induced by 20E and may participate in pupal development. According to the ecdysterone titer level of Bombyx mori [42], this gene is upregulated in the late pupae and moth stages, while the ecdysone titer is at a low level during this period (Fig. S1). Therefore, the expression of Blimp-1 in the silkworm may be activated by other factors.
In the GO analysis of possible direct target genes of the BmBlimp-1 protein, we found that genes were most enriched in the binding term. This term had 63 genes; these genes were mainly predicted to be transcription factors. This suggests that the BmBlimp-1 gene acts as an upstream factor in a regulatory network for pupal wing development. In the qRT-PCR verification of candidate target genes, although no binding sites of the BmBlimp-1 protein within 1 kb upstream and downstream of WCP1a, WCP4, WCP5 and WCP6 genes were found, their expression levels changed significantly. There are several possibilities. First, those genes, except for WCP6, are serially arranged on chromosome 22, and thus, they may share the same cis-regulatory element. Second, we only retrieved the 1 kb upstream and downstream regulatory sites and neglected the longer positions and introns. Third, it is possible that the protein may indirectly regulate the expression levels of these genes by regulating other TFs. In addition, several wing cuticular protein genes were mainly expressed in the wing discs of early pupae, and the expression level declined by day 3 of the pupa [43]. Knocking down the expression of the BmBlimp-1 gene caused several WCP genes to be remarkably upregulated, and the knockdown also resulted in significant downregulation of the WCP9 and WCP10 genes. This implies that the BmBlimp-1 gene may act to activate and inhibit expression as a dual-function regulator. Cuticular proteins and chitin are the major components of the epidermis of the exoskeleton and wings. Many cuticular proteins have a chitin-binding domain, and diverse groups of these complexes are orderly cross-linked to form a stable structure. When one or more of the cuticular proteins is disordered, this can lead to structural changes or tissue abnormalities [44,45]. The changes in the expression of wing cuticular protein genes and chitin synthases A in the RNAi group may result in an abnormal wing. Meanwhile, the wing disc pattern genes such as Wnt1, en, ci and SRF were significantly down-regulated in the RNAi group. These genes play important roles in wing disc development in the embryonic stages, as shown in previous wing studies with flies. However, these genes are also expressed in the pupal wings of the silkworm, and as candidate target genes of BmBlimp-1, they were markedly down-regulated in RNAi individuals. We suggest that these genes may be directly regulated by BmBlimp-1, and that they participate in pupal wing development.
In multiple species aligning, we found that the amino acid sequence of Blimp-1 was highly conserved; in 15 insect species examined, the amino acid residues of the zinc finger motif were very similar. This indicated the existence of similar binding sites of Blimp-1 in those species. The discovery of conserved binding sites downstream of the Wnt1 gene in seven insects also fits this hypothesis.
This study has identified the function of the BmBlimp-1 gene in pupal wing development of the silkworm. Combined with the analysis of direct target genes, we believe that the BmBlimp-1 gene may regulate the expression of several WCP genes along with the wing disc development-related genes Wnt1, en, ci and SRF to participate in pupal wing development in the silkworm. The results serve as motivation for studying the conservative C2H2 zinc finger protein TFs, which are involved in the wing development of the silkworm and other species of Lepidoptera. The candidate target genes analyzed by our screening can provide references for further functional research.
Materials and Methods
Silkworm strain and tissue collection
The wild-type strain Dazao was obtained from the Silkworm Gene Bank of Southwest University, Chongqing, China. Silkworms were reared on mulberry leaves at 25°C in 75% relative humidity under stable conditions (12-h light: 12-h dark). The wing discs were removed using 0.7% normal saline solution and then stored at -80°C until RNA extraction was performed.
Gene cloning and RT-PCR
Total RNA was isolated from the silkworm Dazao strain and wing discs using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and E.Z.N.A.® MicroElute Total RNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer's protocol. The primers that were used for qRT-PCR are listed in Supplementary Table S4. The qRT-PCR was performed using a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with SYBR Green qRT-PCR Mix (Bio-Rad). The cycling parameters were as follows: 95°C for 3 min followed by 40 cycles at 95°C for 10 s and annealing for 30 s. The relative expression levels were analyzed using the classical R= 2-ΔΔCt method. The gene for ribosomal protein L3 (rpL3) of the silkworm was used as an internal control to normalize for equal sample loading.
Phylogenetic analysis
To determine whether BGIBMGA000319 (BmBlimp-1) orthologues existed in species other than B. mori, we performed a phylogenetic analysis using genes included in the Blastp hits with E-values ≤10-5 in the NCBI nr database (http://www.ncbi.nlm.nih.gov/BLAST/), with Drosophila melanogaster (http://www.flybase.org/) [46], the Monarch genome (http://monarchbase.umassmed.edu/) [47], the Heliconius melpomene genome (http://butterflygenome.org/) [48], and Tribolium castanenum (http://beetlebase.org/) [49]. Alignment was generated from a multiple sequence alignment of DNA sequences by MUSCLE [50] and was used to construct nucleotide alignments. The phylogenetic tree was constructed using the neighbor joining method using the MEGA7 program [51]. The confidence levels for various phylogenetic lineages were assessed by bootstrap analysis (1,000 replicates).
siRNA for gene knockdown
The siRNAs for BmBlimp-1 were designed to target 5'-GCCAAAUACACCGACUGAATT-3' for gene knockdown, and the scramble negative control 5'-AGAACCAGAUAACCGAUCCTT-3' (Genepharma, China) was used as a negative control. Five microliters of siRNA 1μg/μl was injected from the chest spiracle into the hemolymph at 36 hours after pupation. After injection, the individuals were maintained at 25°C in 75% humidity and 12 hours light and 12 hours dark until eclosion. Quantitative RT-PCR was employed to measure the mRNA levels of target genes of samples that were obtained at day 4 of the pupa.
DNA-Binding Site Prediction and Target Gene Screening
To obtain the DNA-binding site position weight matrices for the BmBlimp-1 protein, an online de-novo prediction website (http://zf.princeton.edu) was used, and the linear expanded predicted method was chosen [37]. The FIMO program of the MEME software suite was used to search for binding sites in the silkworm genome [52]. Screening the range of 1 kb upstream and downstream of genes was done using an R-script. Comparing the screened genes with known pupal wing transcriptome data yielded possible target gene sets. The GO analysis results of those possible target genes were plotted using the WEGO (Web Gene Ontology Annotation Plot) program (http://wego.genomics.org.cn) [53].
Conservation Analysis of the Zinc Finger Structure of Multi-Species Blimp-1
Alignment of the amino acid sequences of the zinc finger structures of 21 species using the MUSCLE program followed by the Espript 3.0 (Easy Sequencing in PostScript) program [54] was used to render sequence similarities and secondary structure information from the aligned sequences. The sequences of Wnt1 genes of seven insect species were downloaded from NCBI. The prediction of Blimp-1 binding sites of Wnt1 genes in different species followed the method described above. The sequence logo of binding sites downstream of Wnt1 in seven insects was generated by the MEME program.
Supplementary Material
Supplementary figures and tables S1, S2, S4.
Table S3.
Acknowledgements
The research was supported by the National Natural Science Foundation of China (Nos. 31830094, 31472153), Fundamental Research Funds for the Central Universities (No. XDJK2019C046), the Hi-Tech Research and Development 863 Program of China Grant (No. 2013AA102507) and Funds of China Agriculture Research System (No. CARS-18-ZJ0102).
Authors' contributions
FY Dai and H Liu conceived and designed the experiments. SY Wu, CL Li, X Ding, ZL Zhang and CY Fang performed the study. SY Wu and XL Tong analyzed the data. D Tan and H Hu bred the silkworm strain. SY Wu wrote the paper. XL Tong, H Liu and FY Dai edited and revised the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gaston KJ. The Magnitude Of Global Insect Species Richness. Conserv Biol. 1991;5:283-296 doi:DOI 10.1111/j.1523-1739.1991.tb00140.x
2. Stork NE. How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth?. Annu Rev Entomol. 2018:63
3. Shyy W, Kang CK, Chirarattananon P. et al. Aerodynamics, sensing and control of insect-scale flapping-wing flight. Proc Math Phys Eng Sci. 2016;472:20150712 doi:10.1098/rspa.2015.0712
4. Ote M, Mita K, Kawasaki H. et al. Microarray analysis of gene expression profiles in wing discs of Bombyx mori during pupal ecdysis. Insect Biochem Mol Biol. 2004;34:775-784 doi:10.1016/j.ibmb.2004.04.002
5. Dubrovsky EB. Hormonal cross talk in insect development. Trends Endocrinol Metab. 2005;16:6-11 doi:10.1016/j.tem.2004.11.003
6. Sin FY. Relationship between 'early' and 'late' protein induced by 20-hydroxyecdysone in imaginal wing discs of Drosophila melanogaster. Mol Biol Rep. 1987;12:7-11
7. Bray S. Drosophila development: Scalloped and Vestigial take wing. Curr Biol. 1999;9:R245-247
8. Doherty D, Feger G, YoungerShepherd S. et al. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand in Drosophila wing formation. Genes Dev. 1996;10:421-434 doi:Doi 10.1101/Gad.10.4.421
9. Dworkin I& Gibson G. Epidermal growth factor receptor and transforming growth factor-beta signaling contributes to variation for wing shape in Drosophila melanogaster. Genetics. 2006;173:1417-1431 doi:10.1534/genetics.105.053868
10. Hepker J, Blackman RK& Holmgren R. Cubitus interruptus is necessary but not sufficient for direct activation of a wing-specific decapentaplegic enhancer. Development. 1999;126:3669-3677
11. Irvine KD& Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595
12. Montagne J, Groppe J, Guillemin K. et al. The Drosophila Serum Response Factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development. 1996;122:2589-2597
13. Pflugfelder GO, Eichinger F& Shen J. T-Box Genes in Drosophila Limb Development. Curr Top Dev Biol. 2017;122:313-354 doi:10.1016/bs.ctdb.2016.08.003
14. Sharma RP& Chopra VL. Effect of the Wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev Biol. 1976;48:461-465
15. Weihe U, Milan M& Cohen SM. Regulation of Apterous activity in Drosophila wing development. Development. 2001;128:4615-4622
16. Zecca M, Basler K& Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121:2265-2278
17. Abbasi R& Marcus JM. A new A-P compartment boundary and organizer in holometabolous insect wings. Sci Rep. 2017;7:16337 doi:10.1038/s41598-017-16553-5
18. Tong X, Hrycaj S, Podlaha O. et al. Over-expression of Ultrabithorax alters embryonic body plan and wing patterns in the butterfly Bicyclus anynana. Dev Biol. 2014;394:357-366 doi:10.1016/j.ydbio.2014.08.020
19. Goldsmith MR, Shimada T& Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 2005;50:71-100 doi:10.1146/annurev.ento.50.071803.130456
20. Sun W, Yu H, Shen Y. et al. Phylogeny and evolutionary history of the silkworm. Sci China Life Sci. 2012;55:483-496 doi:10.1007/s11427-012-4334-7
21. Futahashi R, Okamoto S, Kawasaki H. et al. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1138-1146
22. Iconomidou VA, Willis JH& Hamodrakas SJ. Unique features of the structural model of 'hard' cuticle proteins: implications for chitin-protein interactions and cross-linking in cuticle. Insect Biochem Mol Biol. 2005;35:553-560 doi:10.1016/j.ibmb.2005.01.017
23. Tong X, He S, Chen J. et al. A novel laminin β gene BmLanB1-w regulates wing-specific cell adhesion in silkworm, Bombyx mori. Sci Rep. 2015;5:12562
24. Chang DH, Angelin-Duclos C& Calame K. BLIMP-1: trigger for differentiation of myeloid lineage. Nat Immunol. 2000;1:169-176
25. Shapiro-Shelef M, Lin KI, Mcheyzer-Williams LJ. et al. Blimp-1 Is Required for the Formation of Immunoglobulin Secreting Plasma Cells and Pre-Plasma Memory B Cells. Immunity. 2003;19:607-620
26. Turner CA, Mack DH& Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297-306
27. Chang DH, Cattoretti G& Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117:305-309
28. Yasuhide O, Bernhard P, Dónal OC. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207-213
29. Duan J, Li R, Cheng D. et al. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:D453-456 doi:10.1093/nar/gkp801
30. Ponting CP, Schultz J, Milpetz F. et al. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;27:229-232
31. Cui X, De Vivo I, Slany R. et al. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331-337 doi:10.1038/ng0498-331
32. Kim JR, Mathew SO& Mathew PA. Blimp-1/PRDM1 regulates the transcription of human CS1 (SLAMF7) gene in NK and B cells. Immunobiology. 2016;221:31-39 doi:10.1016/j.imbio.2015.08.005
33. Xia Q, Cheng D, Duan J. et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8:R162 doi:10.1186/gb-2007-8-8-r162
34. Shimomura M, Minami H, Suetsugu Y. et al. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 2009;10:486 doi:10.1186/1471-2164-10-486
35. Klug A. The Discovery of Zinc Fingers and Their Applications in Gene Regulation and Genome Manipulation. Annu Rev Biochem, Vol 79. 2010;79:213-231 doi:10.1146/annurev-biochem-010909-095056
36. Wolfe SA, Nekludova L& Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183-212 doi:10.1146/annurev.biophys.29.1.183
37. Persikov AV& Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97-108 doi:10.1093/nar/gkt890
38. Bailey TL, Johnson J, Grant CE. et al. The MEME Suite. Nucleic Acids Res. 2015;43:W39-49 doi:10.1093/nar/gkv416
39. Ou J, Deng HM, Zheng SC. et al. Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC Genomics. 2014;15:820 doi:10.1186/1471-2164-15-820
40. Yasuo A, Moustafa S, Yuji K. et al. Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol Cell Biol. 2007;27:8739
41. Ng T, Yu F& Roy S. A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Dev Genes Evol. 2006;216:243
42. Mizoguchi A, Ohashi Y, Hosoda K. et al. Developmental profile of the changes in the prothoracicotropic hormone titer in hemolymph of the silkworm Bombyx mori: correlation with ecdysteroid secretion. Insect Biochem Mol Biol. 2001;31:349-358
43. Takeda M, Mita K, Quan GX. et al. Mass isolation of cuticle protein cDNAs from wing discs of Bombyx mori and their characterizations. Insect Biochem Mol Biol. 2001;31:1019-1028
44. Andersen SO, Hojrup P& Roepstorff P. Insect cuticular proteins. Insect Biochem Mol Biol. 1995;25:153-176
45. Zhu KY, Merzendorfer H, Zhang W. et al. Biosynthesis, Turnover, and Functions of Chitin in Insects. Annu Rev Entomol. 2016;61:177-196 doi:10.1146/annurev-ento-010715-023933
46. Gramates LS, Marygold SJ, dos Santos G. et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45:D663-D671 doi:10.1093/nar/gkw1016
47. Zhan S& Reppert SM. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 2013;41:D758-763 doi:10.1093/nar/gks1057
48. Davey JW, Chouteau M, Barker SL. et al. Major Improvements to the Heliconius melpomene Genome Assembly Used to Confirm 10 Chromosome Fusion Events in 6 Million Years of Butterfly Evolution. G3 (Bethesda). 2016;6:695-708
49. Kim HS, Murphy T, Xia J. et al. BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010;38:D437-442 doi:10.1093/nar/gkp807
50. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-1797 doi:10.1093/nar/gkh340
51. Kumar S, Stecher G& Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870-1874 doi:10.1093/molbev/msw054
52. Bailey TL, Johnson J, Grant CE. et al. The MEME Suite. Nucleic Acids Res. 2015;43:W39-W49
53. Ye J, Zhang Y, Cui H. et al. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018:46
54. Xavier R& Patrice G. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320
Author contact
![]() Corresponding authors: Fang-Yin Dai, E-mail: fydaiedu.cn, Tel.: +86-023-6825-0793 or Huai Liu, E-mail: liuhuaiedu.cn, Tel.: +86-023-68250490.
Corresponding authors: Fang-Yin Dai, E-mail: fydaiedu.cn, Tel.: +86-023-6825-0793 or Huai Liu, E-mail: liuhuaiedu.cn, Tel.: +86-023-68250490.

 Global reach, higher impact
Global reach, higher impact