10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(13):2936-2947. doi:10.7150/ijbs.38000 This issue Cite
Research Paper
MiR-4319 induced an inhibition of epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1
1. Department of Hepatobiliary Surgery, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277 Yanta West Road, Xi'an 710061, China
2. Department of oncology, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277 Yanta West Road, Xi'an 710061, China
#Equate contributions
Received 2019-6-29; Accepted 2019-10-15; Published 2019-11-15
Abstract
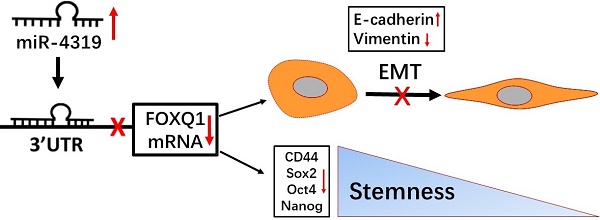
The heterogeneity existing in tumours is responsible for the poor response to treatment. Therefore, elucidating the molecular mechanisms of intratumoural heterogeneity in hepatocellular carcinoma (HCC) is vital for the discovery of new therapeutic methods for improving the prognosis of patients. Of note, cancer stem cells (CSCs) existing in HCC may explain the pathological properties of heterogeneity and recurrence. An increasing number of studies have confirmed that abnormally expressed microRNAs (miRNAs) take part in the carcinogenesis as well as the aggravation of HCC. However, little information is currently available about the specific miR-4319 in HCC. Herein, we demonstrated that the level of miR-4319 was remarkably decreased in HCC specimens and cells compared to that in normal counterparts and that the depression of miR-4319 in tumour specimens correlates with tumour size, histological grade and venous invasion. Through a series of functional experiments, we illustrated that miR-4319 repressed cell proliferation, accelerated apoptosis, inhibited epithelial-mesenchymal transition (EMT) and prevented cancer stemness in HCC cells by targeting FOXQ1 (Forkhead box Q1). An in vivo tumourigenesis assay uncovered that depletion of miR-4319 in Hep3B cells increased tumour growth and elevated the expression of EMT and CSC markers in comparison to those of the control group. Restoration of FOXQ1 expression also partially reversed the miR-4319-induced biological effects on HCC cells. Thus, miR-4319, as a posttranscriptional regulator, plays a profound role in suppressing the malignant progression of HCC, and our study highlights the miR-4319/FOXQ1 cascade as a potential therapeutic target for conquering HCC.
Keywords: miR-4319, FOXQ1, EMT, cancer stemness, HC
Introduction
Hepatocellular carcinoma (HCC) is one of the most aggressive malignancies in the world [1], displaying a dismal prognosis due to the high heterogeneity and high rate of recurrence and metastasis [2, 3, 4]. Although surgical resection is the optimal choice for early-stage HCC, most patients suffering from HCC are often diagnosed at an advanced stage [5]. Therefore, it is imperative to investigate the underlying molecular mechanism participating in the initiation and malignant progression of HCC and urgent to discover new therapeutic targets for HCC patients.
Notably, recent investigations have uncovered that HCC has a heterogeneous cell population named cancer stem cells which display stemness features that are pivotal for tumourigenesis, distant dissemination and chemoresistance of HCC [6-8]. Hepatocellular carcinoma harbours a population of cancer stem cells or possesses the high expression of a panel of genes with stemness markers, which may be a signal for predicting poor treatment outcomes for HCC patients [9]. Moreover, cancer stem cells have gradually become promising therapeutic targets of HCC [6, 10-12], which indicates that finding novel genes modulating cancer stemness could elevate the treatment outcome of HCC. Epithelial-mesenchymal transition (EMT) has been identified as a vital process in the early metastatic dissemination of cancer cells by endowing them with a more invasive phenotype [13]. Intriguingly, a tight link exists between acquiring cancer stem-like traits and EMT induction [14]. EMT induction in mammary epithelial cells generates populations of cells that possess cancer stem-like traits, as determined by mammosphere formation, tumour-seeding capacity, and expression of surface marker profiles of stemness [14, 15]. In addition, CSCs are prone to elevate the expression of multiple factors involved with mesenchymal transdifferentiation, such as vimentin fibronectin and EMT-related transcription factors [14].
As a set of evolutionarily conserved small non-coding RNAs, microRNAs (miRNAs) have been identified as post-transcriptional regulators at the level of gene expression, which is achieved by the combination of complementary sequences with the 3'-untranslated regions (UTRs) of corresponding mRNA of genes encoding for proteins, leading to mRNA degradation or translation cessation[16, 17]. A growing number of studies have indicated that miRNAs are vital in diverse biological processes in HCC, including cell proliferation, the cell cycle, apoptosis, differentiation, metastasis and cancer stemness, by regulating tumour suppressors or oncogenes [18, 19]. Therefore, specific miRNAs are known to be potential therapeutic, diagnostic, and prognostic biological markers in HCC [20]. MiR-4319, a newly identified cancer-related microRNA, was aberrantly expressed and became a predictor of patient survival in cancers [21]. Previous studies have revealed that miR-4319 elicits inhibitory effects on the proliferation of breast cancer and prostate cancer [22, 23], and miR-4319 could repress cancer stemness in triple-negative breast cancer through targeting E2F2 [22]. MiR-4319 also exhibits tumour suppressor activities in NSCLC (non-small-cell lung cancer) through targeting LIN28/RFX5/YAP cascades to mitigate cell migration and proliferation and facilitate cell apoptosis [24]. Moreover, miR-4319 is transcriptionally modulated by PLZF and acts as a tumour suppressor in CRC (colorectal cancer) via targeting ABTB1 [21]. MiR-4319 suppresses the development of thyroid cancer by regulating FUS (fused in sarcoma)-stabilized SMURF1 (SMAD-specific E3 ubiquitin protein ligase 1) [25]. Nevertheless, the biological effects and the latent regulatory mechanism of miR-4319 in HCC remain obscure and need to be further elucidated. The current study investigated the effects and modulatory mechanism of miR-4319 in HCC.
Herein, we uncovered that miR-4319 was reduced in HCC and correlated with adverse prognostic features. Moreover, miR-4319 impeded cell proliferation, accelerated apoptosis, inhibited epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1.
Materials and Methods
HCC specimens and cell culture
In total, 83 HCC specimens and the corresponding neighbouring non-tumour samples were gathered in our hospital from January 2008 to December 2011 with informed consent from all patients. None of the patients had received any prior radiotherapy or chemotherapy. This study was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University, according to the Declaration of Helsinki.
The human immortalized normal hepatocyte cell line LO2 and a group of HCC cell lines (HepG2, Hep3B, SMMC-7721, MHCC-97L and MHCC-97H) (Chinese Academy of Sciences, Shanghai, China) were cultured in DMEM (Invitrogen, Carlsbad, USA) with 10 μl of 10% FBS (Gibco, Grand Island, USA) and incubated at 37 °C and 5% CO 2.
RNA extraction and qRT-PCR
Total RNA in the tissue samples and HCC cell lines was extracted using TRIzol reagent. The quantity and quality of isolated total RNA were evaluated as previously reported [26]. The primers used for qRT-PCR are displayed as follows: miR-4319 (forward) 5′-GCACAGCTCCCTGAGCAA-3′ and (reverse) 5′-CAGTGCGTGTCGTGGAGT-3′; U6 (forward) 5′-CTCGCTTCGGCAGCACAT-3′ and (reverse) 5′-TTTGCGTGTCATCCTTGCG-3′; and FOXQ1 (forward) 5′-GATTTCTTGCTATTGACCGATGC-3′ and (reverse) 5′-CTAATAAAGCTGTAGCCCGTTGC-3′. The primers against GAPDH (HQP006940) were obtained from Genecopoeia (Guangzhou, China). qRT-PCR was conducted as reported previously[27]. The 2-ΔΔCt method was utilized to calculate the relative gene expression.
Cell transfection
MiR-4319 mimic (#4464066), miR-4319 inhibitor (#4464084) or corresponding negative control oligonucleotides (NC mimic and NC inhibitor) were all obtained from Invitrogen. The validated small interfering RNA (siRNA) targeting FOXQ1 and negative control siRNA were purchased from Ambion as previously reported [28]. The FOXQ1 expression plasmid (pcDNA3.1-FOXQ1) was purchased from GenePharma (Shanghai, China). HCC cells were transfected with oligonucleotides and vectors using Lipofectamine 2000 (Invitrogen) based on the manufacturer's instructions. Cells were harvested 48 h post-transfection for further experiments.
Western blot analysis
RIPA buffer (Beyotime, Shanghai, China) was used to lyse the total protein derived from cultured HCC cells. Protein concentrations were determined using a BCA protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). Equal amounts of protein (20 μg/lane) were separated on SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad Laboratories, Hercules, CA, USA). The primary antibodies used in this study are displayed in Supplementary Table 1. The membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (NXA931-1ML and NA934-1ML, GE Healthcare Life Sciences, Beijing, China). Protein bands were visualized using an enhanced chemiluminescence detection kit (Pierce Biotechnology). Blots were semi-quantified by ImageJ software (1.46; National Institutes of Health, Bethesda, MD, USA).
Colony formation assay
After completing the designated intervention, Hep3B and MHCC-97H cells (1000 cells per well) were seeded in a 6-well plate and cultured for two weeks. Then, the colonies were fixed with 4% paraformaldehyde and stained with crystal violet solution after washing three times with PBS. The number of visible colonies was counted under a microscope.
MTT assay
The cell viability of HCC cells was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoliumbromide (MTT) assays. HCC cells were seeded in a 96-well plate at a density of 5×103 cells/well and transfected with miR-4319 mimics, miR-4319 inhibitor, pcDNA3.1-FOXQ1 and si-FOXQ1 for 24, 48, 72, or 96 h. Then, 10 μl of MTT (Sigma‑Aldrich; Merck KGaA) was administered to cultured cells and incubated for 4 h at 37˚C. The supernatant was replaced with 100 μl of DMSO incubated for 15 min, and the absorbance was determined at 490 nm using a multiwell microplate reader.
Correlation between the clinicopathologic characteristics and miR-4319 and FOXQ1 expression in HCC (n = 83).
| Clinical parameters | Cases | Expression level | P value | Expression level | P value | ||
|---|---|---|---|---|---|---|---|
| MiR-4319high (n=42) | MiR-4319low (n=41) | FOXQ1high (n=42) | FOXQ1low (n=41) | ||||
| Age(years) | |||||||
| <65 years | 55 | 27 | 28 | 0.699 | 30 | 25 | 0.314 |
| ≥65 years | 28 | 15 | 13 | 12 | 16 | ||
| Gender | |||||||
| Male | 71 | 37 | 34 | 0.503 | 36 | 35 | 0.964 |
| Female | 12 | 5 | 7 | 6 | 6 | ||
| Tumor size (cm) | |||||||
| <5cm | 38 | 26 | 12 | 0.017* | 13 | 25 | 0.154 |
| ≥5cm | 45 | 19 | 26 | 27 | 28 | ||
| Tumor number | |||||||
| solitary | 68 | 38 | 30 | 0.114 | 33 | 35 | 0.451 |
| multiple | 15 | 5 | 10 | 9 | 6 | ||
| Edmondson | |||||||
| Ⅰ+Ⅱ | 54 | 32 | 22 | 0.031* | 20 | 34 | 0.029* |
| Ⅲ+Ⅳ | 29 | 10 | 19 | 18 | 11 | ||
| TNM stage | |||||||
| Ⅰ+Ⅱ | 62 | 40 | 22 | 0.081 | 27 | 35 | 0.027* |
| Ⅲ+Ⅳ | 21 | 9 | 12 | 15 | 6 | ||
| Capsular | |||||||
| Present | 58 | 30 | 28 | 0.756 | 31 | 27 | |
| Absent | 25 | 12 | 13 | 15 | 10 | ||
| Venous invasion | |||||||
| Present | 23 | 5 | 18 | 0.001* | 16 | 7 | 0.032* |
| Absent | 60 | 37 | 23 | 26 | 34 | ||
| AFP | |||||||
| <400ng/ml | 22 | 13 | 9 | 0.353 | 8 | 14 | 0.195 |
| ≥400ng/ml | 61 | 29 | 32 | 32 | 29 | ||
| HBsAg | |||||||
| positive | 68 | 32 | 36 | 0.294 | 33 | 35 | 0.896 |
| negative | 15 | 6 | 9 | 7 | 8 | ||
HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; TNM, tumor-node-metastasis. *Statistically significant.
EdU assay
An ethynyl deoxyuridine (EdU) incorporation assay was conducted with an EdU kit according to the manufacturer's protocols. The images of EdU staining were visualized by a Zeiss fluorescence photomicroscope (Carl Zeiss, Oberkochen, Germany) and quantified via counting at least five random fields.
Transwell invasion assay
After completing the designated transfection, HCC cells were resuspended with serum-free medium and seeded into the upper chamber coated with Matrigel (Sigma). Culture medium containing 10% FBS served as a chemoattractant in the lower chamber. After being cultured for 48 hours at 37°C, cells that did not invade via the pore of the filter were carefully wiped out with cotton wool. Then, the invasive cells were fixed with 100% methanol and stained with crystal violet.(Sigma). Cell number was counted under a microscope by randomly selecting 5 fields.
Tumoursphere formation assay
HCC cells transfected with miR-4319 mimics, miR-4319 inhibitor, pcDNA3.1-FOXQ1 and si-FOXQ1, were plated in 6‑well ultra‑low-attachment plates (Corning Incorporated, Corning, NY, USA) at a density of 5×103 cells/well in serum‑free DMEM/F12 medium. The DMEM/F12 medium was supplemented with 1% B27, 20 ng/ml human EGF and 20 ng/ml human FGF. Cells were subsequently cultured at 37˚C for 2 weeks to allow formation of tumourspheres. After 14 days, tumourspheres were counted under a light microscope (Nikon Corporation) at a magnification of ×200, and the number was recorded.
Dual-luciferase reporter assay
The 3'-UTR binding sequence of FOXQ1 and the associated mutated sequences were generated and inserted into the pmiR-GLO dual luciferase miRNA target expression vector (Promega, Madison, WI, USA). The dual-luciferase reporter assay was performed according to previous instructions [18].
In vivo tumourigenesis assay
A 4-to-6-week-old female BALB/c nude mouse (obtained from the Experimental Animal Center of Xi'an Jiaotong University School of Medicine, Xi'an) was utilized to establish a subcutaneously implanted tumour model. The xenograft tumours were generated using the Hep3B cell line stably depleting miR-4319 or its corresponding controls. The stable miR-4319-depleting Hep3B cells were generated by infection with lentiviral vector based on the manufacturer's instructions (miR-4319: pLV-[hsa-mir-4319] plasmid; negative control plasmid: pLV-[mir-control], Biosettia), which were in accord with previously described methods [28]. After establishing a stable expression cell line, 5×106 cells were mixed into 150 μL of Matrigel and injected subcutaneously into the flanks of nude mice. The tumour volume was then monitored by detecting its two dimensions and then calculated by the following formula: V (tumour volume: mm 3) = 0.5 × [W (width: mm)] 2 × L (long diameter: mm). Four weeks later, the mice were sacrificed, and the xenograft tumour tissue was weighed. These tumour tissues were then fixed for further histological analysis. The immunohistochemistry procedure was performed as previously reported, and the percentages of stained area were calculated using ImageJ software [29]. All programmes were authorized by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University.
Statistical analysis
To avoid systemic errors, each experiment was repeated more than three times. The results are displayed as the mean ± standard deviation. Student's t-test or one-way ANOVA (one-way analysis of variance) followed by the LSD post hoc test was conducted to compare the differences between two groups or more than two groups, respectively, with SPSS (SPSS 18.0; SPSS Inc., Chicago, IL, USA). A P value<0.05 was considered to be statistically significant.
Results
The level of miR-4319 expression was depressed in HCC compared with that in noncancerous tissues and correlated with adverse prognostic features
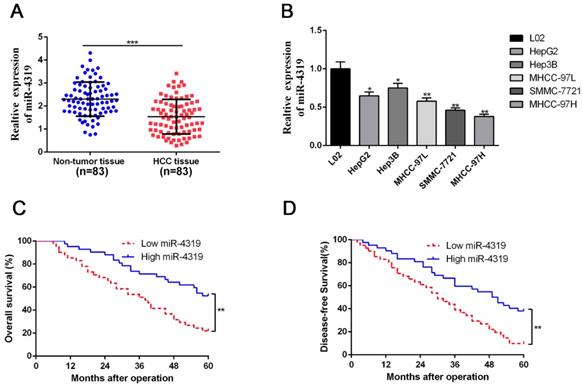
Due to the unclear biological role of miR-4319 in HCC, we first performed qRT-PCR analysis to examine its expression level in 83 pairs of HCC samples and corresponding pericarcinomatous tissues. The expression level of miR-4319 was markedly reduced in HCC samples in comparison to that in the corresponding adjacent nontumour tissues (P <0.01, Figure 1A). As shown in Table 1, the depression of miR-4319 was related to large tumour size (≥ 5 cm; P=0.017), high histological grade (Edmondson-Steiner grade III + IV; P=0.031) and venous invasion (P=0.001). Likewise, the expression of miR-4319 was obviously lower in the group of HCC cell lines compared to in the physiological liver cell line LO2 (P < 0.05, Figure 1 B). We selected MHCC-97H (relatively low expression of miR-4319) and Hep3B (relatively high expression of miR-4319) for further experiments. Furthermore, the overall survival and disease-free survival of HCC patients in the miR-4319 low-expression group was poorer than that of patients in the high-expression group (Figure 1C-D).
MiR-4319 inhibits cell proliferation and accelerates apoptosis in HCC cells
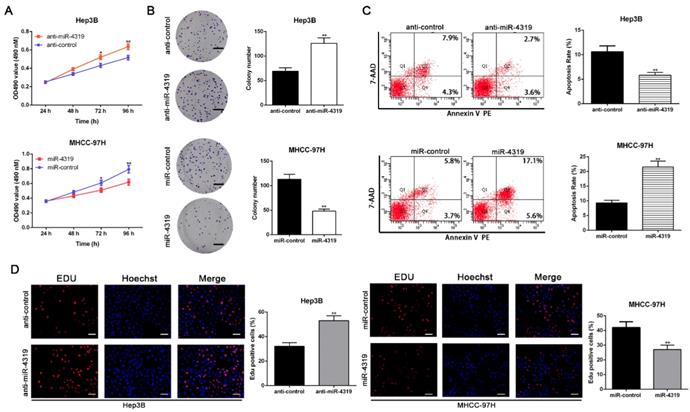
To disclose whether miR-4319 plays a specific role in HCC progression, a series of functional experiments were conducted using a lentivirus system to stably overexpress MHCC-97H cells and knockdown Hep3B cells (Figure S1A-B, P<0.05). As evident from the colony formation assay, MTT assay and EdU incorporation assay, the over-expression of miR-4319 remarkably suppressed the proliferation of MHCC-97H cells in comparison to that of control cells (P<0.01, Figure 2A, 2B and 2D). Conversely, miR-4319 depletion in Hep3B cells enhanced proliferation with elevated cell colonies, increased the O.D value of MTT assay and upregulated the proportion of Edu-positive cells (P<0.01, Figure 2A, 2B and 2D). Additionally, miR-4319 overexpression prominently induced apoptosis of MHCC-97H cells, whereas Hep3B cells with miR-4319 depletion had a lower percentage of apoptotic cells than did the control cells (P < 0.01, Figure 2C). Collectively, these data illustrated that miR-4319 inhibits cell proliferation and accelerates apoptosis in HCC cells.
Downregulation of miR-4319 induces EMT and confers cancer stem properties in HCC
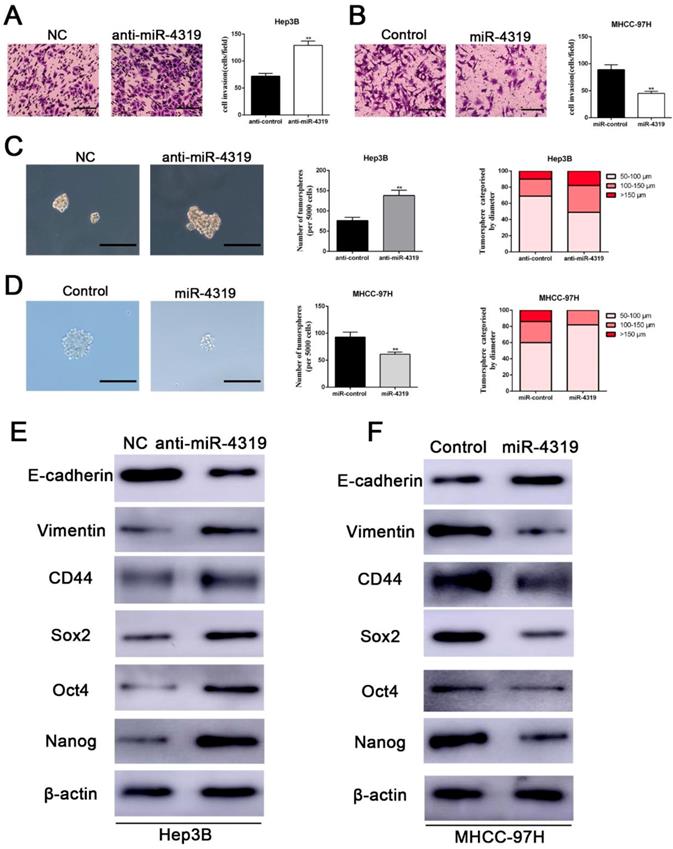
We next examined whether the downregulation of miR-4319 in HCC also impacts EMT and tumour mammosphere formation. Silencing miR-4319 in Hep3B cells increased the invasive capacities of Hep3B cells, as confirmed by the Matrigel invasion assay (P < 0.01, Figure 3A). Moreover, tumoursphere formation, which is recognized as a representative trait of CSCs, was assayed to assess the sphere-forming capacity of HCC cells. The results displayed that both the number and the diameter of tumourspheres in Hep3B cells were greatly enhanced after knockdown of miR-4319 in Hep3B cells (Figure 3C). However, the Matrigel invasion assay and the tumoursphere formation assay indicated that the invasive capacities and the tumoursphere formation abilities of miR-4319-overexpressing MHCC-97H cells were repressed compared with those of control cells (Figure 3B, 3D). Moreover, the results of western blotting showed that the downregulation of miR-4319 in Hep3B cells induced the expression of EMT and CSC markers, as revealed by decreased E-cadherin expression and increased vimentin, CD44, CD133, CD90, EPCAM, Sox2, Oct4, and Nanog expression (Figure 3E and Figure S2). Analogously, overexpression of miR-4319 in MHCC-97H cells restrained the expression of EMT and CSC markers (Figure 3F and Figure S2). Thus, these results suggest that the downregulation of miR-4319 induces EMT and confers cancer stem properties in HCC.
MiR-4319 is reduced in HCC and predicts poor prognosis. (A) The expression of miR-4319 was reduced in 83 HCC tissues compared to adjacent noncancerous tissues as determined by qRT-PCR. P < 0.0001 by t-test. (B) The differences in miR-4319 expression among HCC cell lines (HepG2, Hep3B, MHCC97L, SMMC-7721 and MHCC97H) and the human hepatocyte cell line (LO2). n=3 independent experiments, *P < 0.05 by ANOVA. (C-D) The HCC patients were divided into miR-4319 low-expression (n=41) and miR-4319 high-expression groups (n=42), with the median value of miR-4319 expression as a cut-off value. The overall survival and disease-free survival of HCC patients in the miR-4319 low-expression group were poorer than those in the high-expression group. P < 0.01 by log-rank test.
MiR-4319 modulates HCC cell proliferation and apoptosis. Hep3B cells transfected with miR-4319 inhibitor and MHCC-97H cells transfected with miR-4319 mimics were subjected to further experiments. Downregulation of miR-4319 promoted cell viability (A), colony formation (B) and proliferation (D) and impeded apoptosis (C) in Hep3B cells, while overexpression of miR-4319 repressed cell viability (A), colony formation (B) and proliferation (D) and accelerated apoptosis (C) in MHCC-97H cells. n=3 independent experiments, **P < 0.01 by t-test. Magnification of EdU is ×200, and scale bars = 50 μm. The scale bars of colony formation =1 cm.
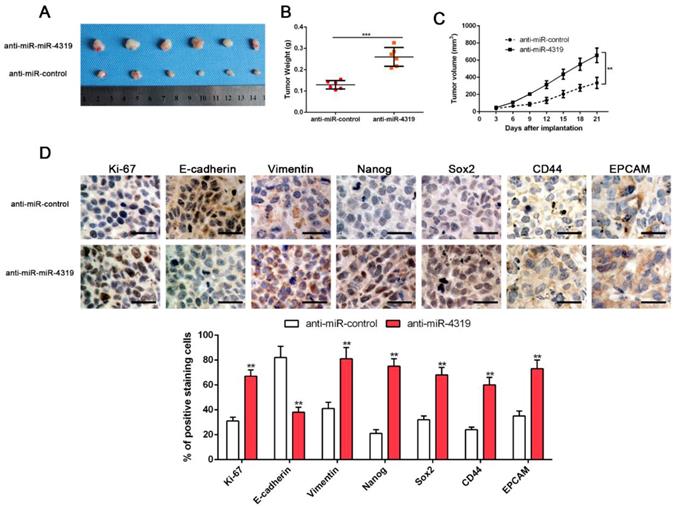
MiR-4319 suppresses tumourigenesis in vivo
CSCs display stem cell-like properties, including self-renewal, tumour initiation, proliferation, invasion and metastasis [10, 30]. Therefore, we investigated whether miR-4319 could repress in vivo tumourigenesis by subcutaneously injecting Hep3B cells with or without depletion of miR-4319. Hep3B cells with anti-miR-4319 generated a higher tumour weight and volume in comparison to those of the control group (Figure 4A-C). Moreover, immune-histochemistry staining and semiquantification analysis of the IHC data showed that subcutaneous tumours arising from depletion of miR-4319 in Hep3B cells increased the expression of EMT (E-cadherin and vimentin) and CSC (Nanog, Sox2, CD44 and EPCAM) markers in comparison to that in the control group (Figure 4D). It is worth noting that miR-4319 suppresses tumourigenesis and cancer stemness in vivo.
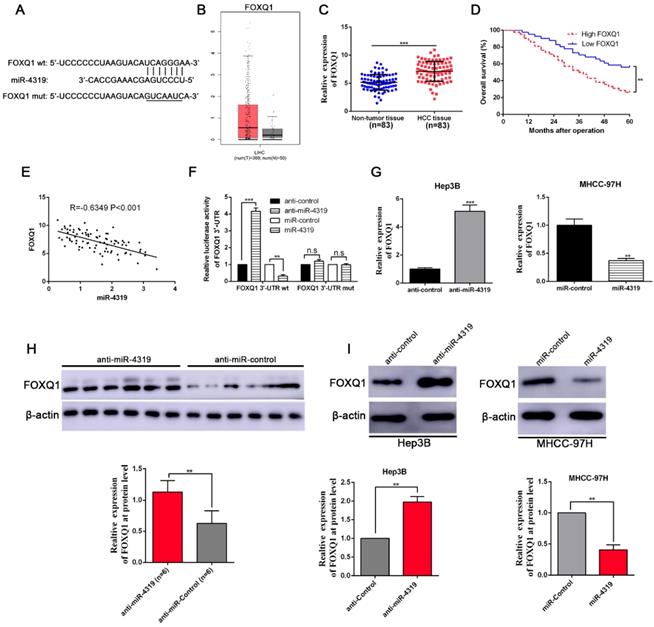
FOXQ1 is identified as a direct target of miR-4319
To uncover the underlying mechanisms by which miR-4319 impeded cell proliferation and repressed EMT and cancer stem traits in HCC, we performed bioinformatics analysis to predict the potential targets of miR-4319. Based on starBase V3.0 and TargetScan algorithms, FOXQ1, frequently reported as an oncogene to participate in the malignant progression of a variety of cancers[31], was selected as a candidate interacting target of miR-4319 (Figure 5A). Previous studies have identified that FOXQ1 promotes proliferation, EMT, and cancer stemness[32]; thus, we first examined the expression of FOXQ1 in HCC. According to the data derived from GEPIA [33] and 83 paired samples of HCC and corresponding pericarcinomatous tissues, the expression level of FOXQ1 was elevated in HCC (Figure 5B-C). The OS and DFS of HCC patients in the FOXQ1 high-expression group were poorer than those in the low-expression group (displayed in Figure 5D and Figure S1C). As shown in Table 1, FOXQ1 overexpression was correlated with high histological grade (Edmondson-Steiner grade III + IV; P=0.029), late tumour stage (TNM stage III + IV; P=0.027) and venous invasion (P=0.032). Furthermore, we determined that there was a negative association between miR-4319 and FOXQ1 mRNA by Spearman's analysis in HCC samples (P<0.05, Figure 5E). Notably, the dual-luciferase reporter assay illuminated that enhanced expression of miR-4319 effectively repressed the relative luciferase activity of wild-type (wt) FOXQ1 instead of the mutant (mt) FOXQ1 in HEK-293T cells (P<0.05, Figure 5F). The expression of FOXQ1 and the potential downstream effectors of FOXQ1, Sox12 [34] (sex determining region Y-box 12) and NDRG1 [31] (N-myc downstream-regulated gene 1), were remarkably reduced at the protein levels in MHCC-97H cells with miR-4319 overexpression but enhanced in Hep3B cells with miR-4319 depletion (Figure 5G and 5I, Figure S3). Interestingly, we found that depletion of FOXQ1 in MHCC-97H cells reduced the expression of Sox12 and NDRG1, whereas overexpression of FOXQ1 in Hep3B cells enhanced the expression level of Sox12 and NDRG1 (Fig S3). Additionally, the expression of FOXQ1 protein in tumour tissues from miR-4319 knockdown-injected mice was significantly higher than that in control mice (Figure 5H). Altogether, FOXQ1 was a direct target of miR-4319.
Downregulation of miR-4319 induces EMT and confers cancer stem properties in HCC. (A) Transwell-invasion and tumoursphere formation assays revealed that downregulation of miR-4319 enhanced invasive ability, increased the number of tumourspheres and elevated the percentage of tumourspheres with larger diameters (C) in Hep3B cells, while overexpression of miR-4319 inhibited cell invasion (B), decreased the number of tumourspheres and elevated the percentage of tumourspheres with smaller diameters (D) in MHCC-97H cells. The results of western blot analysis (E-F) also showed that downregulation of miR-4319 induced EMT and facilitated the expression of cancer stemness markers (CD44, Sox2, Oct4, Nanog) in Hep3B cells, while overexpression of miR-4319 in MHCC-97H cells restrained EMT and cancer stemness. n=3 independent experiments, **P < 0.01 by t-test. The scale bars of transwell assay=50 μm. Magnification of tumoursphere is ×200, and scale bars = 50 μm.
MiR-4319 suppresses tumourigenesis in vivo. Stable miR-4319-depleting Hep3B cells were generated by infection with a lentiviral vector, and miR-4319-depleting Hep3B cells and control vector cells were implanted into nude mice via subcutaneous injection. (A) Representative images of HCC xenografts from depleting Hep3B cells and control vector cells. (B) The tumour weight in the miR-4319-depleting Hep3B group was significantly heavier than that in the control group. n=6, ***P < 0.001 by t-test. (C) The tumour volume in the miR-4319-depleting Hep3B group (n=6) was obviously larger than that in the control group (n=6). **P < 0.01 by ANOVA. (D) The percentage of Ki-67, vimentin, Nanog Sox2, CD44 and EPCAM staining in tumour cells in the miR-4319-depleted Hep3B group was prominently higher than that in the control group, while the percentage of E-cadherin-stained tumour cells in the miR-4319 knockdown group was prominently lower than that in the control group. n=3 fields of 6 tissue sections, **P < 0.01 by t-test. Scale bar: 50 μm.
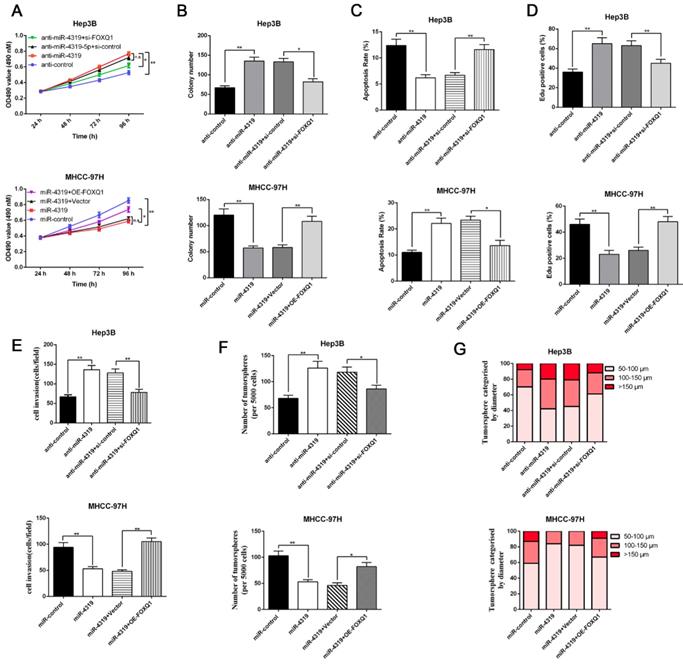
Restoration of FOXQ1 expression partially reversed the miR-4319-induced biological effects on HCC cells
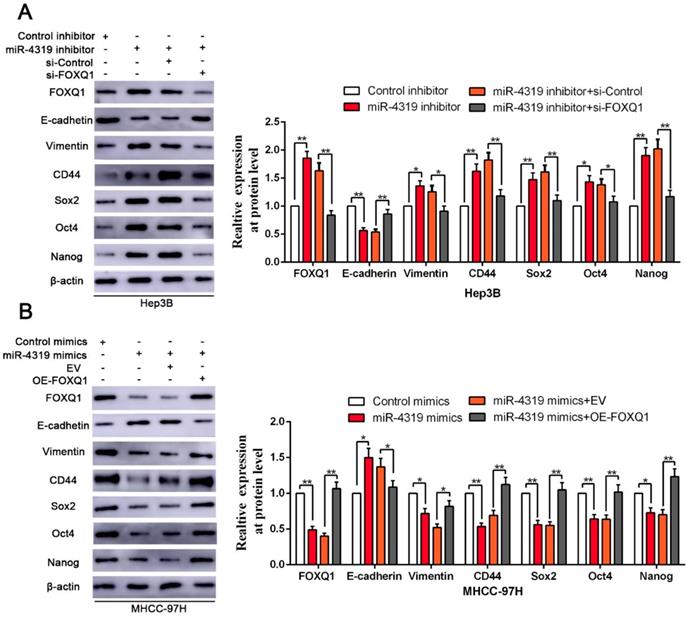
Since FOXQ1 contributes to cancer stem traits and tumourigenesis in HCC, we aimed to investigate whether FOXQ1 mediated the biological effects of miR-4319 in HCC. FOXQ1 was overexpressed in miR-4319-overexpressing MHCC-97H cells (P<0.05, Figurre 6A), revealing that FOXQ1 restoration abolished the potency of miR-4319, leading to a remarkable increase of cell viability, proliferation and colony formation and a decrease in apoptosis (P<0.05, Figure 6A-D). The Matrigel invasion assay and the tumoursphere formation assay verified that FOXQ1 restoration attenuated the suppressive role of miR-4319 in the invasive capacities and tumoursphere formation abilities of MHCC-97H cells (P < 0.01, Figure 6E-G). Western blot analysis also indicated that reintroduction of FOXQ1 rescued the miR-4319-induced inhibition of the expression of EMT and CSC markers in MHCC-97H cells (P < 0.05, Figure 7A-7B and Figure S4A-S4B). In contrast, FOXQ1 knockdown in miR-4319-suppressive Hep3B cells partly ceased the potency of anti-miR-4319 on tumour cell proliferation, colony formation, apoptosis, invasion and cancer stem traits (P<0.05, Figure 6A-G, Figure 7A-7B and Figure S4A-S4B). Thus, the data indicated that FOXQ1 was a functional target of miR-4319 in HCC.
Discussion
Hepatocellular carcinoma (HCC) commonly displays dismal outcomes because of its high possibility of metastatic relapse and chemotherapy resistance. The heterogeneity existing in tumours is responsible for the poor response to treatment [35, 36]. Therefore, elucidating the molecular mechanisms of intratumoural heterogeneity in HCC is vital for the discovery of new therapeutic methods for improving the prognosis of patients. Of note, cancer stem cells (CSCs) existing in HCC may well explain the pathological properties of heterogeneity and recurrence [37]. Herein, we demonstrated that the level of miR-4319 was remarkably decreased in HCC specimens and cells and that miR-4136 induced inhibition of the epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1. Thus, miR-4136, as a post transcriptional regulator, plays a profound role in suppressing the cancer stemness of HCC, which is in accord with findings that miR-4319 exerted an essential role as a tumour suppressor in the repression of cancer stemness in triple-negative breast cancer via downregulation of E2F2[22].
It is well known that FOX family members play a pivotal role in cancer initiation and progression by modulating the cell cycle, DNA damage repair and cancer stem properties [38]. Among FOX family members, FOXQ1 has also been reported to be upregulated and facilitate proliferation, EMT, and distant dissemination in a variety of human cancers, including HCC [34, 39, 40]. Some studies have also unveiled an emerging role of FOXQ1 in modulating cancer stemness. For example, FOXQ1 maintains stemness and drives chemoresistance through its direct targets PDGFRα and β in breast cancer[32]; FOXQ1 also elevates tumour re-initiation in targeted organs and enhances the colonization of disseminated metastatic cells via LAMA4[41]. Moreover, FOXQ1 plays essential roles in mediating crosstalk between CAFs and HCC cells. CAFs induce expression of FOXQ1 in HCC cells, and N-myc downstream-regulated gene 1 (NDRG1) is subsequently trans-activated by FOXQ1 to enhance HCC initiation. Intriguingly, pSTAT6/C-C motif chemokine ligand 26 (CCL26) signalling is also modulated by the FOXQ1/NDRG1 axis, therefore recruiting CAFs to form a positive feedback loop in HCC [31].
FOXQ1 is a direct target of miR-4319. (A) The putative binding sequences between miR-4319 and the 3'-UTR of FOXQ1 mRNA are shown. Mutated sequences in the 3'-UTR of FOXQ1 mRNA are underlined. (B) The expression pattern of FOXQ1 in HCC was derived from GEPIA. (C) The expression of FOXQ1 was increased in 83 HCC tissues compared to that in adjacent noncancerous tissues as determined by qRT-PCR. P < 0.0001 by t-test. (D) The HCC patients were divided into FOXQ1 low-expression (n=41) and miR-4319 high-expression (n=42) groups, with the median value of miR-4319 expression as a cut-off value. The overall survival of HCC patients in the FOXQ1 high-expression group was poorer than that in the low-expression group. P < 0.01 by log-rank test. (E) An inverse correlation between miR-4319 and FOXQ1 mRNA expression was observed in 83 HCC samples. P < 0.001 by Spearman's correlation test. (F) Overexpression of miR-4319 decreased while downregulation of miR-4319 increased the luciferase activity of vectors containing wt 3'-UTR of FOXQ1 rather than mut 3'-UTR of FOXQ1 in HEK-293T cells. n=3 independent experiments, **P < 0.01 by t-test. (G and I) miR-4319 overexpression reduced the levels of FOXQ1 mRNA and protein in MHCC-97H cells, while miR-4319 knockdown increased FOXQ1 abundance in Hep3B cells. n=3 independent experiments, **P < 0.01, *** P < 0.001 by t-test. (H) The expression of FOXQ1 in xenograft tissues. Xenograft tissues arising from the miR-4319 knockdown group (n = 6) and the control group (n = 6) were subjected to immunoblotting analysis to detect FOXQ1 expression at the protein level.
Restoration of FOXQ1 attenuates the tumour suppressive effects of miR-4319. MTT assay (A), colony formation assay (B) and EdU assay (D) revealed that the proliferation of Hep3B cells was enhanced by miR-4319 depletion and subsequently rescued by FOXQ1 knockdown, while overexpression of FOXQ1 abolished the inhibition of proliferation induced by miR-4319 overexpression in MHCC-97H cells. (C) The apoptosis assay uncovered that the apoptosis of Hep3B cells was reduced by miR-4319 depletion and subsequently rescued by FOXQ1 knockdown, while overexpression of FOXQ1 abolished the promotion of apoptosis induced by miR-4319 overexpression in MHCC-97H cells. (E-G) Invasion and tumoursphere formation were increased by miR-4319 depletion and subsequently rescued by FOXQ1 restoration, while invasion, and oncosphere formation ability were dramatically impaired by overexpression of miR-4319. n=3 independent experiments, **P < 0.01 by t-test.
During the past decade, miRNAs have emerged as major players in the complex network of gene regulation, and dysregulation of miRNA expression has been implicated in carcinogenesis [42]. Previously published studies have highlighted that specific miRNAs contribute to HCC progression, and several of these can be used as biomarkers for diagnosis, prognosis, and metastasis predictions in HCC patients [40, 43]. In fact, a few studies have shown that miRNAs may be able to regulate specific FOX genes in different cancers, including oesophageal cancer [44], hepatocellular carcinoma [45], and colorectal cancer [28]; however, such an interaction between miRNAs and FOX proteins remains largely unclear and has not been interrogated in hepatocellular carcinoma. In this study, we corroborated through a series of gain- and loss-of-function assays that miR-4319 impeded cell proliferation, exacerbated cell apoptosis, dampened EMT and mitigated cancer stem traits. As a tumour suppressor, miR-4319 directly targets the 3-UTR of FOXQ1 and controls its expression in HCC. We also confirmed that FOXQ1 expression was enhanced in HCC tissues and negatively correlated with miR-4319 expression. Moreover, alteration of FOXQ1 levels could partially rescue the physiological roles of miR-4319 in HCC cells. Thus, our study highlights the miR-4319/FOXQ1 cascade as a potential therapeutic target for conquering HCC.
Collectively, this study shed light on the functional role and modulatory mechanism of miR-4319 in the malignant progression of HCC. We revealed that miR-4319 directly dampened the expression level of FOXQ1 to mitigate malignant progression of HCC through impeding cell proliferation, accelerating apoptosis, inhibiting epithelial-mesenchymal transition and preventing cancer stemness of HCC, which offered insight into the molecular mechanism underlying miR-4319 in HCC development. Moreover, the miR-4319/FOXQ1 cascade may be a novel target via dampening cancer stem traits to ameliorate heterogeneity and improve the treatment outcomes of HCC.
Abbreviations
HCC: hepatocellular carcinoma; CSCs: cancer stem cells; miRNAs: microRNAs; FOXQ1: Forkhead box Q1; EMT: epithelial-mesenchymal transition; NSCLC: non-small cell lung cancer; EdU: Ethynyl deoxyuridine; IHC: immunohistochemistry; MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; ANOVA: one way analysis of variance; Sox12: sex determining region Y-box 12; NDRG1: N-myc downstream-regulated gene.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81874069, 81602566). SH, YS and QL conceived and designed the experiments; SH, YS and LS performed the experiments; ZL analyzed the data; TS contributed reagents/materials/analysis tools; SH, YS and QL wrote the paper. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
Reintroduction of FOXQ1 expression partially reversed the miR-4319-induced inhibition on EMT and cancer stemness of HCC cells. (A-B) FOXQ1 knockdown in miR-4319-suppressive Hep3B cells partly ceased the promotion of anti-miR-4319 on EMT and cancer stemness, whereas reintroduction of FOXQ1 rescued the miR-4319-induced inhibition on EMT (E-cadherin and Vimentin) and the CSC markers (CD44, Sox2, Oct4 and Nanog) of MHCC-97H cells. n=three independent experiments, **P < 0.01 by t-test.
References
1. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666-81
2. Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH. et al. A decade's studies on metastasis of hepatocellular carcinoma. Journal of cancer research and clinical oncology. 2004;130:187-96
3. Desai JR, Ochoa S, Prins PA, He AR. Systemic therapy for advanced hepatocellular carcinoma: an update. Journal of gastrointestinal oncology. 2017;8:243-55
4. Finn RS, Zhu AX, Farah W, Almasri J, Zaiem F, Prokop LJ. et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: A systematic review and meta-analysis. Hepatology. 2018;67:422-35
5. Diaz-Gonzalez A, Reig M, Bruix J. Treatment of Hepatocellular Carcinoma. Dig Dis. 2016;34:597-602
6. Zhao W, Wang L, Han H, Jin K, Lin N, Guo T. et al. 1B50-1, a mAb raised against recurrent tumor cells, targets liver tumor-initiating cells by binding to the calcium channel alpha2delta1 subunit. Cancer cell. 2013;23:541-56
7. Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY. et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-24
8. Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P. et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer cell. 2008;13:153-66
9. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL. et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499-511
10. Sun L, Wang Y, Wang L, Yao B, Chen T, Li Q. et al. Resolvin D1 prevents epithelial-mesenchymal transition and reduces the stemness features of hepatocellular carcinoma by inhibiting paracrine of cancer-associated fibroblast-derived COMP. Journal of experimental & clinical cancer research: CR. 2019;38:170
11. Lee TK, Cheung VC, Lu P, Lau EY, Ma S, Tang KH. et al. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology. 2014;60:179-91
12. Zhang Y, Guan DX, Shi J, Gao H, Li JJ, Zhao JS. et al. All-trans retinoic acid potentiates the chemotherapeutic effect of cisplatin by inducing differentiation of tumor initiating cells in liver cancer. Journal of hepatology. 2013;59:1255-63
13. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
14. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
15. Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature medicine. 2009;15:1010-2
16. Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annual review of medicine. 2009;60:167-79
17. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
18. Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y. et al. Methylation-mediated repression of microRNA-129-2 suppresses cell aggressiveness by inhibiting high mobility group box 1 in human hepatocellular carcinoma. Oncotarget. 2016;7:36909-23
19. Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y. et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350-65
20. Xu Q, Liu X, Liu Z, Zhou Z, Wang Y, Tu J. et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Molecular cancer. 2017;16:103
21. Huang L, Zhang Y, Li Z, Zhao X, Xi Z, Chen H. et al. MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United European gastroenterology journal. 2019;7:517-28
22. Chu J, Li Y, Fan X, Ma J, Li J, Lu G. et al. MiR-4319 Suppress the Malignancy of Triple-Negative Breast Cancer by Regulating Self-Renewal and Tumorigenesis of Stem Cells. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;48:593-604
23. Lin X, Wang Y. Re-expression of microRNA-4319 inhibits growth of prostate cancer via Her-2 suppression. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2018;20:1400-7
24. Yang Y, Li H, Liu Y, Chi C, Ni J, Lin X. MiR-4319 hinders YAP expression to restrain non-small cell lung cancer growth through regulation of LIN28-mediated RFX5 stability. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;115:108956
25. Bian S. miR-4319 inhibited the development of thyroid cancer by modulating FUS-stabilized SMURF1. Journal of cellular biochemistry. 2019
26. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629-41
27. Sun L, Cao J, Chen K, Cheng L, Zhou C, Yan B. et al. Betulinic acid inhibits stemness and EMT of pancreatic cancer cells via activation of AMPK signaling. International journal of oncology. 2019;54:98-110
28. Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 Are Promising Prognostic Biomarkers and Novel Targets of Tumor-Suppressive miR-342 in Human Colorectal Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:4947-57
29. Chen K, Qian W, Li J, Jiang Z, Cheng L, Yan B. et al. Loss of AMPK activation promotes the invasion and metastasis of pancreatic cancer through an HSF1-dependent pathway. Molecular oncology. 2017;11:1475-92
30. Manrique I, Nguewa P, Bleau AM, Nistal-Villan E, Lopez I, Villalba M. et al. The inhibitor of differentiation isoform Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer letters. 2015;356:899-909
31. Luo Q, Wang CQ, Yang LY, Gao XM, Sun HT, Zhang Y. et al. FOXQ1/NDRG1 axis exacerbates hepatocellular carcinoma initiation via enhancing crosstalk between fibroblasts and tumor cells. Cancer letters. 2018;417:21-34
32. Meng F, Speyer CL, Zhang B, Zhao Y, Chen W, Gorski DH. et al. PDGFRalpha and beta play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer research. 2015;75:584-93
33. Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98-W102
34. Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K. et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920-33
35. Chan LH, Luk ST, Ma S. Turning hepatic cancer stem cells inside out-a deeper understanding through multiple perspectives. Molecules and cells. 2015;38:202-9
36. Muramatsu S, Tanaka S, Mogushi K, Adikrisna R, Aihara A, Ban D. et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology. 2013;58:218-28
37. Wang Y, He L, Du Y, Zhu P, Huang G, Luo J. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell stem cell. 2015;16:413-25
38. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nature reviews Cancer. 2013;13:482-95
39. Xia L, Huang W, Tian D, Zhang L, Qi X, Chen Z. et al. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958-73
40. Peng XH, Huang HR, Lu J, Liu X, Zhao FP, Zhang B. et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Molecular cancer. 2014;13:186
41. Ross JB, Huh D, Noble LB, Tavazoie SF. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nature cell biology. 2015;17:651-64
42. Weng W, Feng J, Qin H, Ma Y, Goel A. An update on miRNAs as biological and clinical determinants in colorectal cancer: a bench-to-bedside approach. Future Oncol. 2015;11:1791-808
43. Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L. et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. Journal of experimental & clinical cancer research: CR. 2018;37:49
44. Sun Y, Yu X, Bai Q. miR-204 inhibits invasion and epithelial-mesenchymal transition by targeting FOXM1 in esophageal cancer. International journal of clinical and experimental pathology. 2015;8:12775-83
45. Zhang J, Yang Y, Yang T, Yuan S, Wang R, Pan Z. et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology. 2015;61:561-73
Author contact
![]() Corresponding author: Dr Qingguang Liu, Department of Hepatobiliary Surgery, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277 Yanta West Road, Xi'an 710061, China. E-mail: liuqingguangsina.com Phone: +086-029-85323905 Fax: +086-029-85323209
Corresponding author: Dr Qingguang Liu, Department of Hepatobiliary Surgery, the First Affiliated Hospital of Xi'an Jiaotong University, No. 277 Yanta West Road, Xi'an 710061, China. E-mail: liuqingguangsina.com Phone: +086-029-85323905 Fax: +086-029-85323209

 Global reach, higher impact
Global reach, higher impact