Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(13):2763-2773. doi:10.7150/ijbs.38859 This issue Cite
Research Paper
Protein arginine methyltransferase 1 is required for maintenance of normal adult hematopoiesis
1. Department of clinical laboratory, The Second Affiliated Hospital, Zhejiang University School of Medicine, No. 88 Jiefang Road, Hangzhou, 310009, Zhejiang, China.
2. Department of Hematological Malignancies Translational Science, Gehr Family Center for Leukemia Research, Hematologic Malignancies and Stem Cell Transplantation Institute, Beckman Research Institute, City of Hope Medical Center, Duarte, CA 91010.
3. Department of Science and Development, The Second Affiliated Hospital, Zhejiang University School of Medicine, No. 88 Jiefang Road, Hangzhou, 310009, Zhejiang, China.
4. Department of Hematology, The Second Affiliated Hospital, Zhejiang University School of Medicine, No. 88 Jiefang Road, Hangzhou, 310009, Zhejiang, China.
*These authors contributed equally to this work.
Received 2019-7-29; Accepted 2019-9-15; Published 2019-10-23
Abstract
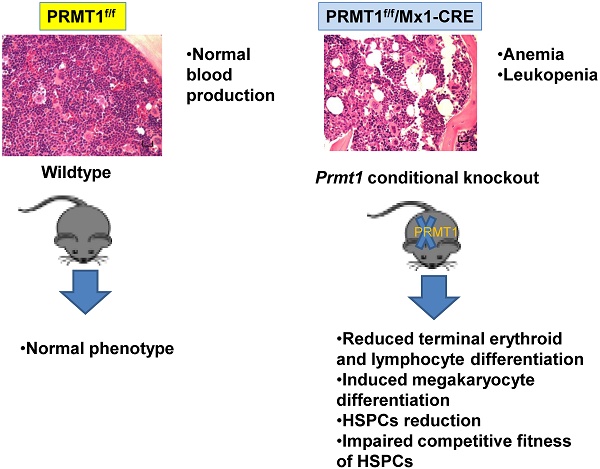
Protein arginine methyltransferase 1 (PRMT1) is the predominant asymmetric (type I) methyltransferase in mammalian cells. Mounting evidence suggested that PRMT1 is essential to embryonic development and tumor pathogenesis, but its role in normal adult hematopoiesis is less studied. We used a Prmt1 conditional knockout (KO) mouse model to identify the role of PRMT1 in normal adult hematopoiesis. The results indicated that deletion of PRMT1 results in anemia and leukopenia, reducing terminal erythroid and lymphocyte differentiation. Additionally, we found a significant decrease of megakaryocyte progenitors (MkPs) compared with similarly treated littermate control mice. The frequency of short-term hematopoietic stem cells (ST-HSCs) and granulocyte-macrophage progenitors (GMPs) populations were significantly lower in PRMT1f/f/Mx1-CRE bone marrow (BM) compared with littermate control mice. Importantly, in-vitro replating assays and BM transplantation results revealed that PRMT1 KO results in reduced hematopoietic stem and progenitor cells (HSPCs) self-renewal capacity. Thus, we conclude that PRMT1 is required for hematopoietic differentiation and the competitive fitness of HSPCs, and we believed that PRMT1 serves as a key epigenetic regulator of normal hematopoiesis that occurs throughout life.
Keywords: PTMT1, HSPCs, normal hematopoiesis
Introduction
Protein arginine methylation is one of the most common posttranslational modifications that affects the function of many proteins, including both histone and non-histone proteins. A family of enzymes, namely, protein arginine methyltransferases (PRMTs), catalyze the formation of mono- or dimethyl arginine on a variety of protein substrates [1], thereby regulating many cellular processes including RNA processing, gene transcription, DNA damage repair, signal transduction and chromosome translocation [2]. PRMT1 is the predominant asymmetric (type I) methyltransferase in mammalian cells, responsible for over 85% of arginine methylation activity. Mounting evidence in cancers, including our own results in leukemia, indicates that aberrantly overexpressed PRMT1 could serve as a potential therapeutic target [3-10]. Homozygous PRMT1 deletion is embryonically lethal [11, 12], but its role in normal adult hematopoiesis is less studied. Others have reported that PRMT1 function is involved in normal B cell differentiation [13]. Some reported that PRMT1, functioning through p38a, serves as a novel molecular switch in lineage-specific differentiation toward megakaryocytes or erythrocytes [14, 15]. While the detailed function of PRMT1 in adult hematopoietic stem and progenitor cells (HSPCs) self-renewal and differentiation is still largely unknown. In this study, we identified the role of PRMT1 in normal adult hematopoiesis using a Prmt1 conditional knockout (KO) mouse model (PRMT1f/f/Mx1-Cre). Overall, PRMT1 deletion in adult mice leads to anemia and leukopenia, thereby disrupting normal hematopoiesis. Although PRMT1 KO barely affected the primary mouse LSK frequency, BM transplantation studies revealed that PRMT1 KO results in reduced competitive fitness of HPSC.
Materials and Methods
Mice
Prmt1 conditional KO mice (PRMT1f/f/Mx1-Cre) were generated by crossing PRMT1f/f mice [16] with Mx1-Cre mice. To induce Prmt1 deletion, 6- to 8-week-old PRMT1f/f /Mx1-Cre mice were injected intraperitoneally with 14 mg/kg per dose of polyinosinic-polycytidylic acid (pIpC; InvivoGen) every other day for 7 doses. Similarly treated littermates lacking Mx1-cre alleles were used as control. All mice were maintained and all procedures were performed in accordance with federal and state government guidelines and established institutional guidelines and protocols approved by the Institutional Animal Care and Use Committee at City of Hope.
Flow Cytometry
Femurs and tibias were crushed with a mortar and pestle to collect BM cells. Spleens were crushed with the end of a plunger. Cells were resuspended in 5 mL phosphate buffered saline (PBS) plus 0.5% bovine serum albumin (BSA), then filtered through a 70- μm filter (BD Biosciences) following by red blood cell lysis. Antibodies were used for flow-cytometry analyses as follows: CD117 (ckit, clone ACK2, eBioscience), Ly-6A/E (Sca-1, clone D7,BioLegend), CD150 (SLAM, clone mShad150, BioLegend,), CD48 (clone HM48.1, BioLegend), CD34 (clone MEC14.7, eBioscience), CD16/CD32 (clone 93, eBioscience), CD127 (IL7R, clone A7R34,eBioscience), CD135 (Flk-2, Flt-3, Ly-72, clone A2F10, eBioscience),CD45.1 (clone A20, BioLegend), CD45.2 (clone 104,BioLegend), CD11b (clone M1/70, eBioscience), Ter119 (cloneTER-119, BioLegend), B220(clone RA3-6B2, eBioscience ), CD3 (clone 17A2, eBioscience) and Ly-6G/Ly-6C (Gr1, cloneRB6-8C5, BioLegend). The lineage antibody cocktail included the following biotin-conjugated anti-mouse antibodies: CD19 (clone eBio1D3),NK-1.1 (clone PK136), B220 (clone RA3-6B2), IgM (clone II/ 41), CD3 (clone 17A2), CD4 (clone GK1.5), CD8 (clone s3-6.7), Gr1 (clone RB6-8C5), CD127 (clone A7R34) at 1 µg/mL,CD11b (clone M1/70),CD11c(clone N418) at 2 µg/mL, and Ter119 (clone Ter119, from BioLegend) at 3 µg/mL. Secondary antibody for the analysis was streptavidin-FITC (BioLegend). Flow cytometry was performed using a 5-laser, 15-detector Foressa X20 (BD Biosciences). Acquired data were analyzed by Flowjo software (TriSTAR).
Phenotypic populations were defined as LSK(Lin-/ckit+/Sca1+), long-term HSCs (LT-HSCs) (Lin-/ckithi/Sca1+/Flt3-/CD150+/CD48-), short-term HSCs (ST-HSCs) (Lin-ckithi/Sca1+/Flt3-/CD150-/CD48-), multipotent progenitors (MPPs) (Lin-/ckithi/Sca1+/CD150+/-/CD48+), lymphoid-primed multipotent progenitors (LMPPs)(Lin-/ckithi/Sca1+/Flt3+/CD150+/-/CD48+), common lymphoid progenitors (CLPs)(Lin-/IL7R+/ckitlo/Sca1lo), myeloid progenitors (MPs) (Lin-/ckit+/Sca1-), common myeloid progenytors (CMPs) (Lin-/ckit+/Sca1-/CD34+/FcgRlo), granulocyte-macrophage progenitors (GMPs) (Lin-/ckit+/Sca1-/CD34+/FcgRhi), and megakaryocyte-erythroid progenitors (MEPs) (Lin-/ckit+/Sca1-/CD34-/FcgRlo).
Colony-forming cell assays
Myeloid/erythroid colony-forming units (CFUs; CFU-GEMMs, CFU-GMs, and BFU-Es) were enumerated using MethoCult containing stem-cell factor (SCF), IL-3, IL-6, and erythropoietin (EPO) (M3434; Stem Cell Technologies). Briefly, 1 105 cells per mL per dish were seeded in MethoCult and were counted at day 7 according to the manufacturer's protocol. For replating assays, cells from each plate were harvested and replated at 1
105 cells per mL per dish were seeded in MethoCult and were counted at day 7 according to the manufacturer's protocol. For replating assays, cells from each plate were harvested and replated at 1  105 cells per mL per dish after 7 days CFC culture.
105 cells per mL per dish after 7 days CFC culture.
Transplantation Experiments
For competitive repopulation experiments, primary donor BM cells (1  106) isolated from PRMT1f/f/Mx1-Cre or PRMT1f/f mice (which are CD45.2), mixed with equal numbers of CD45.1 competitor cells and injected via tail vein into lethally irradiated (11 Gy; 2 split doses) 6- to 8-week-old C57BL/6 congenic CD45.1+ recipient mice. After robust engraftment of donor cells around 4 weeks post-bone marrow transplant (BMT), mice were injected intraperitoneally 7 doses (14 mg/kg per dose) of pIpC. Donor chimerism in peripheral blood (PB) was evaluated over time till 12 weeks for BM engraftment analysis.
106) isolated from PRMT1f/f/Mx1-Cre or PRMT1f/f mice (which are CD45.2), mixed with equal numbers of CD45.1 competitor cells and injected via tail vein into lethally irradiated (11 Gy; 2 split doses) 6- to 8-week-old C57BL/6 congenic CD45.1+ recipient mice. After robust engraftment of donor cells around 4 weeks post-bone marrow transplant (BMT), mice were injected intraperitoneally 7 doses (14 mg/kg per dose) of pIpC. Donor chimerism in peripheral blood (PB) was evaluated over time till 12 weeks for BM engraftment analysis.
For noncompetitive BM primary transplantation, 2 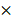 106 primary donor BM cells isolated from PRMT1f/f/Mx1-Cre or PRMT1f/f mice (both are CD45.2+) were injected via tail vein into lethally irradiated congenic CD45.1+ recipient mice. Mice were euthanized for phenotypic analysis at indicated time points.
106 primary donor BM cells isolated from PRMT1f/f/Mx1-Cre or PRMT1f/f mice (both are CD45.2+) were injected via tail vein into lethally irradiated congenic CD45.1+ recipient mice. Mice were euthanized for phenotypic analysis at indicated time points.
Statistics
Statistical analyses were performed with Student's t test or ANOVA for normal distribution. Mann-Whitney U tests were performed when normal distribution was not satisfied. P value less than 0.05 was considered statistically significant.
Results
Generation of PRMT1 conditional knockout mice
To determine PRMT1 function in normal hematopoiesis, we first measured Prmt1 gene expression levels in multiple hematopoietic subpopulations from C57BL/6 mice (6- to 8-week-old). We sorted different subpopulations according to phenotypic markers and then Prmt1 expression levels were determined by quantitative RT-qPCR. There is no statistical significant difference of Prmt1 mRNA levels among CMP, MEP and GMP, and the level of Prmt1 mRNA in the phenotypic LSK was lower relative to GMP (Fig. 1A).
To determine PRMT1 function in hematopoiesis, we have developed conditional Prmt1 KO mouse (PRMT1f/f/Mx1-Cre) by crossing Mx1-Cre transgenic mouse with Prmt1-floxed (Prmt1f) mice as previously described [7]. The PRMT1 conditional allele consists of exons 4 and 5, which encode part of the methyltransferase domain, flanked by loxP sites. Deletion of these exons creates a frameshift, resulting in a functionally null allele (Fig. 1B). We induced Prmt1 deletion of adult hematopoietic cells by injection of 7 doses of pIpC (14 mg/kg every other day, injection, i.p.), and simultaneously, pIpC-treated littermates (Mx1-Cre-negative) were included as controls. PRMT1 flox alleles were confirmed by PCR analysis (Fig. 1C). PRMT1 deletion at protein levels was confirmed by Western blot analysis (Fig. 1D).
Deletion of PRMT1 results in anemia and leukopenia
Overall, deletion of PRMT1 had a prominent effect on adult hematopoiesis in 6- to 8-week-old mice. Analysis of the peripheral blood of these mice revealed significant decreases in red blood cells (RBCs), hemoglobin (Hb) and white blood cells (WBCs) at 12 weeks post pIpC induction and the absolute number of neutrophils and lymphocytes were decreased (Fig. 1E). Notably, RBC and Hb counts were starting to decrease as early as 4 weeks after pIpC induction (Supplemental Fig. 1) compared with littermate controls. Importantly, BM cellularity of PRMT1-KO mice was decreased compared with littermate controls (Fig. 1F left, histologic examination was shown in Fig. 1G) but, PRMT1 deletion did not have obvious effects on spleen cellularity at 12 weeks post pIpC induction (Fig. 1F right). Moreover, the Prmt1 heterozygous KO mice had normal peripheral blood counts and normal BM, spleen cellularity (data not shown). Intriguingly, approximately one out of eight of the PRMT1f/f/Mx1-CRE mice died within 5 months, whereas none of the PRMT1f/f mice (n=6) died at the end point of our evaluation (Supplemental Fig. 2).
Loss of PRMT1 affects terminal hematopoietic differentiation
To evaluate the effects of PRMT1 deletion on adult hematopoiesis, we first determined the frequency of erythroid (Ter119+), mature myeloid (Mac1+/Gr1+), B cells (B220) and T cells (CD3) in the BM of PRMT1-deleted mice and found that Ter119+ and B220+ subsets were significantly decreased compared with littermate controls, while the myeloid and T cell populations were relatively intact (Fig. 2A). Importantly, we also calculated the absolute numbers of erythroid, myeloid, T and B cells on the basis of their frequencies and BM cellularity and observed overall decreases in all lineage cells (Fig. 2B). The observation of B cell maturation defects is consistent with other reports using B cell specific PRMT1 KO mice [17-19].
Moreover, CD71 and Ter119 markers were used to define the maturation stages of erythroid progenitors (Fig. 2C). In PRMT1 KO mice, we observed significantly enriched primitive erythroid subsets including S0 (CD71-/Ter119-) as well as S1 (CD71hi/Ter119-) populations, accounting for a relative decrease of mature erythroid cells (S5, CD71-Ter119hi, Fig. 2D), thereby indicating erythroid maturation block.
Interestingly, our histology analysis of BM showed hypocellularity with megakaryocytosis. Clusters of megakaryocytes were commonly observed in BM of PRMT1 KO mice (Fig. 2E, F), indicating more differentiation to the lineage of megakaryocytes. Additionally, our analysis also showed a significant decrease of megakaryocyte progenitors (MkPs, defined as Lin-ckit+scal-CD41+CD150+) compared with similarly treated littermate control mice (Fig. 2G,H). These results are consistent with previous results [15, 20], confirming the important role of PRMT1 in antagonizing megakaryocyte differentiation.
Deletion of PRMT1 results in anemia and leukopenia. (A) Relative Prmt1 mRNA levels in indicated HSPCs populations, detected by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR; n=3, each performed in duplicate).*p<0.05, p value was determined by one way ANOVA. (B) Generation of a PRMT1 flox allele in mice. Representation of the floxed exons are the blue boxes, and the line represents introns. The black triangles denote loxP sites, and the small arrows denote the primers used for PCR analysis. The PRMT1 conditional allele consists of exons 4 and 5, which encode part of the methyltransferase domain, flanked by loxP sites. (C) Genomic DNA was purified from tails for further PCR analysis. Tissue from 3 different PRMT1f/f and PRMT1f/f/Mx1-CRE mice was used (n=3). Performed in duplicate from 3 independent experiments. (D) PRMT1 protein levels were determined by Western blot analysis using PRMT1f/f or PRMT1f/f/Mx1-CRE mouse BM cells (n=3). Performed in duplicate from 3 independent experiments. (E) Complete blood count (CBC) analysis of peripheral blood cell RBC, Hb, WBC, neutrophil, lymphocyte and Plt counts at 12 weeks after pIpC injection are shown(n=10).**p<0.01,***p<0.001. p values were determined by Mann-Whitney U tests. (F) Number of total BM and spleen cells from PRMT1f/f and PRMT1f/f/Mx1-CRE mice (n=10). ***p<0.001. p value was determined by Mann-Whitney U tests. (G) Representative H&E- staining image shows cross sections of femurs isolated from the PRMT1f/f and PRMT1f/f/Mx1-CRE mice (scale bars, 100µm).
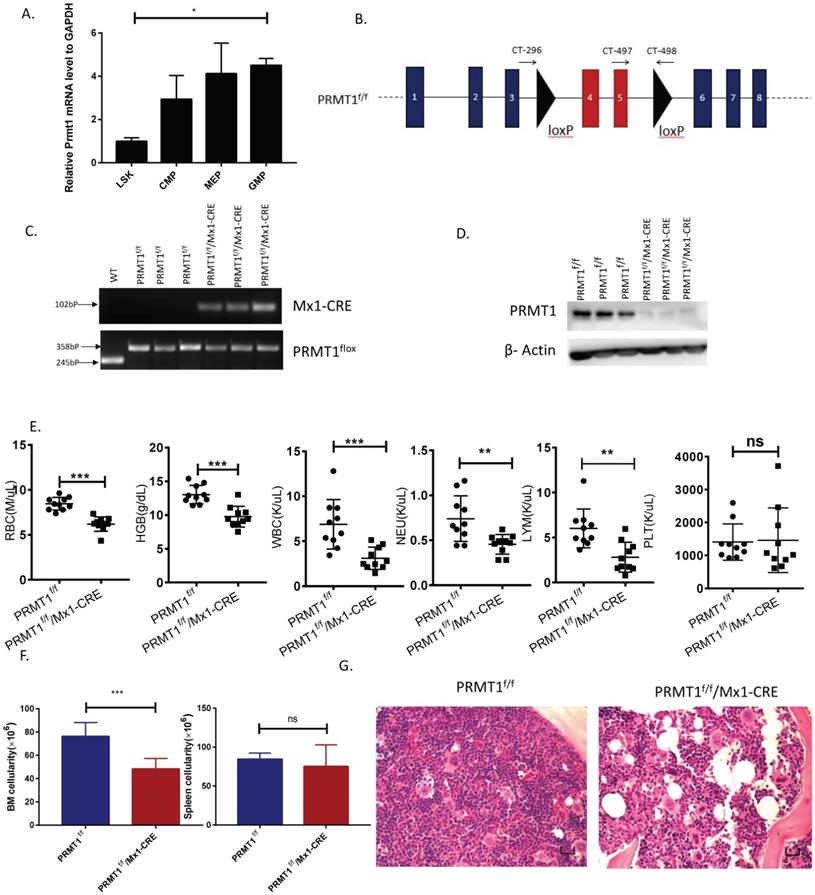
Loss of PRMT1 affects terminal hematopoiesis differentiation. (A) Frequency of CD3/B220, Mac1/Gr1 and Ter119 population in PRMT1f/f/Mx1-CRE and PRMT1f/f mouse BM cells (n=10). **p<0.01. p values were determined by Mann-Whitney U tests. (B) Number of CD3/B220, Mac1/Gr1 and Ter119 population in PRMT1f/f/Mx1-CRE and PRMT1f/f mouse BM cells (n=10). The absolute number of each subpopulation is calculated through multiply total BM cell number by BM percentage. *p<0.05, **p<0.01, ***p<0.001. p values were determined by Mann-Whitney U tests. (C) Representative FACS plots showing various erythroid progenitor subsets defined by the expression of Ter119 and CD71 in PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM analyzed by flow cytometry. (D) Representative FACS plots showing gating strategy and the frequency of each phenotypic erythroid subset (S0-S5) in Lin- BM (n=10).*p<0.05, **p<0.01. p values were determined by Student's t test. (E) H&E staining of the femur section of PRMT1f/f/Mx1-CRE and of PRMT1f/f mice, megakaryocytes are marked with arrows (scale bars, 100µm). (F) Number of megakaryocytes per high power field (hpf) in the BM evaluated in PRMT1f/f/Mx1-CRE and PRMT1f/f mice (n=5). ***p<0.001. p value was determined by Student's t test. (G) Representative FACS plots showing gating strategy and frequency of MkP populations from PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM. (H) Frequency and numbers of MkP in total BM cells (n=5). *p<0.05. p value was determined by Student's t test.
PRMT1 deletion result in adult mouse HSPCs reduction
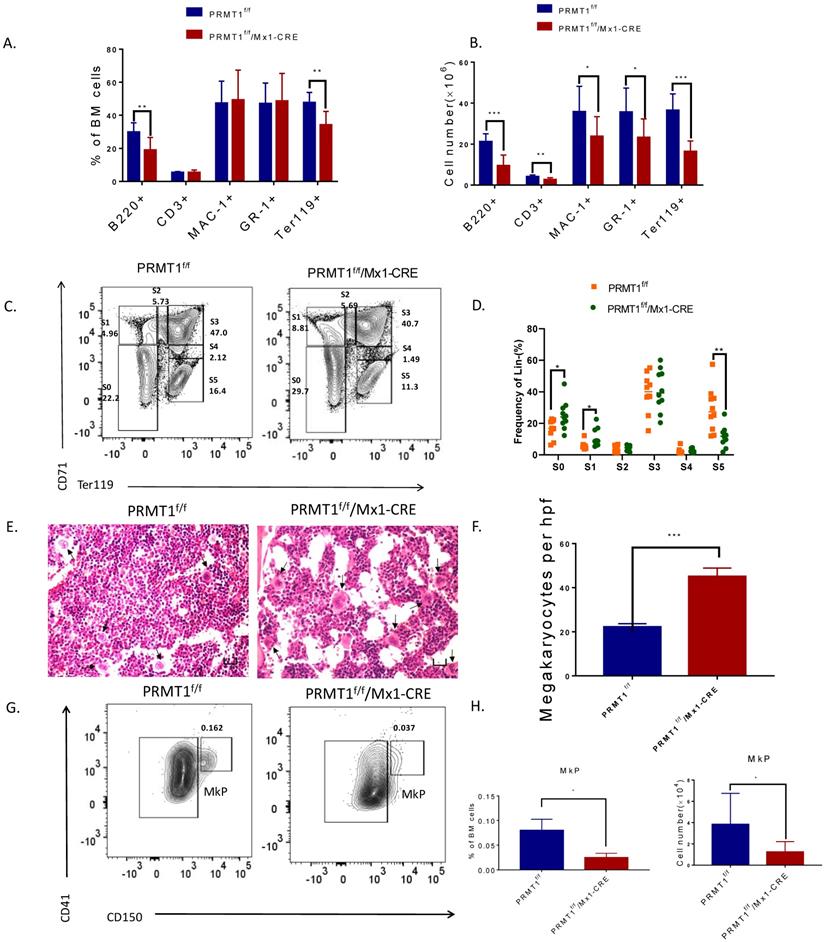
To explore the biology of PRMT1-deficient HSPC, we first analyzed the frequency and absolute cell number of BM HSPC populations from PRMT1-KO mice by flow cytometry. PRMT1 deletion did not affect the frequency of lineage-negative cells (that do not express Ter119, Mac1, Gr1, B220, CD3e, CD4, CD5, CD8 or CD127) (Fig. 3A). The frequencies of phenotypic progenitor subsets in BM, including LSK (Lin-/cKithi/Sca1+),MPP and LMPP was not affected upon PRMT1 KO (Fig. 3B). Notably, the frequency of the ST-HSC population and GMPs was significantly lower in PRMT1f/f/Mx1-CRE BM compared with littermate control mice (Fig. 3B, C). We also noticed that the frequency of MEPs was higher in PRMT1 KO compared with littermate controls (Fig. 3C). Moreover, the absolute cell numbers of ST-HSCs and the GMPs subset was significantly decreased compared with littermate control mice (Fig. 3D, E).
We next evaluated the function of PRMT1 KO hematopoietic progenitor cells using an in vitro colony-forming cell (CFC) assay. After pIpC induction, total BM cells (1×105 cells) from PRMT1f/f/Mx1-CRE or PRMT1f/f mice were seeded in semisolid methylcellulose culture media supplemented with SCF, IL-3, IL-6, and EPO. PRMT1f/f/Mx1-CRE progenitors generated lower numbers of CFUs, also characterized with smaller size at first plating (Fig. 3F). We then performed serial replating every 7 days to assess their capacity to maintain progenitor activity over time. Significantly reduced numbers of CFUs were produced from PRMT1f/f/Mx1-CRE progenitor cells in secondary or tertiary replating (Fig. 3F). Differential counts of colony morphology revealed that PRMT1f/f/Mx1-CRE cells produced a reduced frequency of BFU-E compared with control cells, especially in the first plating (Fig. 3G).
PRMT1 deficiency impairs competitive fitness
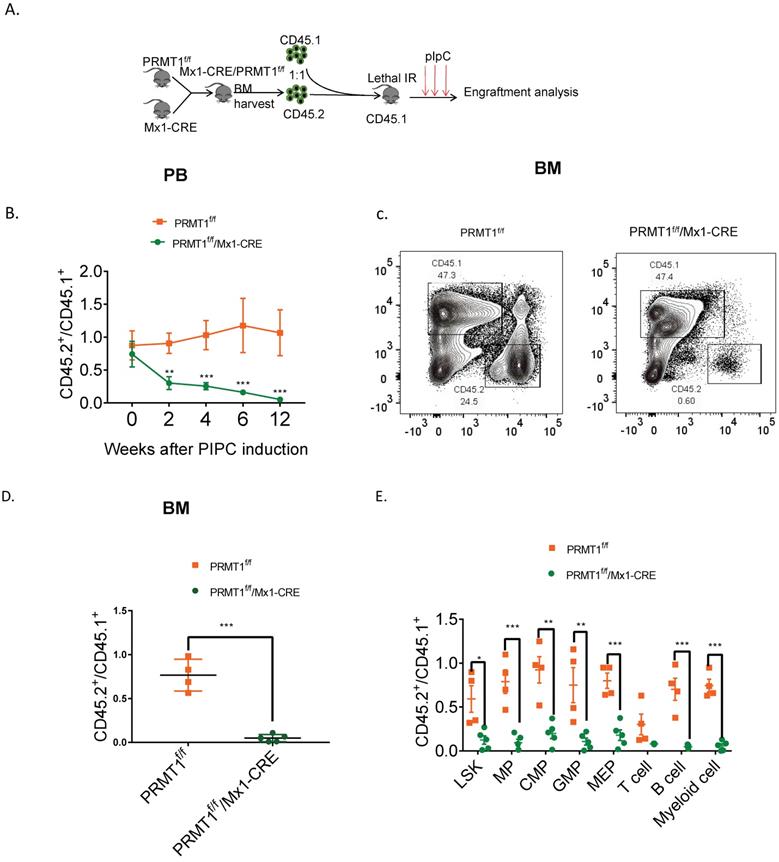
To further assess the long-term consequences of Prmt1 deficiency in adult mouse hematopoiesis, we performed a competitive repopulation assay using unfractionated BM cells (Fig. 4A). We transplanted approximately 1 × 106 BM cells isolated from Prmt1f/f, or PRMT1f/f/Mx1-CRE mice (which are CD45.2), together with equal numbers of WT CD45.1 competitor cells, into lethally irradiated CD45.1 recipient mice. Post pIpC induction, PRMT1-deleted CD45.2 cells were dramatically reduced compared to control donor cells, in both the peripheral blood and BM of recipient mice (Fig. 4B, C, D). Consistently, the chimerism contribution of PRMT1-deficient HSPCs or lineages was reduced relative to PRMT1-intact cells (Fig. 4E).
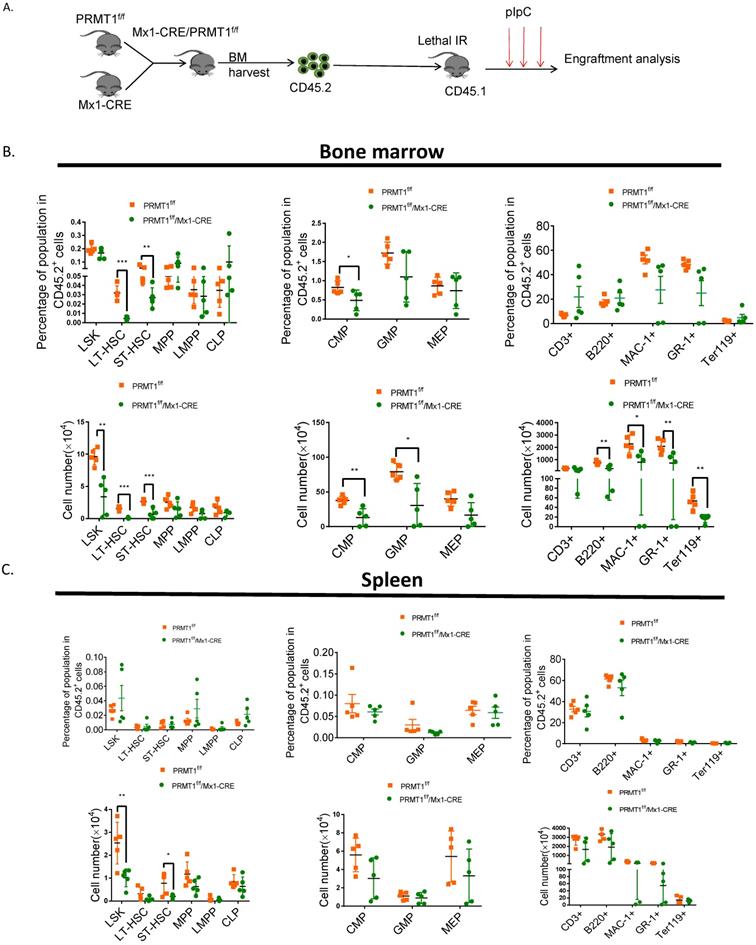
To evaluate the long-term repopulation capacity of Prmt1 null HSCs, we also performed noncompetitive BM transplantation. BM cells (2× 106 cells) isolated from primary donor mice in which PRMT1 deletion was not induced, littermate controls were also included as a separate group. Total BM cells from PRMT1f/f/Mx1-CRE or control mice were harvested and transplanted into lethally irradiated CD45.1+ congenic recipients (Fig. 5A). Four weeks post BMT, PRMT1 deletion was induced by pIpC induction. Thereafter, we monitored donor cells in PB. Notably, CD45.2 cells were significantly reduced in PB of mice receiving PRMT1 KO BM cells (date not shown). We did CBC analysis at 4 weeks after pIpC injection, showing WBC was slightly decreased (Supplemental Fig. 3). While analyzing BM at 16 weeks post BMT, we found PRMT1 KO markedly reduced CD45.2+ donor chimerism of multiple lineages as well as HSPCs subsets relative to those of controls (Fig. 5B). In contrast, PRMT1 deletion had no marked effect on the frequency of CD45.2 cells in the spleen (Fig. 5C). In BM, the absolute numbers of HSPCs subsets and lineages were markedly reduced compared to littermate control mice (Fig. 5B). In spleen, the absolute numbers of LSK and ST-HSCs showed a significant decrease compared with similarly treated littermate control mice (Fig. 5C).
Discussion
PRMT1 is known to be responsible for at least 85% of all arginine methylation catalyzation in mammalian cells [21]. PRMT1 can regulate myeloid differentiation and lymphocyte development [22, 23], however, its function in HSC self-renewal as well as adult hematopoiesis has been unclear. In the present study, we generated a PRMT1 conditional KO mouse model. Using the Cre/lox-conditional system, we characterize the role of PRMT1 in regulating adult mouse HSC function. We found that the loss of PRMT1 expression leads to cytopenia and affects HSC repopulation and multilineage differentiation.
Our studies indicate that PRMT1 deficiency leads to a significantly impaired normal hematopoiesis. Adult HSCs are maintained in a quiescent state, which is important for their longevity and function [24]. Doubling time analysis at homeostasis has shown that ST-HSCs and MPPs divide more frequently than LT-HSCs [25, 26]. Moreover, our chimeric BM transplant experiments demonstrate that the PRMT1 deletion indeed decreased HSPCs in vivo repopulating capacity.
PRMT1 deletion results in adult mouse HSPCs reduction. (A) Representative FACS plots showing gating strategy and frequency of phenotypic populations including LSKs, MPs, CMPs, GMPs and MEPs from PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM. (B) Frequency of LSK, LT-HSC, ST-HSC, MPP, LMPP and CLP populations in PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM (n=10). *p<0.05. p value was determined by Student's t test. (C) Frequency of CMP, GMP and MEP subsets in PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM (n=10).**p<0.01. p value was determined by Student's t test. (D) Number of LSK, LT-HSC, ST-HSC, MPP, LMPP and CLP populations in PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM (n=10). **p<0.01. p value was determined by Mann-Whitney U tests. (E) Number of CMP, GMP and MEP subsets in PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM (n=10). ***p<0.001. p value was determined by Mann-Whitney U tests. (F) Colony-forming progenitor (CFU) assays were performed using PRMT1f/f/Mx1-CRE and PRMT1f/f mouse bone marrow cells. CFU- number was shown. Cells were replated over 2 rounds of weekly successive replating (P2, P3). Performed in duplicate from 3 independent experiments.*p<0.05, **p<0.01. p values were determined by Student's test. (G) Percentage of BFU-E, CFU-GM, CFU-GEMM, CFU-G and CFU-M colonies at 1st plating. *p<0.05. Performed in duplicate from 3 independent experiments. p value was determined by Student's test.
Competitive repopulation assay to assess the hematopoietic consequences of PRMT1 deficiency in vivo. (A) Scheme of the competitive repopulation assay. (B) Ratio of CD45.2/CD45.1 chimerism in the PB 2-12 weeks after competitive repopulation assay (n=5). **p<0.01, ***p<0.001. p values were determined by two-way ANOVA. (C) Representative FACS plots showing gating CD45.1 and CD45.2 population from PRMT1f/f/Mx1-CRE and of PRMT1f/f mouse BM. (D) Ratio of CD45.2/CD45.1 chimerism in the BM of indicated recipient PRMT1f/f/Mx1-CRE (n=5) and PRMT1f/f mice (n=4). ***p<0.001. p value was determined by Student's t test. (E) CD45.2 to CD45.1 ratio in LSK, MPs, CMP, GMP, MEP, T cell, B cell and myeloid cells of primary recipients at 12 weeks. Each dot represents an individual mouse transplanted with PRMT1f/f (n=4) or PRMT1f/f/Mx1-CRE mouse (n=5) BM cells. *p<0.05, **p<0.01, ***p<0.001. p value were determined by Student's t test.
In addition, we found an erythroid maturation block in PRMT1f/f/ Mx1-CRE mice, in agreement with other's findings that, mediated through p38a, PRMT1 could promote lineage-specific differentiation toward erythrocytes [14, 15]. Further supporting our results, others have demonstrated that inhibition of PRMT1 through RNAi depleted levels of the activating histone mark H4R3me and increased levels of the repressive H3K9me and H3K27me marks at the β-globin locus in erythrocytes[27], contributing to an anemia phenotype. Regarding megakaryocyte maturation, we noticed that the frequency of MEPs was higher in PRMT1 KO compared with littermate controls. Additionally, through histology analysis of BM, we did find increased number of megakaryocytes. Our result is in agreement with others study, showing PRMT1 overexpression antagonized megakaryocytic differentiation [28]. However, in our mouse studies, we found PRMT1 KO did not affect platelet level. Through mice femur histology analysis, we could not distinguish granular megakaryocyte or platelet-forming megakaryocyte. And the phenotypic marker of platelet-forming megakaryocyte is still not clear. Thus, we postulate that the platelet-forming megakaryocytes did not increase; conversely, the granular megakaryocyte may increase.
Noncompetitive BM transplantation to assess the hematopoietic consequences of PRMT1 deficiency in vivo. (A) Scheme of experimental design. (B) Percentage and number of CD45.2+ donor-derived population in the BM of PRMT1f/f/Mx1-CRE or PRMT1f/f recipient mice 16 weeks after BM transplantation (n=5). *p<0.05, **p<0.01, ***p<0.001. p values were determined by Student's t test. (C) Percentage and number of CD45.2+ donor-derived population in the spleen of PRMT1f/f/Mx1-CRE or PRMT1f/f recipient mice 16 weeks after BM transplantation (n=5). *p<0.05, **p<0.01. p values were determined by Student's t test.
PRMT1 is an active component of signal transduction pathways including those from the B-cell receptor (BCR) [4, 18, 29-32]. Our flow-cytometry analysis showed a significant decrease in the frequency and absolute number of mature B cells in PRMT1f/f/Mx1-CRE mouse BM, and, consistent with other's results [17], found that mice with a B-cell-specific deletion of PRMT1 display a partial block in B cell development at the pre-B cell stage, confirming that PRMT1 is required for lymphocyte development. Infantino et al [19] undertook an immunology analysis on mice with Prmt1 deletion in mature B cells. Their analysis revealed that PRMT1-catalyzed protein substrate methylation is essential for normal B cell proliferation, differentiation and survival. Taken together, our results show that PRMT1 is required for normal B cell development.
In the transplant context, PRMT1-deletion reduced HSPCs and lineage cell number evidenced by engraftment analysis (Fig. 5B), indicating that the hematopoietic changes in PRMT1 KO animals were caused by cell autonomous. Consistently, others also used transplant assay to evaluate the cell autonomous effect [33-35]. But we can't exclude the possible non-cell autonomous effects.
Although knockout of Prmt1 in mice can result in embryonic lethality at embryonic day 7.5[11, 21], but Prmt1-conditional KO adult mice didn't die till pIpC induction 16 weeks (supplemental Fig. 2). These results suggested that PRMT1 inhibitors could be safely applied to hematological malignancies.
In conclusion, PRMT1 is required at multiple stages of hematopoietic differentiation. PRMT1 function is important for HSPCs' competitive fitness. Our results indeed identify that PRMT1 serves as a key epigenetic regulator of normal hematopoiesis that occurs throughout life.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported in part by National Institutes of Health, National Heart, Lung, and Blood Institute grant R01 HL141336, a Research Scholar Grant (RSG-19-036-01-LIB) from the American Cancer Society, St. Baldrick's Foundation Research Grant Award #632462. The Medical and Health Research Foundation of Zhejiang Province (grant no. 2017KY369). We also thanked the Analytical Cytometry Core assisted by Lucy Brown and Animal Resource Center at City of Hope, supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institutes of Health. We thank Stéphane Richard for supplying us with PRMT1f/f mice.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA. et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Molecular & Cellular Proteomics. 2014;13:372-87
2. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Molecular cell. 2009;33:1-13
3. Baldwin RM, Morettin A, Côté J. Role of PRMTs in cancer: Could minor isoforms be leaving a mark? World journal of biological chemistry. 2014;5:115
4. Cheung N, Chan LC, Thompson A, Cleary ML, So CWE. Protein arginine-methyltransferase-dependent oncogenesis. Nature cell biology. 2007;9:1208
5. Kim Y-R, Lee BK, Park R-Y, Nguyen NTX, Bae JA, Kwon DD. et al. Differential CARM1 expression in prostate and colorectal cancers. BMC cancer. 2010;10:197
6. Mathioudaki K, Scorilas A, Ardavanis A, Lymberi P, Tsiambas E, Devetzi M. et al. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumor Biology. 2011;32:575-82
7. He X, Zhu Y, Lin Y-C, Li M, Du J, Dong H. et al. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD+ Acute Myeloid Leukemia. Blood. 2019;134:548-60
8. Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI. et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. International journal of cancer. 2011;128:562-73
9. Shia W-J, Okumura AJ, Yan M, Sarkeshik A, Lo M-C, Matsuura S. et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119:4953-62
10. Zou L, Zhang H, Du C, Liu X, Zhu S, Zhang W. et al. Correlation of SRSF1 and PRMT1 expression with clinical status of pediatric acute lymphoblastic leukemia. Journal of hematology & oncology. 2012;5:42
11. Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Molecular and cellular biology. 2000;20:4859-69
12. Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR. et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. Journal of Biological Chemistry. 2000;275:7723-30
13. Hata K, Yanase N, Sudo K, Kiyonari H, Mukumoto Y, Mizuguchi J. et al. Differential regulation of T-cell dependent and T-cell independent antibody responses through arginine methyltransferase PRMT 1 in vivo. FEBS letters. 2016;590:1200-10
14. Hua W-K, Chang Y-I, Yao C-L, Hwang S-M, Chang C-Y, Lin W-J. Protein arginine methyltransferase 1 interacts with and activates p38α to facilitate erythroid differentiation. PloS one. 2013;8:e56715
15. Chang Y-I, Hua W-K, Yao C-L, Hwang S-M, Hung Y-C, Kuan C-J. et al. Protein-arginine methyltransferase 1 suppresses megakaryocytic differentiation via modulation of the p38 MAPK pathway in K562 cells. Journal of Biological Chemistry. 2010;285:20595-606
16. Yu Z, Chen T, Hebert J, Li E, Richard S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Molecular and cellular biology. 2009;29:2982-96
17. Dolezal E, Infantino S, Drepper F, Börsig T, Singh A, Wossning T. et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nature immunology. 2017;18:911
18. Infantino S, Benz B, Waldmann T, Jung M, Schneider R, Reth M. Arginine methylation of the B cell antigen receptor promotes differentiation. Journal of Experimental Medicine. 2010;207:711-9
19. Infantino S, Light A, O'Donnell K, Bryant V, Avery DT, Elliott M. et al. Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nature communications. 2017;8:891
20. Zhang L, Tran N-T, Su H, Wang R, Lu Y, Tang H. et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. elife. 2015;4:e07938
21. Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. Journal of Biological Chemistry. 2000;275:19866-76
22. Mizutani S, Yoshida T, Zhao X, Nimer SD, Taniwaki M, Okuda T. Loss of RUNX 1/AML 1 arginine-methylation impairs peripheral T cell homeostasis. British journal of haematology. 2015;170:859-73
23. Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S. et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes & development. 2008;22:640-53
24. Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S. et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell stem cell. 2015;16:302-13
25. Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell stem cell. 2013;13:102-16
26. Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118-29
27. Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes & development. 2005;19:1885-93
28. Jin S, Mi Y, Song J, Zhang P, Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34+ cells. Experimental and therapeutic medicine. 2018;15:2563-8
29. Blanchet F, Cardona A, Letimier FA, Hershfield MS, Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. Journal of Experimental Medicine. 2005;202:371-7
30. Boisvert F-M, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell cycle. 2005;4:1834-41
31. Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP. et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96
32. Mowen KA, Schurter BT, Fathman JW, David M, Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Molecular cell. 2004;15:559-71
33. Jackson JT, Shields BJ, Shi W, Di Rago L, Metcalf D, Nicola NA. et al. Hhex Regulates Hematopoietic Stem Cell Self-Renewal and Stress Hematopoiesis via Repression of Cdkn2a. Stem Cells. 2017;35:1948-57
34. Hua W-K, Qi J, Cai Q, Carnahan E, Ramirez MA, Li L. et al. HDAC8 regulates long-term hematopoietic stem-cell maintenance under stress by modulating p53 activity. Blood. 2017;130:2619-30
35. Jeannet R, Cai Q, Liu H, Vu H, Kuo YH. Alcam regulates long-term hematopoietic stem cell engraftment and self-renewal. Stem Cells. 2013;31:560-71
Author contact
![]() Corresponding authors: Ling Li, Department of Hematological Malignancies Translational Science, Gehr Family Center for Leukemia Research, Hematologic Malignancies and Stem Cell Transplantation Institute, Beckman Research Institute, City of Hope Medical Center, Duarte, CA 91010; or Xiaoying Zhao, Department of Hematology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, 310009, China. E-mail address: lingliorg (Ling Li) (lead contact), zrxzedu.cn (Xiaoying Zhao)
Corresponding authors: Ling Li, Department of Hematological Malignancies Translational Science, Gehr Family Center for Leukemia Research, Hematologic Malignancies and Stem Cell Transplantation Institute, Beckman Research Institute, City of Hope Medical Center, Duarte, CA 91010; or Xiaoying Zhao, Department of Hematology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, 310009, China. E-mail address: lingliorg (Ling Li) (lead contact), zrxzedu.cn (Xiaoying Zhao)

 Global reach, higher impact
Global reach, higher impact